Hyper-Aerotolerant Campylobacter coli from Duck Sources and Its Potential Threat to Public Health: Virulence, Antimicrobial Resistance, and Genetic Relatedness
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of C. coli from Duck Samples
2.2. Analysis of the Aerotolerance Levels of C. coli Isolates
2.3. Virulence genes of C. coli Isolates
2.4. Antimicrobial Resistance of C. coli Isolates
2.5. Clonal Distribution Analysis of C. coli Isolates from Duck Sources Based on MLST Genotypes
2.6. Analysis of the Genetic Relatedness between C. coli Isolates from Duck and Human Sources
2.7. Statistical Analysis
3. Results
3.1. Identification of Aerotolerant C. coli Isolates from Duck Sources
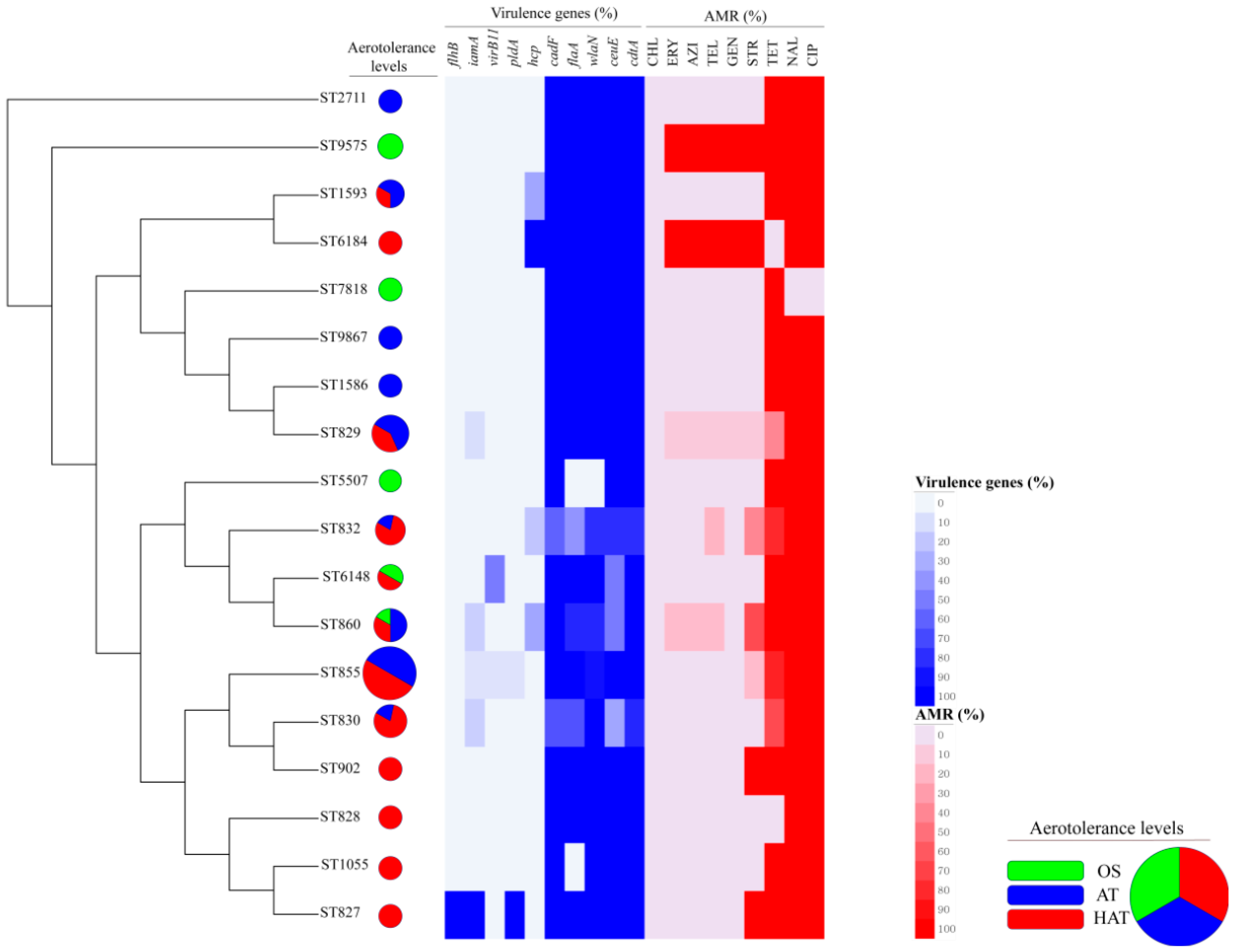
3.2. Virulence Genes in C. coli Isolates
3.3. Antimicrobial Resistance of C. coli Isolates
3.4. Clonal Distribution Analysis of C. coli Isolates Based on MLST Genotypes
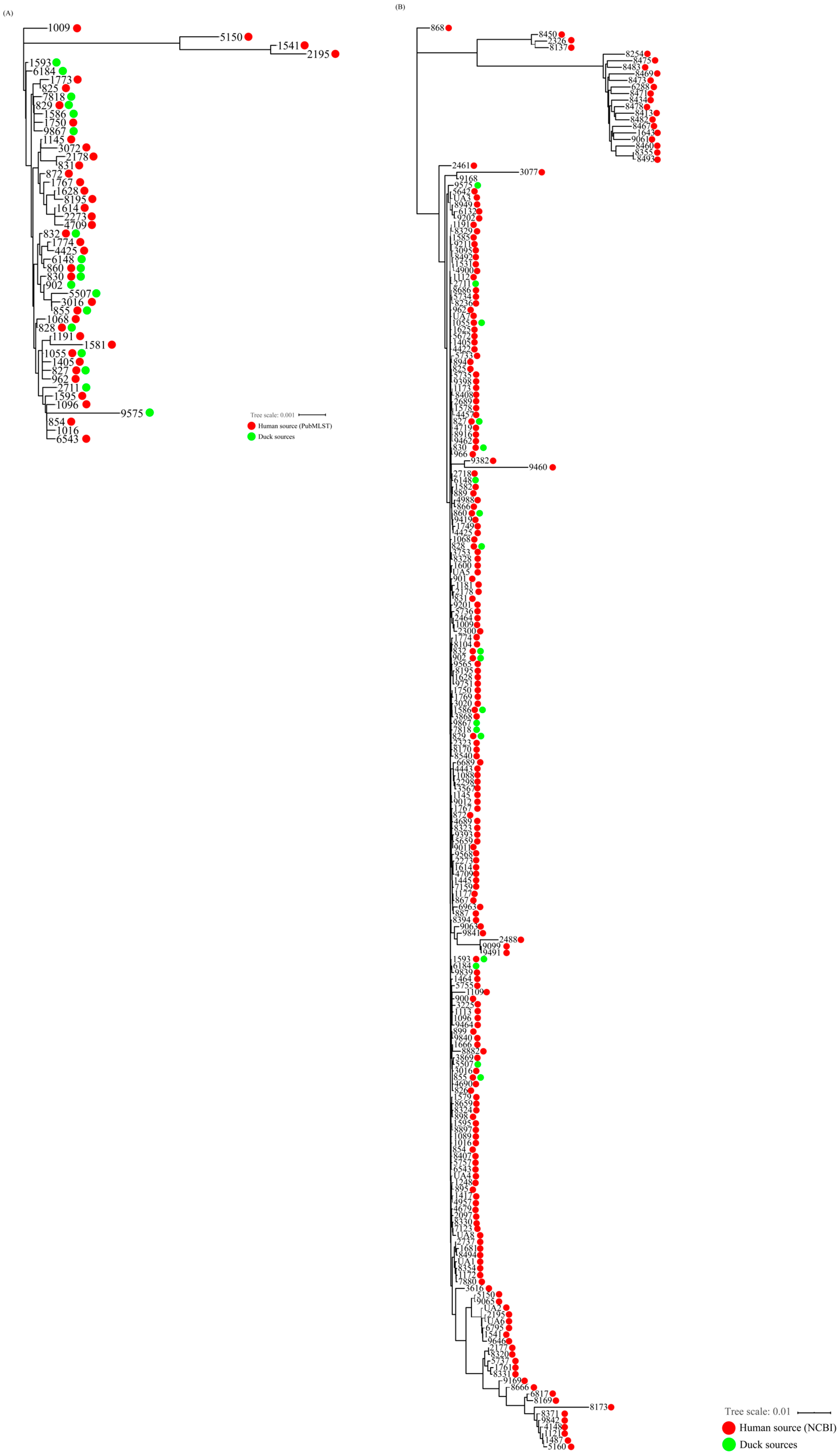
3.5. Genetic Relatedness of C. coli Isolates from Duck and Human Sources
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alfredson, D.A.; Korolik, V. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 2007, 277, 123–132. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef]
- Azrad, M.; Tkhawkho, L.; Isakovich, N.; Nitzan, O.; Peretz, A. Antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli: Comparison between Etest and a broth dilution method. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 23. [Google Scholar] [CrossRef]
- Datta, S.; Niwa, H.; Itoh, K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 2003, 52, 345–348. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Maiden, M.C. The evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb. Perspect. Biol. 2015, 7, a018119. [Google Scholar] [CrossRef]
- An, J.-U.; Ho, H.; Kim, J.; Kim, W.-H.; Kim, J.; Lee, S.; Mun, S.-H.; Guk, J.-H.; Hong, S.; Cho, S. Dairy cattle, a potential reservoir of human campylobacteriosis: Epidemiological and molecular characterization of Campylobacter jejuni from cattle farms. Front. Microbiol. 2018, 9, 3136. [Google Scholar] [CrossRef]
- Gargiulo, A.; Sensale, M.; Marzocco, L.; Fioretti, A.; Menna, L.F.; Dipineto, L. Campylobacter jejuni, Campylobacter coli, and cytolethal distending toxin (CDT) genes in common teals (Anas crecca). Vet. Microbiol. 2011, 150, 401–404. [Google Scholar] [CrossRef][Green Version]
- Giacomelli, M.; Salata, C.; Martini, M.; Montesissa, C.; Piccirillo, A. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb. Drug Resist. 2014, 20, 181–188. [Google Scholar] [CrossRef]
- Sahin, O.; Kassem, I.I.; Shen, Z.; Lin, J.; Rajashekara, G.; Zhang, Q. Campylobacter in Poultry: Ecology and Potential Interventions. Avian Dis. 2015, 59, 185–200. [Google Scholar] [CrossRef]
- Kassem, I.I.; Kehinde, O.; Kumar, A.; Rajashekara, G. Antimicrobial-Resistant Campylobacter in Organically and Conventionally Raised Layer Chickens. Foodborne Pathog. Dis. 2017, 14, 29–34. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Miller, W.G.; De Reuse, H.; Mendz, G.L. Oxygen requirement and tolerance of Campylobacter jejuni. Res. Microbiol. 2007, 158, 644–650. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Pocheron, A.-L.; Hernould, M.; Haddad, N.; Tresse, O.; Cappelier, J.-M. Description of Campylobacter jejuni Bf, an atypical aero-tolerant strain. Gut Pathog. 2015, 7, 30. [Google Scholar] [CrossRef]
- Oh, E.; McMullen, L.; Jeon, B. High Prevalence of Hyper-Aerotolerant Campylobacter jejuni in Retail Poultry with Potential Implication in Human Infection. Front. Microbiol. 2015, 6, 1263. [Google Scholar] [CrossRef]
- Silva, J.; Leite, D.; Fernandes, M.; Mena, C.; Gibbs, P.A.; Teixeira, P. Campylobacter spp. as a foodborne pathogen: A review. Front. Microbiol. 2011, 2, 200. [Google Scholar] [CrossRef]
- Grant, A.; Hashem, F.; Parveen, S. Salmonella and Campylobacter: Antimicrobial resistance and bacteriophage control in poultry. Food Microbiol. 2016, 53, 104–109. [Google Scholar] [CrossRef]
- Unicomb, L.E.; Fullerton, K.E.; Kirk, M.D.; Stafford, R.J. Outbreaks of campylobacteriosis in Australia, 2001 to 2006. Foodborne Pathog. Dis. 2009, 6, 1241–1250. [Google Scholar] [CrossRef]
- Wei, B.; Cha, S.Y.; Kang, M.; Roh, J.H.; Seo, H.S.; Yoon, R.H.; Jang, H.K. Antimicrobial susceptibility profiles and molecular typing of Campylobacter jejuni and Campylobacter coli isolates from ducks in South Korea. Appl. Environ. Microbiol. 2014, 80, 7604–7610. [Google Scholar] [CrossRef]
- Chon, J.W.; Lee, S.K.; Yoon, Y.; Yoon, K.S.; Kwak, H.S.; Joo, I.S.; Seo, K.H. Quantitative prevalence and characterization of Campylobacter from chicken and duck carcasses from poultry slaughterhouses in South Korea. Poult. Sci. 2018, 97, 2909–2916. [Google Scholar] [CrossRef]
- Little, C.L.; Richardson, J.F.; Owen, R.J.; de Pinna, E.; Threlfall, E.J. Prevalence, characterisation and antimicrobial resistance of Campylobacter and Salmonella in raw poultrymeat in the UK, 2003–2005. Int. J. Environ. Health Res. 2008, 18, 403–414. [Google Scholar] [CrossRef]
- Nor Faiza, S.; Saleha, A.A.; Jalila, A.; Fauziah, N. Occurrence of Campylobacter and Salmonella in ducks and duck eggs in Selangor, Malaysia. Trop. Biomed. 2013, 30, 155–158. [Google Scholar]
- Hamed, E.A.; AbdelRahman, M.A.; Shalaby, A.G.; Morsy, M.M.; Nasef, S.A. Antibiotic resistance and polymorphism in the quinolone resistance-determining region of Campylobacter spp. isolated from 1-day-old ducklings. Vet. J. 2016, 211, 100–103. [Google Scholar] [CrossRef]
- Oh, E.; McMullen, L.M.; Chui, L.; Jeon, B. Differential Survival of Hyper-Aerotolerant Campylobacter jejuni under Different Gas Conditions. Front. Microbiol. 2017, 8, 954. [Google Scholar] [CrossRef]
- Konkel, M.E.; Gray, S.A.; Kim, B.J.; Garvis, S.G.; Yoon, J. Identification of the Enteropathogens Campylobacter jejuni and Campylobacter coli Based on the cadF Virulence Gene and Its Product. J. Clin. Microbiol. 1999, 37, 510–517. [Google Scholar]
- Bacon, D.J.; Alm, R.A.; Burr, D.H.; Hu, L.; Kopecko, D.J.; Ewing, C.P.; Guerry, P. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 2000, 68, 4384–4390. [Google Scholar] [CrossRef]
- Linton, D.; Gilbert, M.; Hitchen, P.G.; Dell, A.; Morris, H.R.; Wakarchuk, W.W.; Gregson, N.A.; Wren, B.W. Phase variation of a β-1, 3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 2000, 37, 501–514. [Google Scholar] [CrossRef]
- Corcionivoschi, N.; Gundogdu, O.; Moran, L.; Kelly, C.; Scates, P.; Stef, L.; Cean, A.; Wren, B.; Dorrell, N.; Madden, R.H. Virulence characteristics of hcp+ Campylobacter jejuni and Campylobacter coli isolates from retail chicken. Gut Pathog. 2015, 7, 20. [Google Scholar] [CrossRef]
- Koolman, L.; Whyte, P.; Burgess, C.; Bolton, D. Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 2015, 12, 424–432. [Google Scholar] [CrossRef]
- Guerry, P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007, 15, 456–461. [Google Scholar] [CrossRef]
- Dasti, J.I.; Tareen, A.M.; Lugert, R.; Zautner, A.E.; Gross, U. Campylobacter jejuni: A brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. IJMM 2010, 300, 205–211. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Clinical Breakpoints, Epidemiological Cut-Off (ECOFF) Values and EUCAST Disk Diffusion Methodology for Campylobacter jejuni and Campylobacter coli. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/Campylobacter_wide_consultation_August_2012.pdf (accessed on 1 August 2012).
- Segreti, J.; Gootz, T.D.; Goodman, L.J.; Parkhurst, G.W.; Quinn, J.P.; Martin, B.A.; Trenholme, G.M. High-level quinolone resistance in clinical isolates of Campylobacter jejuni. J. Infect. Dis. 1992, 165, 667–670. [Google Scholar] [CrossRef]
- Engberg, J.; Aarestrup, F.M.; Taylor, D.E.; Gerner-Smidt, P.; Nachamkin, I. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: Resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 2001, 7, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Bae, W.; Kaya, K.N.; Hancock, D.D.; Call, D.R.; Park, Y.H.; Besser, T.E. Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington State. Appl. Environ. Microbiol. 2005, 71, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-S.; Cho, Y.-S.; Yoon, S.-K.; Yu, M.-A.; Kim, C.-M.; Lee, J.-O.; Pyun, Y.-R. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from raw chicken meat and human stools in Korea. J. Food Prot. 2006, 69, 2915–2923. [Google Scholar] [CrossRef]
- Tang, Y.; Sahin, O.; Pavlovic, N.; LeJeune, J.; Carlson, J.; Wu, Z.; Dai, L.; Zhang, Q. Rising fluoroquinolone resistance in Campylobacter isolated from feedlot cattle in the United States. Sci. Rep. 2017, 7, 494. [Google Scholar] [CrossRef]
- Karki, A.B.; Marasini, D.; Oakey, C.K.; Mar, K.; Fakhr, M.K. Campylobacter coli from Retail Liver and Meat Products is More Aerotolerant than Campylobacter jejuni. Front. Microbiol. 2018, 9, 2951. [Google Scholar] [CrossRef]
- Bolton, D.J. Campylobacter virulence and survival factors. Food Microbiol. 2015, 48, 99–108. [Google Scholar] [CrossRef]
- Hendrixson, D.R.; DiRita, V.J. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 2004, 52, 471–484. [Google Scholar] [CrossRef]
- Sato, K.; Bartlett, P.; Kaneene, J.; Downes, F. Comparison of prevalence and antimicrobial susceptibilities of Campylobacter spp. isolates from organic and conventional dairy herds in Wisconsin. Appl. Environ. Microbiol. 2004, 70, 1442–1447. [Google Scholar] [CrossRef]
- Payot, S.; Bolla, J.-M.; Corcoran, D.; Fanning, S.; Mégraud, F.; Zhang, Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006, 8, 1967–1971. [Google Scholar] [CrossRef]
- Ku, B.K.; Kim, H.J.; Lee, Y.J.; Kim, Y.I.; Choi, J.S.; Park, M.Y.; Kwon, J.W.; Nam, H.M.; Kim, Y.H.; Jung, S.C.; et al. Genetic characterization and antimicrobial susceptibility of Campylobacter spp. isolated from domestic and imported chicken meats and humans in Korea. Foodborne Pathog. Dis. 2011, 8, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Wimalasena, S.; De Silva, B.C.J.; Hossain, S.; Pathirana, H.; Heo, G.J. Prevalence and characterisation of quinolone resistance genes in Aeromonas spp. isolated from pet turtles in South Korea. J. Glob. Antimicrob. Resist. 2017, 11, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Na, S.H.; Moon, D.C.; Choi, M.J.; Oh, S.J.; Jung, D.Y.; Sung, E.J.; Kang, H.Y.; Hyun, B.H.; Lim, S.K. Antimicrobial Resistance and Molecular Characterization of Extended-Spectrum beta-Lactamase-Producing Escherichia coli Isolated from Ducks in South Korea. Foodborne Pathog. Dis. 2019. Available online: https://www.liebertpub.com/doi/abs/10.1089/fpd.2019.2644 (accessed on 15 July 2019). [CrossRef] [PubMed]
- Na, S.H.; Moon, D.C.; Choi, M.J.; Oh, S.J.; Jung, D.Y.; Kang, H.Y.; Hyun, B.H.; Lim, S.K. Detection of oxazolidinone and phenicol resistant enterococcal isolates from duck feces and carcasses. Int. J. Food Microbiol. 2019, 293, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Pereira, S.; Sahin, O.; Lin, J.; Huang, S.; Michel, L.; Zhang, Q. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 2005, 102, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef]
- Oh, E.; McMullen, L.; Jeon, B. Impact of oxidative stress defense on bacterial survival and morphological change in Campylobacter jejuni under aerobic conditions. Front. Microbiol. 2015, 6, 295. [Google Scholar] [CrossRef]
- Askoura, M.; Sarvan, S.; Couture, J.F.; Stintzi, A. The Campylobacter jejuni Ferric Uptake Regulator Promotes Acid Survival and Cross-Protection against Oxidative Stress. Infect. Immun. 2016, 84, 1287–1300. [Google Scholar] [CrossRef]
- Elhadidy, M.; Miller, W.G.; Arguello, H.; Alvarez-Ordonez, A.; Dierick, K.; Botteldoorn, N. Molecular epidemiology and antimicrobial resistance mechanisms of Campylobacter coli from diarrhoeal patients and broiler carcasses in Belgium. Transbound. Emerg. Dis. 2019, 66, 463–475. [Google Scholar] [CrossRef]
- Duarte, A.; Santos, A.; Manageiro, V.; Martins, A.; Fraqueza, M.J.; Canica, M.; Domingues, F.C.; Oleastro, M. Human, food and animal Campylobacter spp. isolated in Portugal: High genetic diversity and antibiotic resistance rates. Int. J. Antimicrob. Agents 2014, 44, 306–313. [Google Scholar] [CrossRef]
- Litrup, E.; Torpdahl, M.; Nielsen, E. Multilocus sequence typing performed on Campylobacter coli isolates from humans, broilers, pigs and cattle originating in Denmark. J. Appl. Microbiol. 2007, 103, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Niederer, L.; Kuhnert, P.; Egger, R.; Buttner, S.; Hachler, H.; Korczak, B.M. Genotypes and antibiotic resistances of Campylobacter jejuni and Campylobacter coli isolates from domestic and travel-associated human cases. Appl. Environ. Microbiol. 2012, 78, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Asakura, H.; Sakata, J.; Nakamura, H.; Yamamoto, S.; Murakami, S. Phylogenetic Diversity and Antimicrobial Resistance of Campylobacter coli from Humans and Animals in Japan. Microbes Environ. 2019, 34, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Chui, L.; Bae, J.; Li, V.; Ma, A.; Mutschall, S.K.; Taboada, E.N.; McMullen, L.M.; Jeon, B. Frequent Implication of Multistress-Tolerant Campylobacter jejuni in Human Infections. Emerg. Infect. Dis. 2018, 24, 1037–1044. [Google Scholar] [CrossRef]


| Aerotolerance Levels | No. Isolates | Virulence Genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| flaA | flhB | cadF | pldA | iamA | ceuE | cdtA | wlaN | hcp | virB11 | ||
| OS | 6 | 83.3% | 0% | 100% | 0% | 0% | 83.3% | 100% | 83.3% | 16.7% | 0% |
| AT | 22 | 100% | 0% | 95.5% | 0% | 4.5% | 90.9% | 95.5% | 100% | 13.6% | 0% |
| HAT | 28 | 75% | 3.6% | 89.3% | 7.1% | 14.3% | 78.6% | 96.4% | 89.3% | 3.6% | 7.1% |
| Total | 56 | 85.7% | 1.8% | 92.9% | 3.6% | 8.9% | 83.9% | 96.4% | 92.9% | 8.9% | 3.6% |
| Agents | Aerotolerance Levels | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 6 | 8 | 16 | 32 | 64 | >64 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | OS | 2 | 2 | 2 | |||||||||||
| AT | 3 | 7 | 6 | 3 | 2 | 1 | |||||||||
| HAT | 1 | 7 | 5 | 8 | 5 | 1 | 1 | ||||||||
| CHL | OS | 2 | 3 | 1 | |||||||||||
| AT | 17 | 5 | |||||||||||||
| HAT | 15 | 9 | 4 | ||||||||||||
| CIP | OS | 1 | 1 | 4 | |||||||||||
| AT | 11 | 9 | 2 | ||||||||||||
| HAT | 2 | 10 | 9 | 6 | 1 | ||||||||||
| TET | OS | 2 | 1 | 3 | |||||||||||
| AT | 4 | 1 | 1 | 1 | 5 | 10 | |||||||||
| HAT | 4 | 3 | 1 | 1 | 5 | 14 | |||||||||
| TEL | OS | 2 | 2 | 2 | |||||||||||
| AT | 3 | 5 | 5 | 4 | 1 | 3 | 1 | ||||||||
| HAT | 6 | 7 | 4 | 4 | 4 | 3 | |||||||||
| GEN | OS | 2 | 2 | 2 | |||||||||||
| AT | 4 | 15 | 2 | 1 | |||||||||||
| HAT | 1 | 24 | 2 | 1 | |||||||||||
| AZI | OS | 3 | 1 | 2 | |||||||||||
| AT | 3 | 13 | 5 | 1 | |||||||||||
| HAT | 2 | 13 | 10 | 1 | 1 | 1 | |||||||||
| STR | OS | 2 | 1 | 3 | |||||||||||
| AT | 4 | 15 | 3 | ||||||||||||
| HAT | 7 | 13 | 2 | 6 | |||||||||||
| NAL | OS | 1 | 5 | ||||||||||||
| AT | 18 | 4 | |||||||||||||
| HAT | 22 | 6 |
| AMR Patterns | No. of Isolates | OS | AT | HAT |
|---|---|---|---|---|
| TET | 1 | 1 | 0 | 0 |
| CIP-NAL | 11 | 0 | 4 | 7 |
| CIP-NAL-TET | 30 | 2 | 15 | 13 |
| CIP-NAL-TET-STR* | 8 | 1 | 2 | 5 |
| CIP-NAL-TET-STR-TEL* | 1 | 0 | 0 | 1 |
| CIP-NAL-STR-ERY-TEL-AZI-GEN* | 2 | 0 | 1 | 1 |
| CIP-NAL-TET-STR-ERY-TEL-AZI* | 1 | 0 | 0 | 1 |
| CIP-NAL-TET-STR-ERY-TEL-AZI-GEN* | 2 | 2 | 0 | 0 |
| Aerotolerance Levels | Eight Shared STs * | Not-Shared STs ** | Odds | OR (95% CI) † |
|---|---|---|---|---|
| HAT vs OS | ||||
| Oxygen-sensitive (OS) | 1 (2.4%) | 5 (35.7%) | 0.2 | 30 (2.7, 328.6) |
| Aerotolerant (AT) | 17 (40.5%) | 5 (35.7%) | 3.4 | |
| Hyper-aerotolerant (HAT) | 24 (57.1%) | 4 (28.6%) | 6 | |
| Total | 42 | 14 | ||
| 56 | ||||
| Aerotolerance Levels | Eleven Shared STs * | Not-Shared STs ** | Odds | OR (95% CI) † |
|---|---|---|---|---|
| HAT vs OS | ||||
| Oxygen-sensitive (OS) | 1 (2.1%) | 5 (55.6%) | 0.2 | 65 (4.9, 861.5) |
| Aerotolerant (AT) | 20 (42.6%) | 2 (22.2%) | 10 | |
| Hyper-aerotolerant (HAT) | 26 (55.3%) | 2 (22.2%) | 13 | |
| Total | 47 | 9 | ||
| 56 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guk, J.-H.; Kim, J.; Song, H.; Kim, J.; An, J.-U.; Kim, J.; Ryu, S.; Jeon, B.; Cho, S. Hyper-Aerotolerant Campylobacter coli from Duck Sources and Its Potential Threat to Public Health: Virulence, Antimicrobial Resistance, and Genetic Relatedness. Microorganisms 2019, 7, 579. https://doi.org/10.3390/microorganisms7110579
Guk J-H, Kim J, Song H, Kim J, An J-U, Kim J, Ryu S, Jeon B, Cho S. Hyper-Aerotolerant Campylobacter coli from Duck Sources and Its Potential Threat to Public Health: Virulence, Antimicrobial Resistance, and Genetic Relatedness. Microorganisms. 2019; 7(11):579. https://doi.org/10.3390/microorganisms7110579
Chicago/Turabian StyleGuk, Jae-Ho, Junhyung Kim, Hyokeun Song, Jinshil Kim, Jae-Uk An, Jonghyun Kim, Sangryeol Ryu, Byeonghwa Jeon, and Seongbeom Cho. 2019. "Hyper-Aerotolerant Campylobacter coli from Duck Sources and Its Potential Threat to Public Health: Virulence, Antimicrobial Resistance, and Genetic Relatedness" Microorganisms 7, no. 11: 579. https://doi.org/10.3390/microorganisms7110579
APA StyleGuk, J.-H., Kim, J., Song, H., Kim, J., An, J.-U., Kim, J., Ryu, S., Jeon, B., & Cho, S. (2019). Hyper-Aerotolerant Campylobacter coli from Duck Sources and Its Potential Threat to Public Health: Virulence, Antimicrobial Resistance, and Genetic Relatedness. Microorganisms, 7(11), 579. https://doi.org/10.3390/microorganisms7110579







