Perilla frutescens Leaf Alters the Rumen Microbial Community of Lactating Dairy Cows
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Diets, and Experimental Design
2.2. Sample Collection and Measurements
2.3. DNA Extraction and Sequencing
2.4. Bioinformatic Analysis
2.5. Correlation and Statistical Analysis
3. Results
3.1. Characteristics of PFL
3.2. Milk Performance and Rumen Fermentation
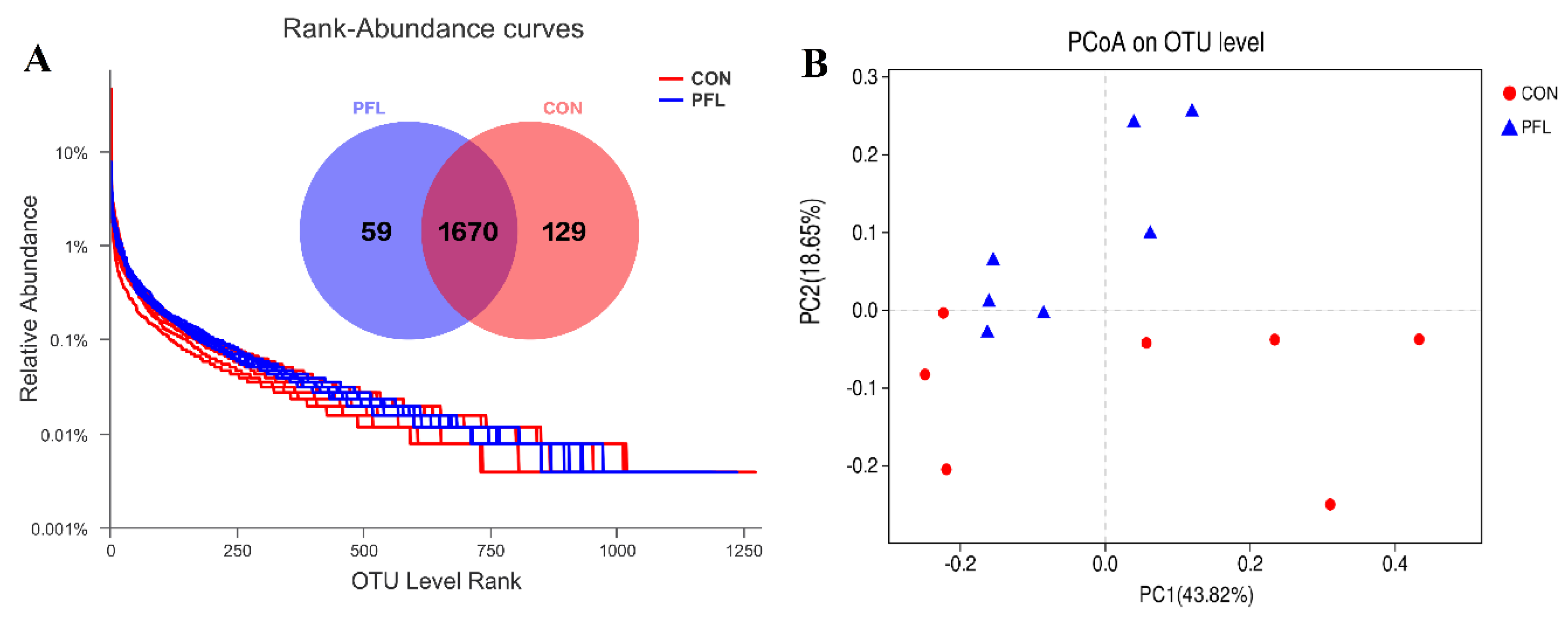
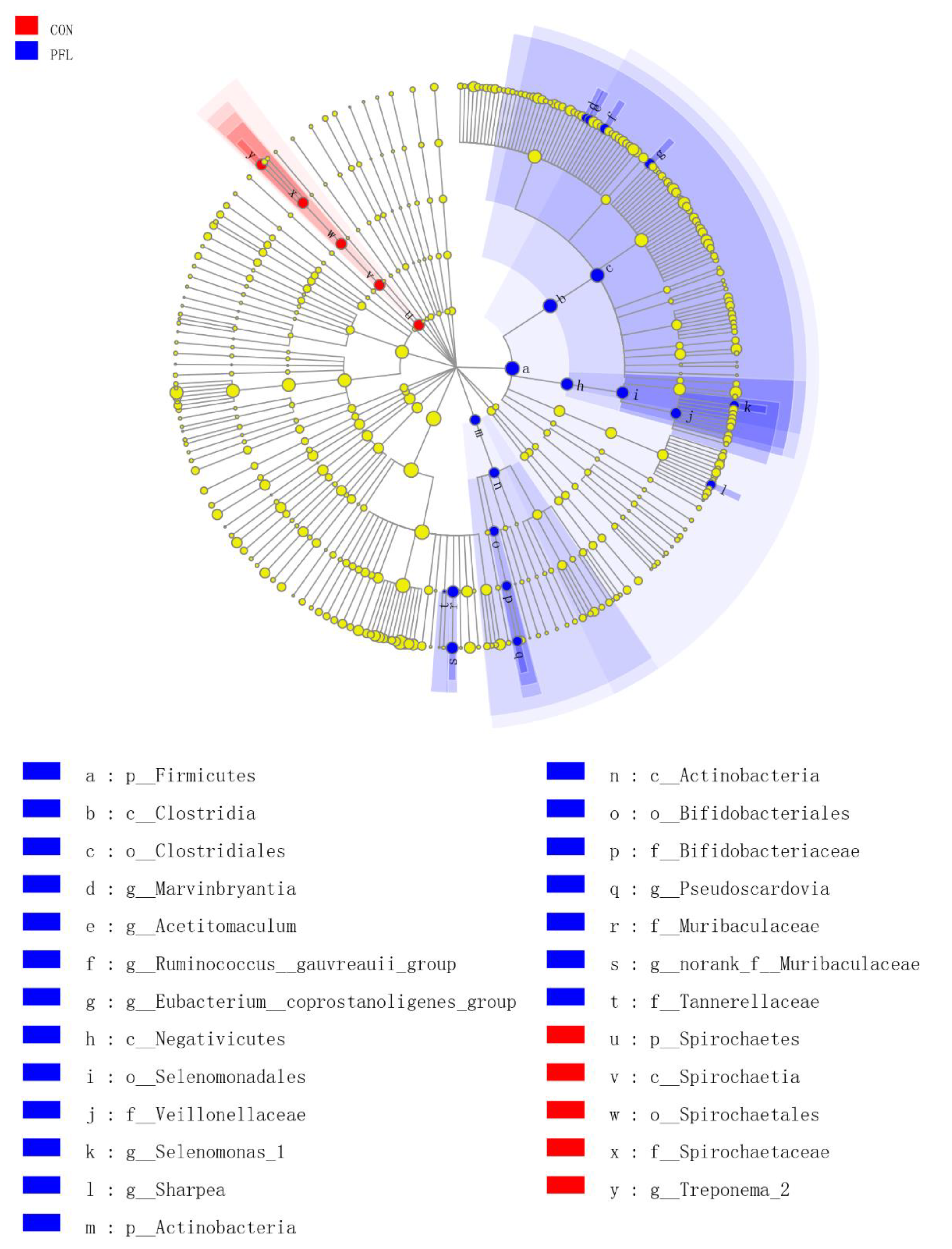
3.3. Change in Ruminal Bacterial Communities
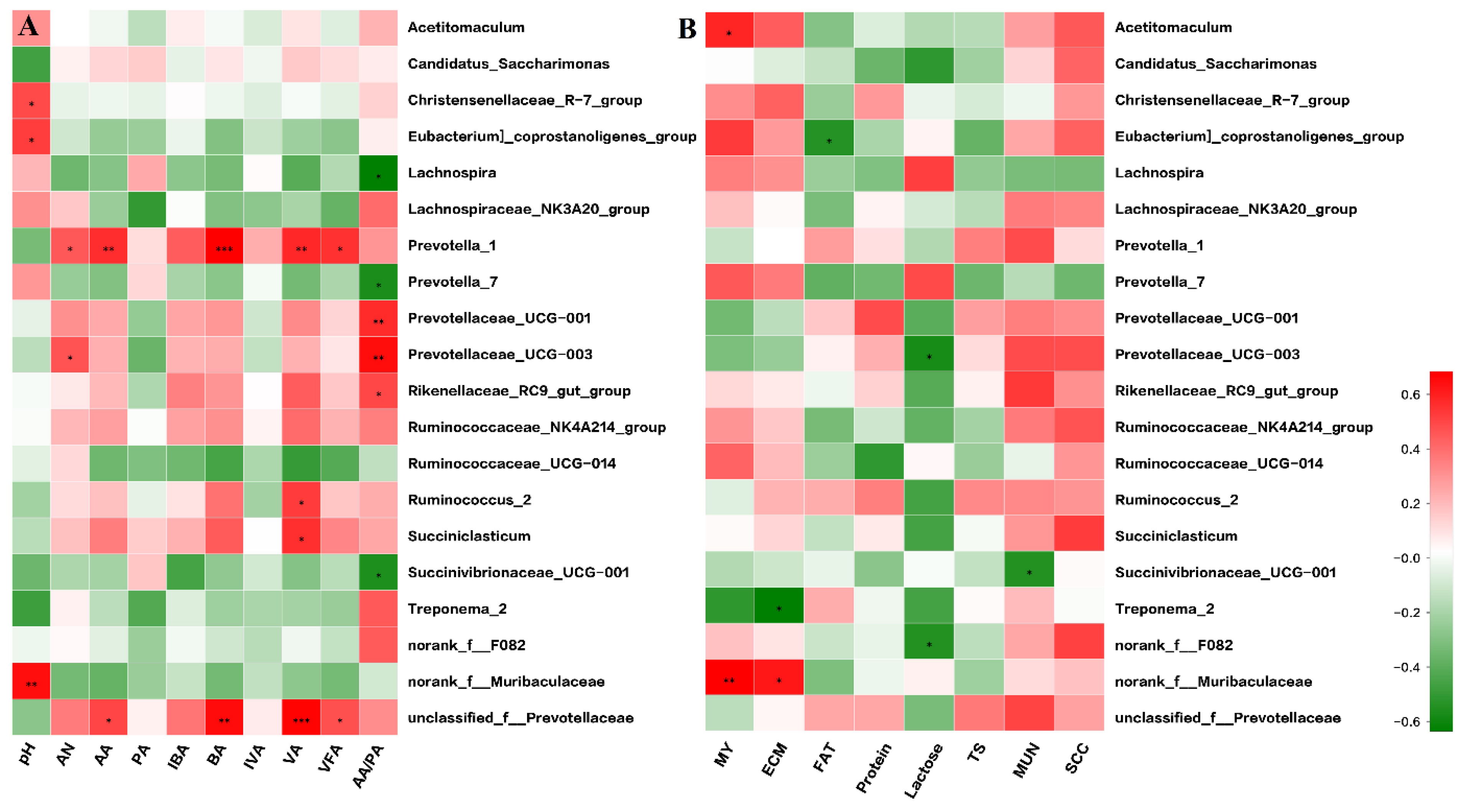
3.4. Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, B.; Ma, M.; Diao, Q.; Tu, Y. Saponin-Induced Shifts in the Rumen Microbiome and Metabolome of Young Cattle. Front. Microbiol. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Kamra, D.; Patra, A.; Chatterjee, P.; Kumar, R.; Agarwal, N.; Chaudhary, L. Effect of plant extracts on methanogenesis and microbial profile of the rumen of buffalo: A brief overview. Aust. J. Exp. Agr. 2008, 48, 175–178. [Google Scholar] [CrossRef]
- Jun, H.-I.; Kim, B.-T.; Song, G.-S.; Kim, Y.-S. Structural characterization of phenolic antioxidants from purple perilla (Perilla frutescens var. acuta) leaves. Food Chem. 2014, 148, 367–372. [Google Scholar] [CrossRef]
- Yu, H.; Qiu, J.-F.; Ma, L.-J.; Hu, Y.-J.; Li, P.; Wan, J.-B. Phytochemical and phytopharmacological review of Perilla frutescens L. (Labiatae), a traditional edible-medicinal herb in China. Food Chem. Toxicol. 2017, 108, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Mertenat, D.; dal Cero, M.; Vogl, C.R.; Ivemeyer, S.; Meier, B.; Maeschli, A.; Hamburger, M.; Walkenhorst, M. Ethnoveterinary knowledge of farmers in bilingual regions of Switzerland–is there potential to extend veterinary options to reduce antimicrobial use? J. Ethnopharmacol. 2019, 246, 112184. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.N.; Oh, J.; Firkins, J.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.; Adesogan, A.; Yang, W.; Lee, C. Special topics—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar]
- Shen, H.; Lu, Z.; Xu, Z.; Chen, Z.; Shen, Z. Associations among dietary non-fiber carbohydrate, ruminal microbiota and epithelium G-protein-coupled receptor, and histone deacetylase regulations in goats. Microbiome 2017, 5, 123. [Google Scholar] [CrossRef]
- Yusuf, A.L.; Adeyemi, K.D.; Samsudin, A.A.; Goh, Y.M.; Alimon, A.R.; Sazili, A.Q. Effects of dietary supplementation of leaves and whole plant of Andrographis paniculata on rumen fermentation, fatty acid composition and microbiota in goats. BMC Vet. Res. 2017, 13, 349. [Google Scholar] [CrossRef] [PubMed]
- Seow, Y.X.; Yeo, C.R.; Chung, H.L.; Yuk, H.G. Plant essential oils as active antimicrobial agents. Crit. Rev. Food Sci. Nutr. 2014, 54, 625–644. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, M.; Wu, Y.; Wang, L.; Liu, J.; Jiang, L.; Yu, Z. Medicinal herbs as a potential strategy to decrease methane production by rumen microbiota: A systematic evaluation with a focus on Perilla frutescens seed extract. Appl. Microbiol. Biot. 2016, 100, 9757–9771. [Google Scholar] [CrossRef] [PubMed]
- Vasta, V.; Daghio, M.; Cappucci, A.; Buccioni, A.; Serra, A.; Viti, C.; Mele, M. Invited review: Plant polyphenols and rumen microbiota responsible for fatty acid biohydrogenation, fiber digestion, and methane emission: Experimental evidence and methodological approaches. J. Dairy Sci. 2019, 102, 3781–3804. [Google Scholar] [CrossRef]
- Shen, J.; Chai, Z.; Song, L.; Liu, J.; Wu, Y. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012, 95, 5978–5984. [Google Scholar] [CrossRef]
- Hu, W.-L.; Liu, J.-X.; Ye, J.-A.; Wu, Y.-M.; Guo, Y.-Q. Effect of tea saponin on rumen fermentation in vitro. Anim. Feed Sci. Technol. 2005, 120, 333–339. [Google Scholar] [CrossRef]
- Broderick, G.; Kang, J. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media1. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.C.; Wang, H.Y.; Gai, X.P. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Wang, B.; Tu, Y.; Zhao, S.; Hao, Y.; Liu, J.; Liu, F.; Xiong, B.; Jiang, L. Effect of tea saponins on milk performance, milk fatty acids, and immune function in dairy cow. J. Dairy Sci. 2017, 100, 8043–8052. [Google Scholar] [CrossRef]
- Klevenhusen, F.; Petri, R.M.; Kleefisch, M.-T.; Khiaosa-ard, R.; Metzler-Zebeli, B.U.; Zebeli, Q. Changes in fibre-adherent and fluid-associated microbial communities and fermentation profiles in the rumen of cattle fed diets differing in hay quality and concentrate amount. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Xue, M.; Sun, H.; Wu, X.; Guan, L.; Liu, J. Assessment of rumen bacteria in dairy cows with varied milk protein yield. J. Dairy Sci. 2019, 102, 5031–5041. [Google Scholar] [CrossRef] [PubMed]
- Zened, A.; Combes, S.; Cauquil, L.; Mariette, J.; Klopp, C.; Bouchez, O.; Troegeler-Meynadier, A.; Enjalbert, F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 2013, 83, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Kamke, J.; Kittelmann, S.; Soni, P.; Li, Y.; Tavendale, M.; Ganesh, S.; Janssen, P.H.; Shi, W.; Froula, J.; Rubin, E.M. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 2016, 4, 56. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.J.; Miller, R.A.; Ericsson, A.C.; Harrison, D.C.; Strong, R.; Schmidt, T.M. Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol. 2019, 19, 130. [Google Scholar] [CrossRef] [PubMed]
- Ormerod, K.L.; Wood, D.L.; Lachner, N.; Gellatly, S.L.; Daly, J.N.; Parsons, J.D.; Dal’Molin, C.G.; Palfreyman, R.W.; Nielsen, L.K.; Cooper, M.A. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 2016, 4, 36. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.; Gálvez, E.J.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef]
- Ragsdale, S.W. Enzymology of the Wood–Ljungdahl pathway of acetogenesis. Ann. N. Y. Acad. Sci. 2008, 1125, 129. [Google Scholar] [CrossRef]
- Martin, C.; Morgavi, D.; Doreau, M. Methane mitigation in ruminants: From microbe to the farm scale. Animal 2010, 4, 351–365. [Google Scholar] [CrossRef]
- Wang, H.; He, Y.; Li, H.; Wu, F.; Qiu, Q.; Niu, W.; Gao, Z.; Su, H.; Cao, B. Rumen fermentation, intramuscular fat fatty acid profiles and related rumen bacterial populations of Holstein bulls fed diets with different energy levels. Appl. Microbiol. Biot. 2019, 1–12. [Google Scholar] [CrossRef]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhao, R. Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef]
- Tong, J.; Zhang, H.; Yang, D.; Zhang, Y.; Xiong, B.; Jiang, L. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS ONE 2018, 13, e0198225. [Google Scholar] [CrossRef] [PubMed]
- Firrman, J.; Liu, L.; Zhang, L.; Argoty, G.A.; Wang, M.; Tomasula, P.; Kobori, M.; Pontious, S.; Xiao, W. The effect of quercetin on genetic expression of the commensal gut microbes Bifidobacterium catenulatum, Enterococcus caccae and Ruminococcus gauvreauii. Anaerobe 2016, 42, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Duda-Chodak, A. The inhibitory effect of polyphenols on human gut microbiota. J. Physiol. Pharmacol. 2012, 63, 497–503. [Google Scholar] [PubMed]
- Watanabe, M.; Kaku, N.; Ueki, K.; Ueki, A. Falcatimonas natans gen. nov., sp. nov., a strictly anaerobic, amino-acid-decomposing bacterium isolated from a methanogenic reactor of cattle waste. Int. J. Syst. Evol. Micr. 2016, 66, 4639–4644. [Google Scholar] [CrossRef]
- Kumar, S.; Treloar, B.P.; Teh, K.H.; McKenzie, C.M.; Henderson, G.; Attwood, G.T.; Waters, S.M.; Patchett, M.L.; Janssen, P.H. Sharpea and Kandleria are lactic acid producing rumen bacteria that do not change their fermentation products when co-cultured with a methanogen. Anaerobe 2018, 54, 31–38. [Google Scholar] [CrossRef]
- Gutierrez, J. Numbers and characteristics of lactate utilizing organisms in the rumen of cattle. J. Bacterial. 1953, 66, 123. [Google Scholar]
- Stanton, T.; Canale-Parola, E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch. Microbiol. 1980, 127, 145–156. [Google Scholar] [CrossRef]
- Bekele, A.Z.; Koike, S.; Kobayashi, Y. Phylogenetic diversity and dietary association of rumen Treponema revealed using group-specific 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 2011, 316, 51–60. [Google Scholar] [CrossRef]
- Luo, D.; Gao, Y.; Lu, Y.; Qu, M.; Xiong, X.; Xu, L.; Zhao, X.; Pan, K.; Ouyang, K. Niacin alters the ruminal microbial composition of cattle under high-concentrate condition. Anim. Nutr. 2017, 3, 180–185. [Google Scholar] [CrossRef]
- Liu, J.; Pu, Y.-Y.; Xie, Q.; Wang, J.-K.; Liu, J.-X. Pectin induces an in vitro rumen microbial population shift attributed to the pectinolytic Treponema group. Curr. Microbiol. 2015, 70, 67–74. [Google Scholar] [CrossRef]
- Homer, K.A.; Manji, F.; Beighton, D. Inhibition of peptidase and glycosidase activities of Porphyromonas gingivalis, Bacteroides intermedius and Treponema denticola by plant extracts. J. Clin. Periodontol. 1992, 19, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Pope, P.; Smith, W.; Denman, S.; Tringe, S.; Barry, K.; Hugenholtz, P.; McSweeney, C.; McHardy, A.; Morrison, M. Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar wallabies. Science 2011, 333, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.d.O.; Thompson, F. The family succinivibrionaceae. Prokaryotes: Gammaproteobacteria 2014, 639–648. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Zingaretti, L.; Popova, M.; Estellé, J.; Bernard, A.; Pons, N.; Bellot, P.; Mach, N.; Rau, A.; Roume, H. Identification of rumen microbial biomarkers linked to methane emission in Holstein dairy cows. J. Anim. Breed. Genet. 2019. [Google Scholar] [CrossRef]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biot. 2007, 75, 165–174. [Google Scholar] [CrossRef]

| Item | CON | PFL | SEM | p-Value | ||
|---|---|---|---|---|---|---|
| Treatment | Week | Treatment × Week | ||||
| Dry matter intake, kg/d | 24.3 | 24.7 | 0.17 | 0.23 | <0.01 | 0.63 |
| Milk yield, kg/d | ||||||
| Raw | 36.8 | 39.2 | 0.77 | 0.04 | <0.01 | 0.12 |
| ECM a | 45.4 | 47.3 | 1.34 | 0.35 | 0.04 | 0.57 |
| Protein | 1.28 | 1.36 | 0.026 | 0.04 | 0.10 | 0.73 |
| Fat | 1.76 | 1.78 | 0.078 | 0.91 | <0.01 | 0.41 |
| Lactose | 1.84 | 2.05 | 0.046 | 0.01 | 0.10 | 0.77 |
| Total solids | 4.98 | 5.27 | 0.127 | 0.12 | 0.07 | 0.66 |
| Milk content, g/100g | ||||||
| Protein | 3.47 | 3.39 | 0.037 | 0.19 | <0.01 | 0.58 |
| Fat | 4.80 | 4.40 | 0.187 | 0.15 | <0.01 | 0.09 |
| Lactose | 5.05 | 5.10 | 0.025 | 0.18 | 0.60 | 0.08 |
| TS | 13.5 | 13.1 | 0.21 | 0.19 | <0.01 | 0.16 |
| Milk urea nitrogen, mg/dL | 16.3 | 16.8 | 0.34 | 0.34 | <0.01 | 0.93 |
| Fat: Protein | 1.38 | 1.30 | 0.051 | 0.30 | <0.01 | 0.16 |
| Somatic cell count, ×103 | 386.6 | 269.9 | 109.74 | 0.46 | 0.89 | 0.99 |
| Feed efficiency b | 1.51 | 1.63 | 0.011 | <0.01 | <0.01 | 0.48 |
| Items | Treatments a | SEM | p-Value | |||
|---|---|---|---|---|---|---|
| CON | SD | PFL | SD | |||
| pH | 6.20 | 0.23 | 6.47 | 0.22 | 0.075 | 0.04 |
| Ammonia-nitrogen, mg/dL | 9.97 | 4.99 | 8.66 | 2.20 | 1.825 | 0.63 |
| Concentration, mmol/L | ||||||
| Total VFA a | 67.8 | 13.1 | 71.5 | 15.5 | 6.33 | 0.70 |
| Acetate | 42.6 | 9.56 | 42.8 | 9.34 | 4.12 | 0.96 |
| Propionate | 15.7 | 2.91 | 18.2 | 3.78 | 1.52 | 0.30 |
| Butyrate | 7.39 | 1.58 | 8.15 | 2.11 | 0.741 | 0.50 |
| Valerate | 0.99 | 0.22 | 1.15 | 0.28 | 0.099 | 0.29 |
| Isobutyrate | 0.41 | 0.09 | 0.43 | 0.12 | 0.052 | 0.79 |
| Isovalerate | 0.7 | 0.22 | 0.69 | 0.25 | 0.12 | 0.94 |
| Molar proportion, % | ||||||
| Acetate | 62.6 | 3.92 | 59.9 | 1.35 | 1.1 | 0.14 |
| Propionate | 23.4 | 3.50 | 25.6 | 1.85 | 1.13 | 0.23 |
| Butyrate | 10.9 | 0.95 | 11.3 | 0.62 | 0.22 | 0.20 |
| Valerate | 1.46 | 0.19 | 1.61 | 0.14 | 0.05 | 0.08 |
| Isobutyrate | 0.6 | 0.07 | 0.6 | 0.14 | 0.05 | 0.98 |
| Isovalerate | 1.03 | 0.20 | 0.96 | 0.31 | 0.129 | 0.73 |
| Acetate: propionate | 2.74 | 0.55 | 2.35 | 0.21 | 0.162 | 0.14 |
| Items | Treatments | SEM a | p-Value | |
|---|---|---|---|---|
| CON | PFL | |||
| Sobs | 1147 | 1171 | 79.538 | 0.62 |
| Shannon | 5.12 | 5.58 | 0.429 | 0.13 |
| Simpson | 0.05 | 0.01 | 0.038 | 0.16 |
| Ace | 1379 | 1385 | 70.5 | 0.87 |
| Chao | 1394 | 1386 | 69.4 | 0.84 |
| Coverage | 0.99 | 0.99 | 0.001 | 0.31 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Yu, Z.; Wang, B. Perilla frutescens Leaf Alters the Rumen Microbial Community of Lactating Dairy Cows. Microorganisms 2019, 7, 562. https://doi.org/10.3390/microorganisms7110562
Sun Z, Yu Z, Wang B. Perilla frutescens Leaf Alters the Rumen Microbial Community of Lactating Dairy Cows. Microorganisms. 2019; 7(11):562. https://doi.org/10.3390/microorganisms7110562
Chicago/Turabian StyleSun, Zhiqiang, Zhu Yu, and Bing Wang. 2019. "Perilla frutescens Leaf Alters the Rumen Microbial Community of Lactating Dairy Cows" Microorganisms 7, no. 11: 562. https://doi.org/10.3390/microorganisms7110562
APA StyleSun, Z., Yu, Z., & Wang, B. (2019). Perilla frutescens Leaf Alters the Rumen Microbial Community of Lactating Dairy Cows. Microorganisms, 7(11), 562. https://doi.org/10.3390/microorganisms7110562





