Acinetobacter Strain KUO11TH, a Unique Organism Related to Acinetobacter pittii and Isolated from the Skin Mucus of Healthy Bighead Catfish and Its Efficacy Against Several Fish Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Cultivation
2.2. Bacterial Cell Morphology Analysis
2.3. DNA Extraction, PCR Amplification and DNA Sequencing
2.4. Phylogenetic Analysis
2.5. Genome Sequencing, Assembly and Annotation
2.6. Genome Sequencing Analysis, ANI, in silico DDH Calculations and Phylogeny
2.7. DNA G+C Content Analysis
2.8. GenBank/EMBL/DDBJ Accession Numbers
2.9. Phenotypic and Chemotaxonomic Analyses
2.10. Cellular Fatty Acid Analysis
2.11. Identification of Antibiotic Resistance Genes
2.12. Antibiotic Susceptibility Test
2.13. Antagonism Against Pathogenic Bacteria
2.14. Quantitative Analysis of Bacterial Coculture Assay Results
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brisou, J.; Prevot, A.R. Studies on bacterial taxonomy. X. The revision of species under Acromobacter group. Ann. Institut Pasteur 1954, 86, 722–728. [Google Scholar]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Guo, Z.B.; Du, Z.M.; Yang, H.Y.; Bi, Y.J.; Wang, G.Q.; Tan, Y.F. Cellular fatty acids as chemical markers for differentiation of Acinetobacter baumannii and Acinetobacter calcoaceticus. Biomed. Environ. Sci. 2012, 25, 711–717. [Google Scholar] [PubMed]
- Harding, C.M.; Tracy, E.N.; Carruthers, M.D.; Rather, P.N.; Actis, L.A.; Munson, R.S., Jr. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. MBio 2013, 4, e00360. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, P.J.; Jeanjean, S. Delineation of new proteolytic genomic species in the genus Acinetobacter. Res. Microbiol. 1989, 140, 291–299. [Google Scholar] [CrossRef]
- Bouvet, P.J.M.; Grimont, P.A.D. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Evol. Microbiol. 1986, 36, 228–240. [Google Scholar]
- Gerner-Smidt, P.; Tjernberg, I.; Ursing, J. Reliability of phenotypic tests for identification of Acinetobacter species. J. Clin. Microbiol. 1991, 29, 277–282. [Google Scholar] [PubMed]
- Vaneechoutte, M.; Nemec, A.; Musilek, M.; van der Reijden, T.J.; van den Barselaar, M.; Tjernberg, I.; Calame, W.; Fani, R.; de Baere, T.; Dijkshoorn, L. Description of Acinetobacter venetianus ex Di Cello et al. 1997 sp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- Cosgaya, C.; Mari-Almirall, M.; van Assche, A.; Fernandez-Orth, D.; Mosqueda, N.; Telli, M.; Huys, G.; Higgins, P.G.; Seifert, H.; Lievens, B.; et al. Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int. J. Syst. Evol. Microbiol. 2016, 66, 4105–4111. [Google Scholar] [PubMed]
- Nemec, A.; Krizova, L.; Maixnerova, M.; Sedo, O.; Brisse, S.; Higgins, P.G. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2015, 65, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Maslunka, C.; Gifford, B.; Tucci, J.; Gürtler, V.; Seviour, R.J. Insertions or deletions (Indels) in the rrn 16S-23S rRNA gene internal transcribed spacer region (ITS) compromise the typing and identification of strains within the Acinetobacter calcoaceticus-baumannii (Acb) complex and closely related members. PLoS ONE 2014, 9, e105390. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.L.; Lee, Y.T.; Kuo, S.C.; Yang, S.P.; Fung, C.P.; Lee, S.D. Rapid identification of Acinetobacter baumannii, Acinetobacter nosocomialis and Acinetobacter pittii with a multiplex PCR assay. J. Med. Microbiol. 2014, 63, 1154–1159. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Lu, C.; Chen, H. Acine to bacter baumannii a novel pathogen of acute epidemic in mandarin fish (Siniperca chuatsi). Wei Sheng Wu Xue Tong Bao 1997, 24, 104–106,183. [Google Scholar]
- Xia, L.; Xiong, D.; Gu, Z.; Xu, Z.; Chen, C.; Xie, J.; Xu, P. Recovery of Acinetobacter baumannii from diseased channel catfish (Ictalurus punctatus) in China. Aquaculture 2008, 284, 285–288. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Cao, X.; Wang, W.; Luo, Y. Isolation, identification and characterisation of an emerging fish pathogen, Acinetobacter pittii, from diseased loach (Misgurnus anguillicaudatus) in China. Antonie Leeuwenhoek 2019. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Chen, H.; Wang, X.; Zou, Y.; Huang, C. Identification and phylogenetic analysis of the pathogenic Acinotobacter baumannii from hybridized prussian carp. Chin. Vet. Sci. 2009, 39, 303–309. [Google Scholar] [CrossRef]
- Carr, E.L.; Kampfer, P.; Patel, B.K.; Gurtler, V.; Seviour, R.J. Seven novel species of Acinetobacter isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2003, 53, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.S.; Jung, J.; Jeon, C.O.; Park, W. Acinetobacter oleivorans sp. nov. is capable of adhering to and growing on diesel-oil. J. Microbiol. 2011, 49, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Krizova, L.; Maixnerova, M.; Sedo, O.; Nemec, A. Acinetobacter bohemicus sp. nov. widespread in natural soil and water ecosystems in the Czech Republic. Syst. Appl. Microbiol. 2014, 37, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Krizova, L.; McGinnis, J.; Maixnerova, M.; Nemec, M.; Poirel, L.; Mingle, L.; Sedo, O.; Wolfgang, W.; Nemec, A. Acinetobacter variabilis sp. nov. (formerly DNA group 15 sensu Tjernberg & Ursing), isolated from humans and animals. Int. J. Syst. Evol. Microbiol. 2015, 65, 857–863. [Google Scholar] [PubMed]
- Li, Y.; Chang, J.; Guo, L.M.; Wang, H.M.; Xie, S.J.; Piao, C.G.; He, W. Description of Acinetobacter populi sp. nov. isolated from symptomatic bark of Populus × euramericana canker. Int. J. Syst. Evol. Microbiol. 2015, 65, 4461–4468. [Google Scholar] [CrossRef] [PubMed]
- Poppel, M.T.; Skiebe, E.; Laue, M.; Bergmann, H.; Ebersberger, I.; Garn, T.; Fruth, A.; Baumgardt, S.; Busse, H.J.; Wilharm, G. Acinetobacter equi sp. nov., isolated from horse faeces. Int. J. Syst. Evol. Microbiol. 2016, 66, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Rooney, A.P.; Dunlap, C.A.; Flor-Weiler, L.B. Acinetobacter lactucae sp. nov., isolated from iceberg lettuce (Asteraceae: Lactuca sativa). Int. J. Syst. Evol. Microbiol. 2016, 66, 3566–3572. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations (FAO). Fishstat Plus; FAO: Rome, Italy, 2017. [Google Scholar]
- Na-Nakorn, U.; Kamonrat, W.; Ngamsiri, T. Genetic diversity of walking catfish, Clarias macrocephalus, in Thailand and evidence of genetic introgression from introduced farmed C. gariepinus. Aquaculture 2004, 240, 145–163. [Google Scholar] [CrossRef]
- Mohapatra, S.; Chakraborty, T.; Kumar, V.; DeBoeck, G.; Mohanta, K.N. Aquaculture and stress management: A review of probiotic intervention. J. Anim. Physiol. Anim. Nutr. 2013, 97, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Hai, N.V. Research findings from the use of probiotics in tilapia aquaculture: A review. Fish Shellfish Immunol. 2015, 45, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Kamlage, B. Methods for General and Molecular Bacteriology; 791 Pages, Numerous Figures and Tables; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1996. [Google Scholar]
- Fredriksson, N.J.; Hermansson, M.; Wilen, B.M. The choice of PCR primers has great impact on assessments of bacterial community diversity and dynamics in a wastewater treatment plant. PLoS ONE 2013, 8, e76431. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Gundi, V.A.; Khamis, A.; Raoult, D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 2006, 44, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Harayama, S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 1995, 61, 1104–1109. [Google Scholar] [PubMed]
- Campanella, J.J.; Bitincka, L.; Smalley, J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Saitou, N. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Medigue, C.; Calteau, A.; Cruveiller, S.; Gachet, M.; Gautreau, G.; Josso, A.; Lajus, A.; Langlois, J.; Pereira, H.; Planel, R.; et al. MicroScope—An integrated resource for community expertise of gene functions and comparative analysis of microbial genomic and metabolic data. Brief. Bioinform. 2017, 20, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rossello-Mora, R.; Glockner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; Klenk, H.-P.; Göker, M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genom. Sci. 2010, 2, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 1956, 178, 703. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, R.C.; Fryer, T.F.; Reiter, B. The production and characterization of lipases from a micrococcus and a pseudomonad. J. Gen. Microbiol. 1967, 48, 401–418. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Whittaker, P.; Fry, F.S.; Curtis, S.K.; Al-Khaldi, S.F.; Mossoba, M.M.; Yurawecz, M.P.; Dunkel, V.C. Use of fatty acid profiles to identify food-borne bacterial pathogens and aerobic endospore-forming bacilli. J. Agric. Food Chem. 2005, 53, 3735–3742. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Disk Susceptibility Tests; National Committee for Clinical Laboratory Standards: Villanova, PA, USA, 1997.
- Koch, A.L. Growth measurement. In Methods for General and Molecular Bacteriology; Gerhardt, P.R., Murray, G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 248–277. [Google Scholar]
- Nemec, A.; Radolfova-Krizova, L.; Maixnerova, M.; Vrestiakova, E.; Jezek, P.; Sedo, O. Taxonomy of haemolytic and/or proteolytic strains of the genus Acinetobacter with the proposal of Acinetobacter courvalinii sp. nov. (genomic species 14 sensu Bouvet & Jeanjean), Acinetobacter dispersus sp. nov. (genomic species 17), Acinetobacter modestus sp. nov., Acinetobacter proteolyticus sp. nov. and Acinetobacter vivianii sp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 1673–1685. [Google Scholar] [PubMed]
- Nemec, A.; Krizova, L.; Maixnerova, M.; van der Reijden, T.J.K.; Deschaght, P.; Passet, V.; Vaneechoutte, M.; Brisse, S.; Dijkshoorn, L. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Microbiol. Res. 2011, 162, 393–404. [Google Scholar]
- Kim, P.S.; Shin, N.R.; Kim, J.Y.; Yun, J.H.; Hyun, D.W.; Bae, J.W. Acinetobacter apis sp. nov., isolated from the intestinal tract of a honey bee, Apis mellifera. J. Microbiol. 2014, 52, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, J.; Anand, S.; Jindal, S.; Rajagopal, R.; Lal, R. Acinetobacter indicus sp. nov., isolated from a hexachlorocyclohexane dump site. Int. J. Syst. Evol. Microbiol. 2012, 62, 2883–2890. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Ino, T.; Iizuka, H. Acinetobacter radioresistens sp. nov. Isolated from cotton and soil. Int. J. Syst. Evol. Microbiol. 1988, 38, 209–211. [Google Scholar] [CrossRef]
- Nemec, A.; Dijkshoorn, L.; Cleenwerck, I.; de Baere, T.; Janssens, D.; van der Reijden, T.J.; Jezek, P.; Vaneechoutte, M. Acinetobacter parvus sp. nov., a small-colony-forming species isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2003, 53, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Dong, X.H.; Tan, B.P.; Yang, Q.H.; Chi, S.Y.; Liu, H.Y.; Liu, X.Q. Effects of dietary probiotic on the growth performance, non-specific immunity and disease resistance of cobia, Rachycentron canadum. Aquac. Nutr. 2012, 18, 46–55. [Google Scholar] [CrossRef]
- Verschuere, L.; Rombaut, G.; Sorgeloos, P.; Verstraete, W. Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 2000, 64, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.X.; Zhao, S.M.; Peng, N.; Xu, C.P.; Wang, J.; Liang, Y.X. Effects of a probiotic (Bacillus subtilis FY99-01) on the bacterial community structure and composition of shrimp (Litopenaeus vannamei, Boone) culture water assessed by denaturing gradient gel electrophoresis and high-throughput sequencing. Aquac. Res. 2016, 47, 857–869. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, Y.; Dong, H.; Wang, Y.; Zheng, X.; Zhang, J. Effect of dietary Clostridium butyricum on growth, intestine health status and resistance to ammonia stress in Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2017, 65, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Li, Y.; Wang, M.; Zeng, Z. Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in china. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.M.; Rong, Y.J.; Zhao, M.X.; Song, B.; Chi, Z.M. Antibacterial activity of the lipopetides produced by Bacillus amyloliquefaciens M1 against multidrug-resistant Vibrio spp. isolated from diseased marine animals. Appl. Microbiol. Biotechnol. 2014, 98, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Tian, X.; Dong, S.; Peng, M.; Wang, D. Effects of dietary Bacillus cereus G19, B. cereus BC-01, and Paracoccus marcusii DB11 supplementation on the growth, immune response, and expression of immune-related genes in coelomocytes and intestine of the sea cucumber (Apostichopus japonicus Selenka). Fish Shellfish Immunol. 2015, 45, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, M.; Li, X.; Han, Y.; Wu, F.; Liu, Y. The effects of feeding Lactobacillus pentosus on growth, immunity, and disease resistance in Haliotis discus hannai Ino. Fish Shellfish Immunol. 2018, 78, 42–51. [Google Scholar] [CrossRef] [PubMed]
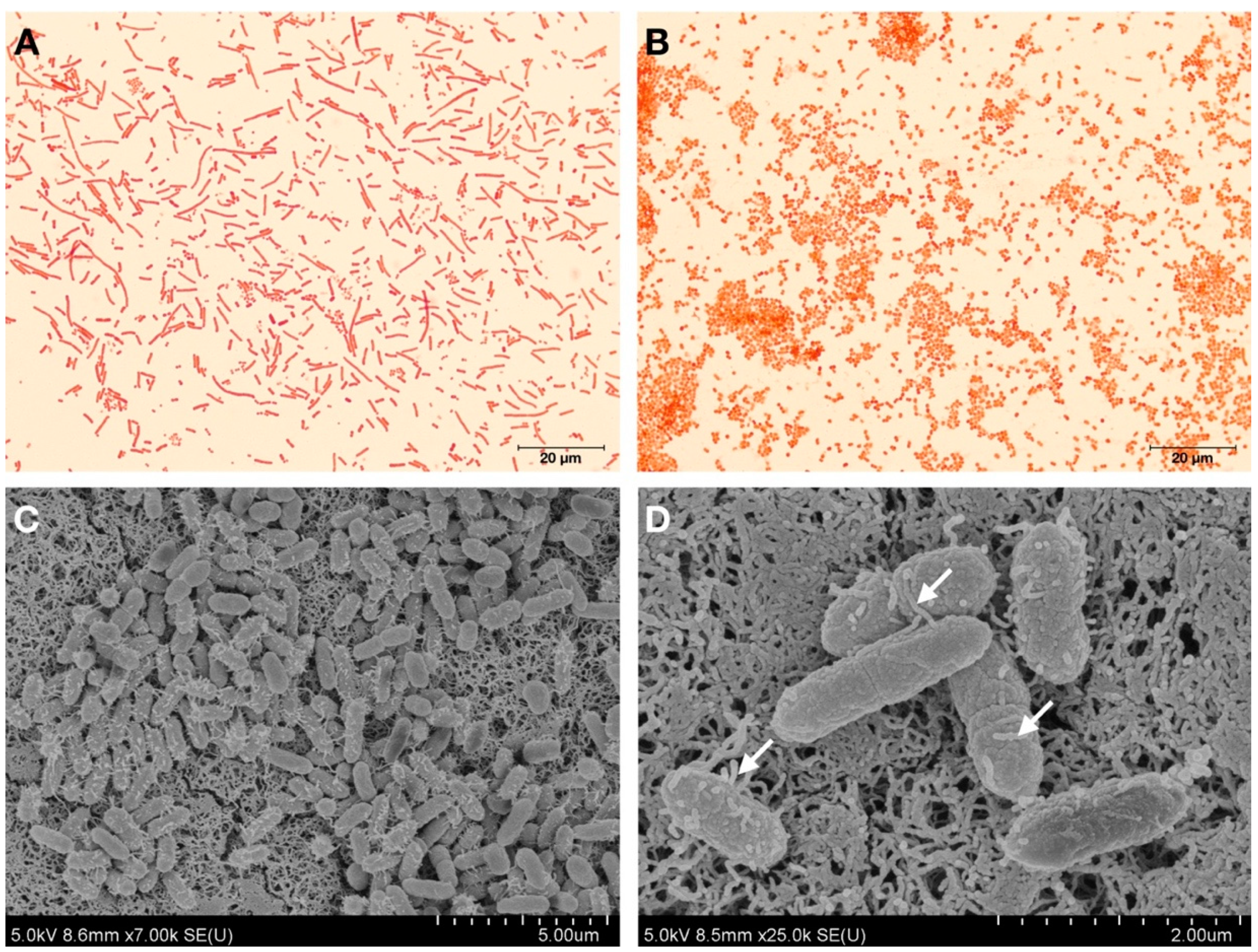


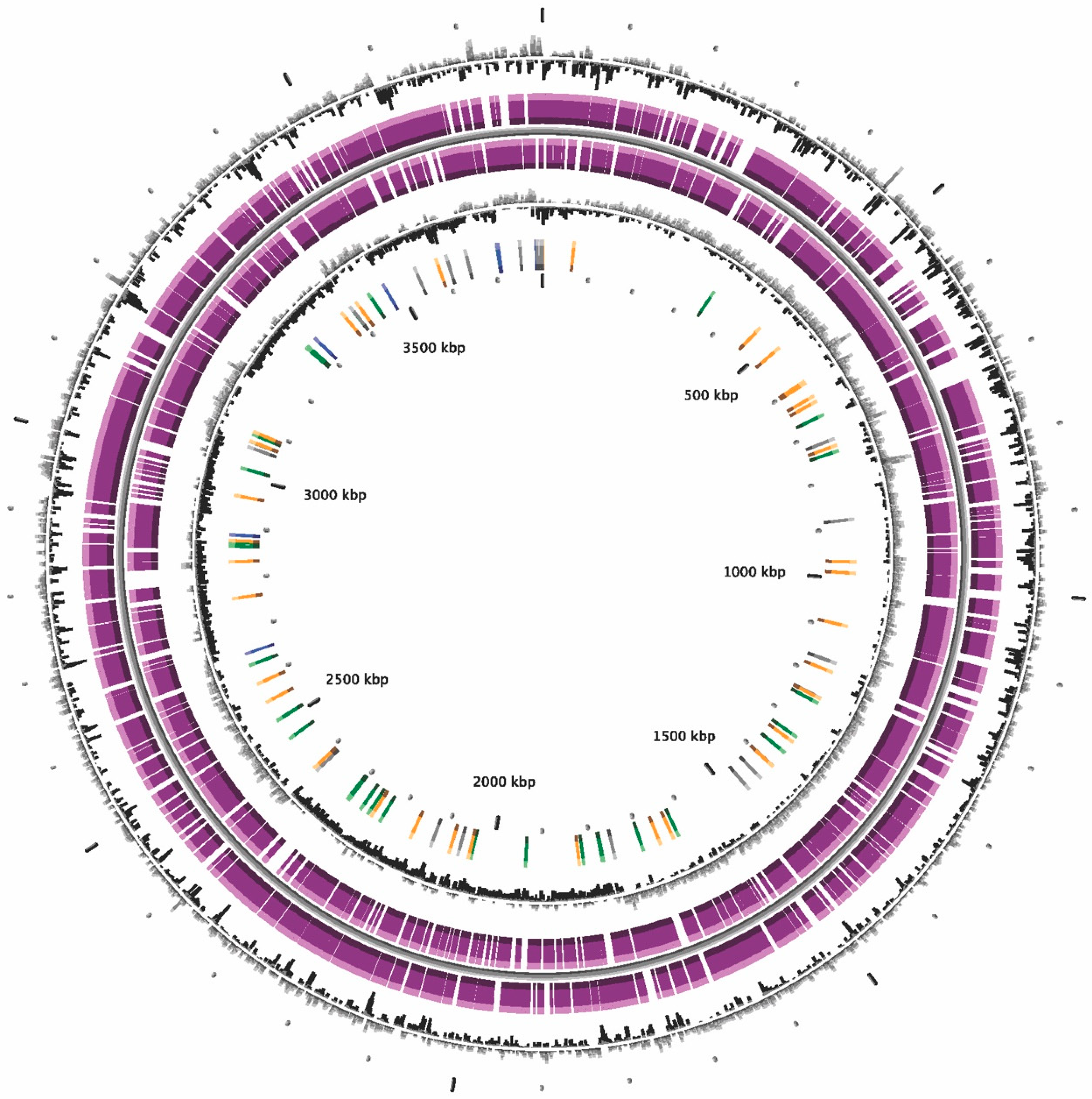
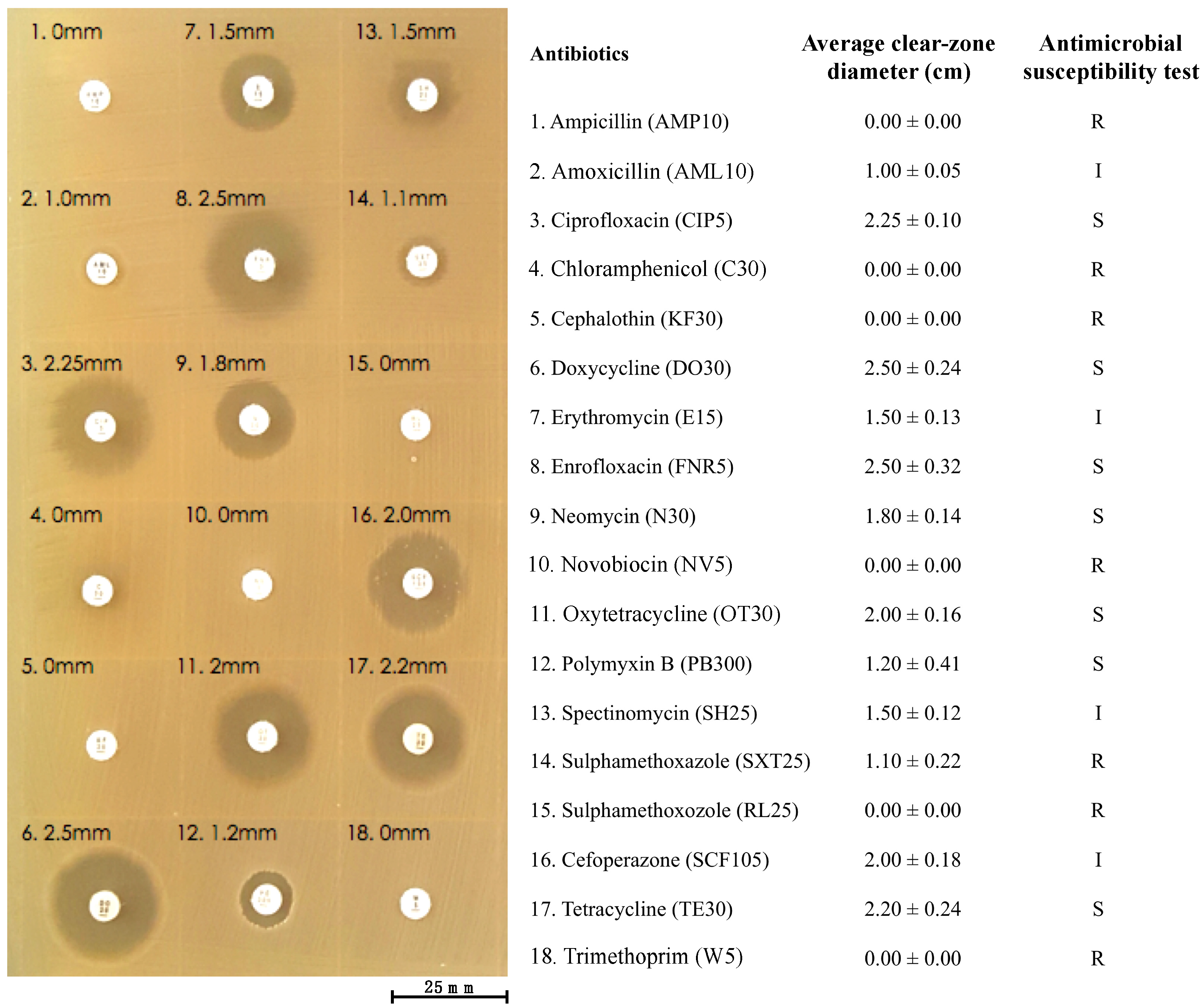

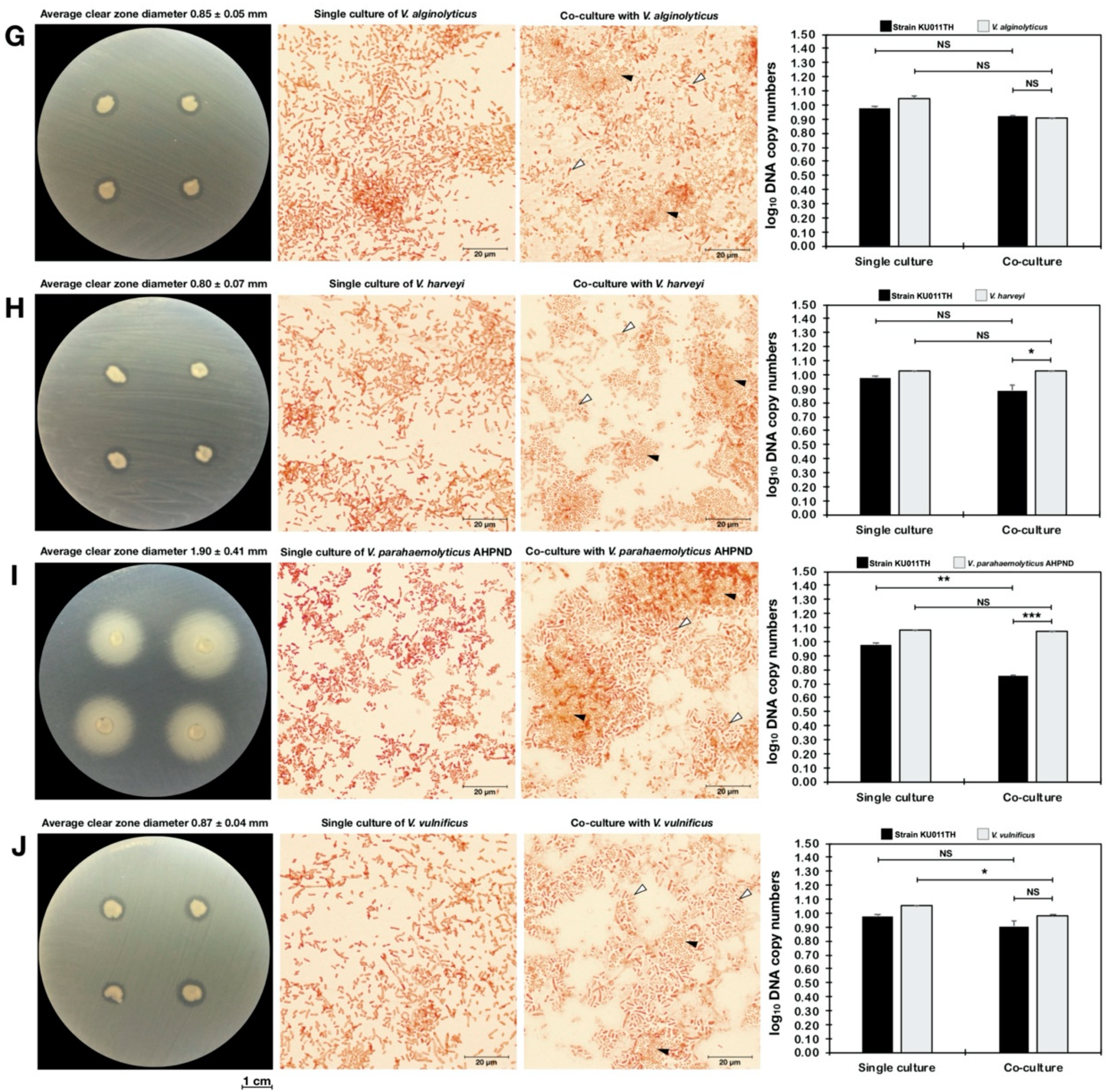
| Bacterial Strains | Primer Names | Nucleotide Sequence (5′->3′) | Amplicon Size (bp) | Accession Number |
|---|---|---|---|---|
| Acinetobacter sp. KU011TH | KU011TH_F KU011TH_R | GGCGTGCGTATTGTTTTACGTGAT CAATACCGTTTTCTGTATCTGCGG | 154 | MG950236 |
| Aeromonas hydrophila | Aeromonas_F Aeromonas_R | CAAGGCTGATATCTCCTATCCCTATG GCCACTCAGGGTCAGGTCAT | 66 | KU196733 |
| Flavobacterium columnare | Flavobac_F Flavobac_R | CCTGTACCTAATTGGGGAAAAGAGG GCGGTTATGGCCTTGTTTATCATAGA | 113 | CP018912 |
| Flectobacillus roseus | Flectoba_F Flectoba_R | AGGGTAGCTACCAGGCAACTGG ATCCCGTTCTTGACGCGGAAC | 202 | MG322214 |
| Streptococcus agalactiae | Strepto_F Strepto_R | GGAAACCTGCCATTTGCGTCT AATCTATTTCTAGATCGTGGAAT | 190 | CP033822 |
| Staphylococcus warneri | Staphylo_F Staphylo_F | TGTAGCTAACTTAGATAGTGTTCCTTCT CCGCCACCGTTATTTCTT | 62 | CP033098 |
| Edwardsiella tarda | Edward_F Edward _R | CAGTGATAAAAAGGGGTGGA CTACACAGCAACGACAACG | 114 | CP023706 |
| Vibrio alginolyticus Vibrio harveyi Vibrio parahaemolyticus Vibrio parahaemolyticus AHPND Vibrio vulnificus | Vibrio_F Vibrio_R | GGCGTAAAGCGCATGCAGGT GAAATTCTACCCCCCTCTACAG | 120 | GQ455007 * GQ455008 * HQ123986 * NR117907 * |
| Bacterial Strain | Sequence Similarity with Strain KU011TH (%) | Origin of Bacterial Isolate | ||
|---|---|---|---|---|
| 16S rRNA Gene | gyrB Gene | rpoB Gene | ||
| Acb complex | ||||
| A. lactucae NRRL B-41902 T | 99.9 | 91.2 | 96.7 | Iceberg lettuce, water |
| A. pittii LMG1035 T | 99.8 | 97.6 | 98.7 | Human, soil, water |
| A. calcoaceticus DSM 30006 T | 99.5 | 90.4 | 93.9 | Human, soil, water |
| A. nosocomialis NIPH2119 | 99.2 | 89.1 | 93.1 | Human |
| A. seifertii NIPH973 T | 99.1 | NR | 87.6 | Clinical specimen |
| A. baumannii ATCC 19606T | 98.0 | 87.6 | 93.6 | Clinical specimen |
| Non-Acb complex | ||||
| A. dispersus ANC 4105 T | 98.0 | 78.7 | 85.1 | Clinical specimen |
| A. proteolyticus NIPH 809 T | 98.0 | 76.4 | 84.6 | Clinical specimen |
| A. apis HYN18 T | 97.6 | NR | 78.2 | Tract of a honey bee |
| A. albensis ANC 4874 T | 97.5 | 72.1 | 81.9 | Natural soil and water |
| A. vivianii NIPH 2168 T | 97.4 | 76.4 | 84.6 | Clinical specimen |
| A. courvalinii ANC 3623 T | 97.1 | 76.6 | 83.3 | Conjunctiva (agama lizard) |
| A. nectaris SAP 763 T | 97.5 | 72.4 | 77.6 | Flower (floral nectar) |
| A. bohemicus ANC 3994 T | 97.5 | 54.0 | 80.3 | Natural soil and water |
| A. boissieri SAP 284 T | 96.7 | 69.3 | 82.9 | Flower (floral nectar) |
| A. guangdongensis 1NM-4 T | 96.6 | 71.7 | 78.7 | Abandoned lead–zinc ore |
| A. modestus NIPH 236 T | 96.8 | 78.3 | 84.1 | Urine / clinical specimen |
| A. hemolyticus DSM6962 T | 96.5 | 75.2 | 81.0 | Clinical specimen |
| A. pragensis ANC 4149 T | 96.5 | 68.6 | 84.1 | Natural soil and water |
| A. puyangensis BQ4-1 T | 96.7 | 65.2 | 73.1 | Bark of Populus × euramericana canker |
| Bacterial Strain of the Acb Complex | Accession Number | ANIb (%) | In silico DDH (%) | G+C Difference (%) |
|---|---|---|---|---|
| Acinetobacter pittii | Range | 94.0–94.6 | 62.4–63.2 | 0.05–0.28 |
| A. pittii XM1570 | AMXH01000001 | 94.7 | 63.2 | 0.26 |
| A. pittii ANC 3678 | APQN01000001 | 94.6 | 62.4 | 0.28 |
| A. pittii DSM 25618 | BBST01000001 | 94.5 | 62.7 | 0.13 |
| A. pittii NBRC 110509 | BBUA01000001 | 94.5 | 63.1 | 0.05 |
| A. pittii LMG1035 | NC_016603 | 94.4 | 63.1 | 0.27 |
| A. pittii CR12-42 | JQNT01000001 | 94.6 | 63.1 | 0.19 |
| Acinetobacter lactucae | range | 92.2–92.7 | 50.1–51.3 | 0.08–0.28 |
| A. lactucae ANC 4052 | APQO01000001 | 92.2 | 50.7 | 0.29 |
| A. lactucae CI78 | AVOE01000001 | 92.5 | 50.5 | 0.16 |
| A. lactucae OTEC-02 | NZ_CP020015 | 92.5 | 50.5 | 0.28 |
| A. lactucae ABBL098 | LLGZ01000001 | 92.4 | 50.7 | 0.27 |
| A. lactucae TG29425 | RFEL01000001 | 92.7 | 51.3 | 0.09 |
| A. lactucae TG41018 | RFES01000001 | 92.6 | 50.1 | 0.08 |
| Acinetobacter calcoaceticus | range | 89.4–90.2 | 38.5–40.8 | 0.01–0.19 |
| A. calcoaceticus RUH2202 | ACPK01000001 | 89.4 | 38.7 | 0.03 |
| A. calcoaceticus NIPH 13 | APOE01000001 | 89.5 | 38.9 | 0.11 |
| A. calcoaceticus ANC 3680 | APQH01000001 | 89.4 | 38.9 | 0.19 |
| A. calcoaceticus DSM 30006 | APQI01000001 | 89.4 | 38.6 | 0.09 |
| 3.5 A. calcoaceticus ANC 3811 | APQJ01000001 | 90.2 | 40.8 | 0.01 |
| 3.6 A. calcoaceticus NCTC12983 | UFSJ01000001 | 89.5 | 38.5 | 0.16 |
| Acinetobacter seifertii | range | 87.3–87.6 | 34.1–34.5 | 0.00–0.09 |
| A. seifertii C917 | APCT01000001 | 87.3 | 34.1 | 0.09 |
| A. seifertii NIPH 973 | APOO01000001 | 87.4 | 34.2 | 0.04 |
| A. seifertii KCJK1723 | LYQI01000001 | 87.4 | 34.3 | 0.04 |
| A. seifertii MI421-133 | PHFF01000001 | 87.4 | 34.5 | 0.08 |
| A. seifertii KCJK7915 | QAYP01000001 | 87.6 | 34.3 | 0.01 |
| A. seifertii SAb133 | SNSA01000001 | 87.6 | 34.3 | 0.00 |
| Acinetobacter baumannii | range | 87.1–87.3 | 33.7–33.9 | 0.44–0.51 |
| A. baumannii AB030 | NZ_CP009257 | 87.1 | 33.8 | 0.48 |
| A. baumannii AB030 | CP009257 | 87.1 | 33.8 | 0.48 |
| A. baumannii ACICU | CP000863 | 87.3 | 33.8 | 0.47 |
| A. baumannii D1279779 | CP003967 | 87.2 | 33.8 | 0.44 |
| A. baumannii ZW85-1 | CP006768 | 87.3 | 33.9 | 0.51 |
| A. baumannii XH386 | CP010779 | 87.1 | 33.7 | 0.51 |
| Acinetobacter nosocomialis | range | 86.8–87.2 | 33.1–33.4 | 0.06–0.34 |
| A. nosocomialis P020 | APCE01000001 | 86.9 | 33.4 | 0.18 |
| A. nosocomialis NBRC 110500 | BBOT01000001 | 86.9 | 33.3 | 0.12 |
| A. nosocomialis LMG 10619 | BBSR01000001 | 87.1 | 33.1 | 0.08 |
| A. nosocomialis 6411 | NZ_CP010368 | 87.2 | 33.3 | 0.18 |
| A. nosocomialis ABBL058 | LLFD01000001 | 86.8 | 33.3 | 0.06 |
| A. nosocomialis AB6 | PXNE01000001 | 87.0 | 33.2 | 0.34 |
| Bacterial Strain | Genome Characteristics | ||||||
|---|---|---|---|---|---|---|---|
| Accession Number | Genome Size (Mb) | G+C Content (mol%) | Contigs | Coding Sequences (CDS) | rRNA Genes | tRNA Genes | |
| Strain KU011TH | PSSN00000000 | 3.79 | 38.56 | 27 | 3619 | 8 | 63 |
| A. pittii LMG1035 T | APQP00000000 | 3.83 | 38.80 | 33 | 3675 | 19 | 78 |
| A. lactucae NRRL B-41902T | LRPE00000000 | 3.92 | 38.60 | 94 | 3735 | 4 | 63 |
| A. calcoaceticus DSM 30006 T | APQI00000000 | 3.92 | 38.70 | 12 | 3808 | 18 | 75 |
| A. seifertii NIPH973 T | APOO00000000 | 4.23 | 38.60 | 26 | 4180 | 13 | 70 |
| A. baumannii ATCC 19606 T | ACQB01000000 | 3.93 | 38.90 | 2 | 3725 | 18 | 71 |
| A. nosocomialis NIPH2119 T | APOP01000001 | 3.91 | 38.70 | 19 | 3730 | 18 | 74 |
| Phenotypic Characteristics | Bacterial Strain in the Acb Complex | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Temperature range (°C) | 4–41 | 37–44 | 15–44 | 25–44 | 25–41 | 37–44 | 25–41 |
| Temperature optimum (°C) | 32 | 30 | 30 | 30 | 30 | 37 | 30 |
| pH range/optimum | 3–10/7 | NR | 6–9/7 | NR | NR | NR | NR |
| NaCl concentration range/optimum (% w/v) | 0–10/1 | NR | 0–5 | NR | NR | NR | NR |
| Acidification of d-glucose | − | + | − | + | + | + | + |
| Hemolysis of sheep blood | − | − | NR | − | − | − | − |
| Adipate/adipic acid | − | + | NR | + | − | + | + |
| Citrate (Simmons)/trisodium citrate | + | + | NR | + | + | + | + |
| d-glucose | + | − | NR | − | − | − | − |
| d-malate/malic acid | − | + | − | − | + | + | + |
| d-ribose | + | − | NR | − | − | + | + |
| dl-lactate/d-lactose | − | + | NR | + | + | + | + |
| Gelatin/gelatinase | − | NR | NR | − | − | NR | NR |
| l-arabinose | + | + | NR | − | − | + | + |
| l-arginine | + | + | NR | + | + | + | + |
| l-sorbose | − | NR | − | NR | NR | NR | NR |
| Fatty Acid | Bacterial Strain | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Acb Complex * | Non-Acb Complex | ||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| C12:0 | 1.8 | 5.4 | 9.7 | 8.3 | 6.4 | 11.2 | 5.1 | 5.9 | 3.8 |
| C14:0 | 0.7 | - | 0.7 | 3.3 | 1.7 | 1.3 | 0.7 | 1.7 | 1.3 |
| C16:0 | 31.2 | 25.2 | 17.6 | 18.0 | 10.6 | 15.6 | 18.2 | 16.8 | 16.3 |
| C17:0 | 1.3 | 1.3 | 2.4 | L | 1.5 | 1.2 | 2.4 | 0.7 | 1.7 |
| C18:0 | 1.8 | 1.5 | 0.9 | 0.5 | 4.6 | 1.5 | 0.7 | 1.0 | 2.3 |
| C16:1 | 8.5 | - | 6.6 | - | 4.4 | 2.4 | 0.4 | - | 0.4 |
| C17:1ω10 | 0.7 | 2.0 | - | - | - | - | - | - | - |
| C18:1ω9t | 3.5 | - | - | - | - | - | - | - | - |
| C18:1ω9c | 35.4 | 36.9 | 34.9 | 6.1 | 19.6 | 25.8 | 25.2 | 38.2 | 28.1 |
| C18:2ω6c | 3.6 | - | - | - | - | 15.8 | - | - | - |
| C20:2 | 6.0 | - | - | - | - | - | - | - | - |
| Gene | Identity (%) | Resistant to |
|---|---|---|
| acrA | 43.10 | aminoglycosides, beta-lactams and fluoroquinolones |
| acrB | 54.08 | panipenem, penems, aztreonam, sulfonamides, azithromycin, novobiocin, meropenem, colistin, ciprofloxacin erythromycin, tetracycline, polymyxin, trimethoprim, aminocoumarin antibiotics, beta-lactams, fluoroquinolones, chloramphenicol, macrolides and tetracycline derivatives |
| alaS | 58.72 | novobiocin and aminocoumarin antibiotics |
| ampC | 93.99 | cephalosporins and beta-lactams |
| bepE | 97.92 | tetracycline, fluoroquinolones and tetracycline derivatives |
| macB | 52.72 | erythromycin and macrolides |
| mfd | 50.61 | sparfloxacin, norfloxacin, nalidixic acid, gatifloxacin, moxifloxacin, levofloxacin, ciprofloxacin and fluoroquinolones |
| nolG | 30.86 | thiamphenicol and chloramphenicol |
| smvA | 35.61 | fluoroquinolones |
| tufB | 68.22 | kirromycin and elfamycin |
| Bacterial Strain | Single Culture | Antagonistic Activity of the Bacterial Strain KU011TH Against Pathogens | |||
|---|---|---|---|---|---|
| Coculture Assay | Dot-Spot Assay | ||||
| DNA Copy Number of Bacterial Cells (copies/mL) | DNA Copy Number of Bacterial Cells (copies/mL) | Clear-Zone Diameter (cm) | |||
| NB Medium | NB Medium with 1.5% NaCl | Strain KU011TH | Pathogen | ||
| Strain KU011TH | 2.37 ± 8.81 × 1011 | 3.51 ± 2.14 × 109 | - | - | - |
| Aeromonas hydrophila | 5.47 ± 3.39 × 1011 | - | 2.85 ± 4.23 × 108 ** | 2.47 ± 1.54 × 106 * | 1.70 ± 0.15 |
| Flavobacterium columnare | 4.62 ± 6.84 × 109 | - | 1.30 ± 2.93 × 108 *** | 2.97 ± 5.01 × 106 ** | 1.72 ± 0.28 |
| Flectobacillus roseus | 6.80 ± 3.23 × 109 | - | 1.72 ± 1.18 × 108 ** | 7.00 ± 9.98 × 105 *** | 1.90 ± 0.16 |
| Streptococcus agalactiae | 3.39 ± 5.37 × 1010 | - | 1.45 ± 2.90 × 108 ** | 4.00 ± 1.39 × 105 ** | 1.32 ± 0.20 |
| Staphylococcus warneri | 1.87 ± 1.51 × 109 | - | 1.59 ± 6.98 × 108 ** | 8.03 ± 8.64 × 105 ** | 2.12 ± 0.10 |
| Edwardsiella tarda | 5.59 ± 3.20 × 1010 | - | 2.80 ± 6.22 × 108 ** | 1.91 ± 5.38 × 106 ** | 2.20 ± 0.21 |
| Vibrio alginolyticus | - | 1.68 ± 3.72 × 1011 | 2.24 ± 3.81 × 108 NS | 4.78 ± 6.52 × 108 NS | 0.85 ± 0.05 |
| Vibrio harveyi | - | 3.56 ± 1.23 × 1010 | 8.46 ± 9.78 × 107 NS | 4.74 ± 7.58 × 1010 NS | 0.80 ± 0.07 |
| Vibrio parahaemolyticus AHPND | - | 1.25 ± 9.69 × 1012 | 5.51 ± 5.13 × 105 ** | 6.44 ± 1.33 × 1011 NS | 1.90 ± 0.41 |
| Vibrio vulnificus | - | 1.72 ± 8.93 × 1011 | 2.10 ± 2.47 × 108 NS | 4.23 ± 2.31 × 109 * | 0.87 ± 0.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bunnoy, A.; Na-Nakorn, U.; Kayansamruaj, P.; Srisapoome, P. Acinetobacter Strain KUO11TH, a Unique Organism Related to Acinetobacter pittii and Isolated from the Skin Mucus of Healthy Bighead Catfish and Its Efficacy Against Several Fish Pathogens. Microorganisms 2019, 7, 549. https://doi.org/10.3390/microorganisms7110549
Bunnoy A, Na-Nakorn U, Kayansamruaj P, Srisapoome P. Acinetobacter Strain KUO11TH, a Unique Organism Related to Acinetobacter pittii and Isolated from the Skin Mucus of Healthy Bighead Catfish and Its Efficacy Against Several Fish Pathogens. Microorganisms. 2019; 7(11):549. https://doi.org/10.3390/microorganisms7110549
Chicago/Turabian StyleBunnoy, Anurak, Uthairat Na-Nakorn, Pattanapon Kayansamruaj, and Prapansak Srisapoome. 2019. "Acinetobacter Strain KUO11TH, a Unique Organism Related to Acinetobacter pittii and Isolated from the Skin Mucus of Healthy Bighead Catfish and Its Efficacy Against Several Fish Pathogens" Microorganisms 7, no. 11: 549. https://doi.org/10.3390/microorganisms7110549
APA StyleBunnoy, A., Na-Nakorn, U., Kayansamruaj, P., & Srisapoome, P. (2019). Acinetobacter Strain KUO11TH, a Unique Organism Related to Acinetobacter pittii and Isolated from the Skin Mucus of Healthy Bighead Catfish and Its Efficacy Against Several Fish Pathogens. Microorganisms, 7(11), 549. https://doi.org/10.3390/microorganisms7110549







