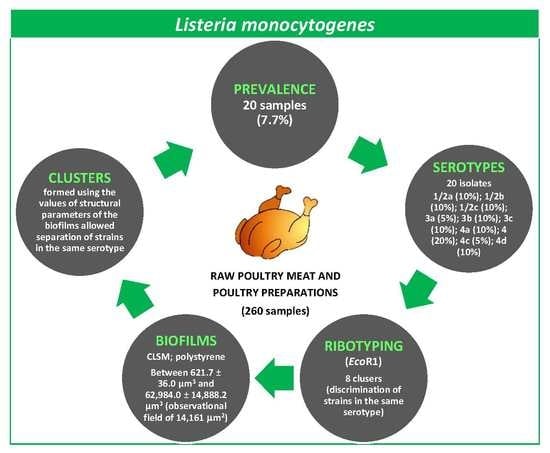
Prevalence, Molecular Typing, and Determination of the Biofilm-Forming Ability of Listeria monocytogenes Serotypes from Poultry Meat and Poultry Preparations in Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Procedure
2.2. Microbiological Analysis
2.3. Serotyping and Ribotyping
2.4. Study of Biofilms
2.5. Data Analysis
3. Results
3.1. Prevalence of L. monocytogenes in Poultry
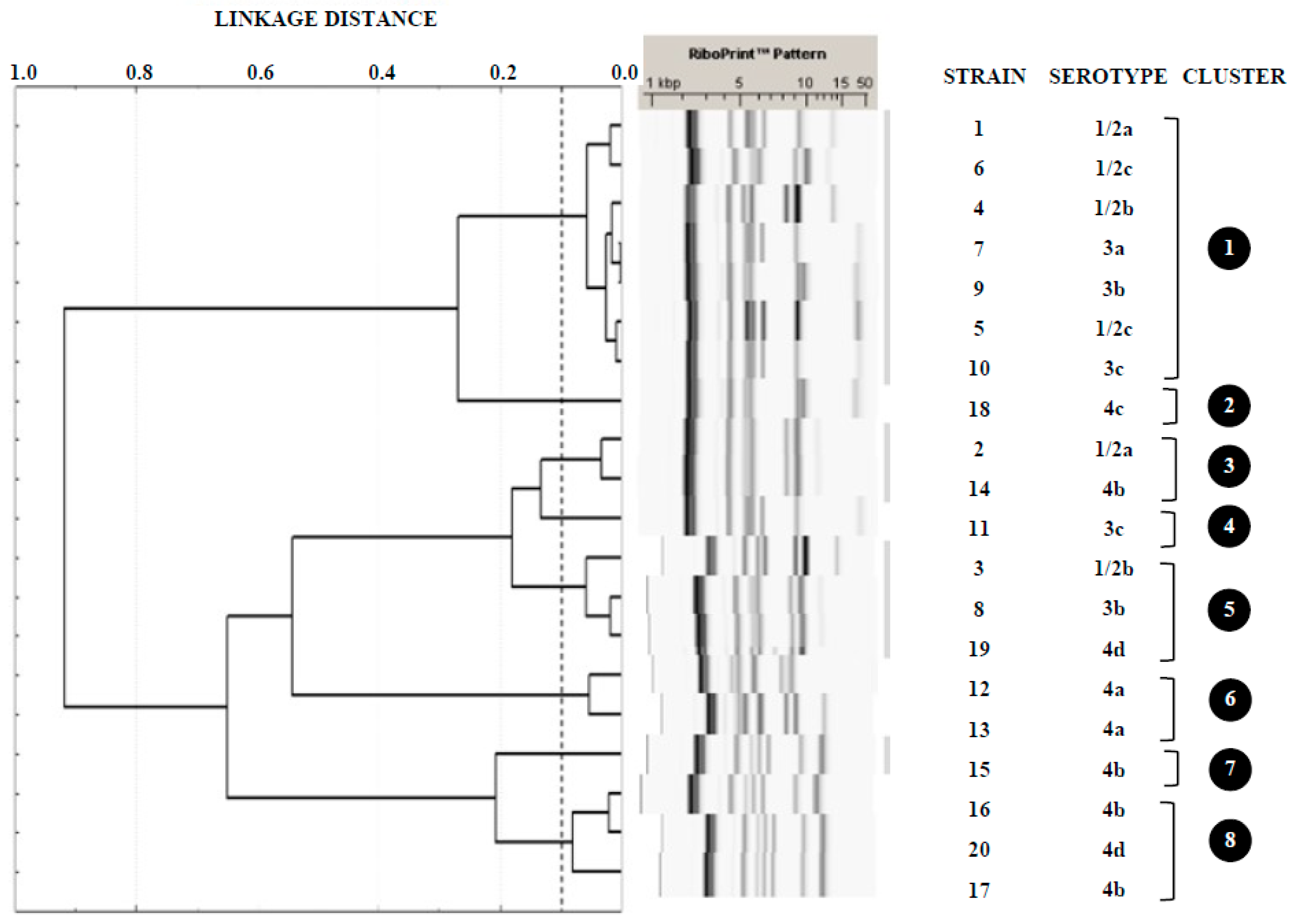
3.2. Serotyping and Ribotyping of L. monocytogenes
3.3. Architecture and Structural Parameters of Biofilms
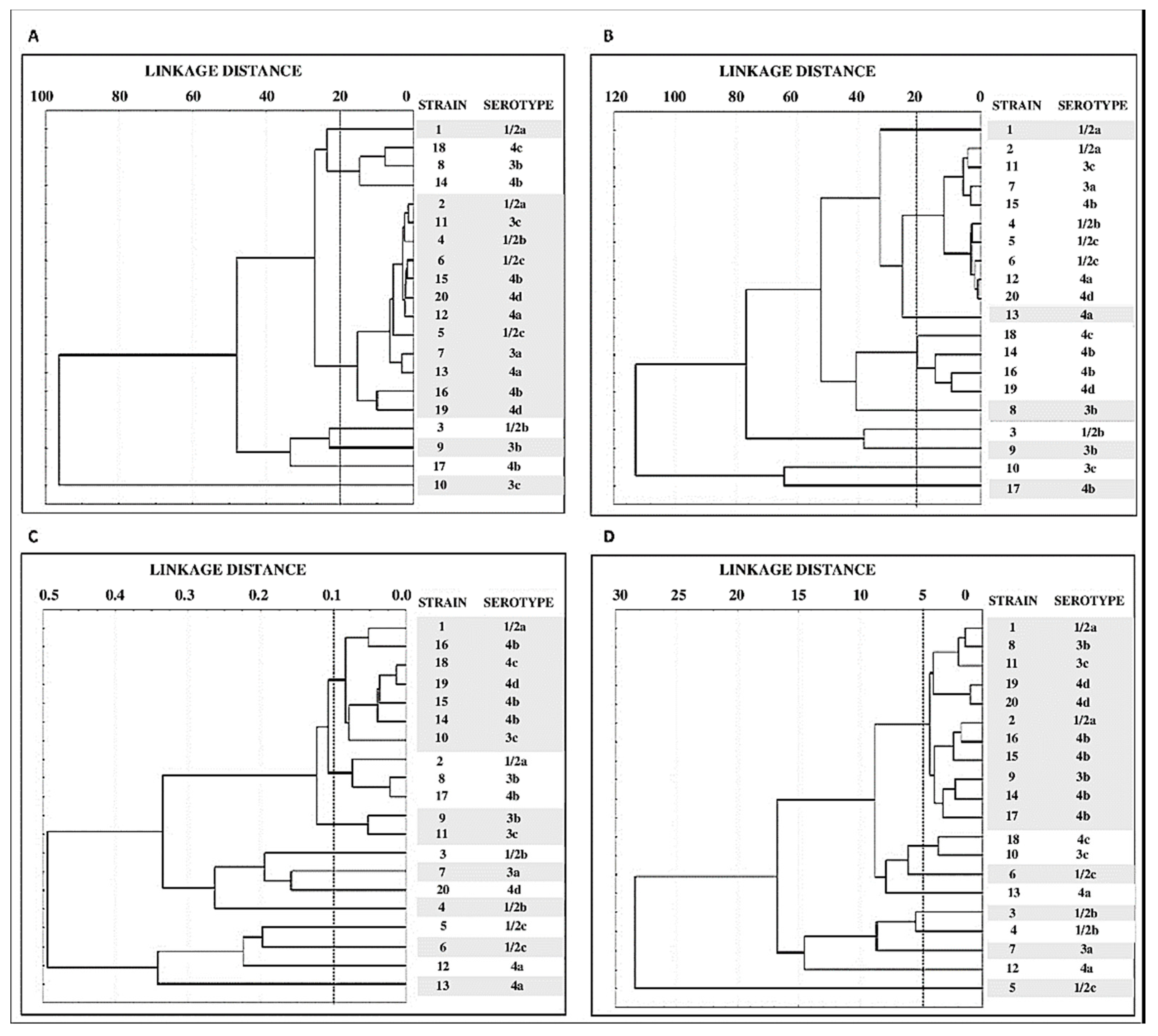
3.4. Grouping of Strains by Structural Parameters of Biofilms
4. Discussion
4.1. Prevalence of L. monocytogenes in Poultry
4.2. Serotyping and Ribotyping of Listeria monocytogenes
4.3. Study of Biofilms
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jamshidi, A.; Zeinali, T. Significance and characteristics of Listeria monocytogenes in poultry products. Int. J. Food Sci. 2019, 7835253. [Google Scholar] [CrossRef] [PubMed]
- Rothrock, M.; Micciche, A.C.; Bodie, A.R.; Ricke, S.C. Listeria occurrence and potential control strategies in alternative and conventional poultry processing and retail. Front. Sustain. Food Syst. 2019, 3, 33. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, 262. [Google Scholar] [CrossRef]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Tompkin, R.B. Control of Listeria monocytogenes in the food-processing environment. J. Food Prot. 2002, 65, 709–725. [Google Scholar] [CrossRef]
- Kathariou, S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef]
- Capita, R.; Riesco-Peláez, F.; Alonso-Hernando, A.; Alonso-Calleja, C. Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 2014, 80, 1268–1280. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Riesco-Peláez, F.; Carballo, J.; García-Fernández, C.; Capita, R.; Alonso-Calleja, C. Structure and viability of 24- and 72-h-old biofilms formed by four pathogenic bacteria on polystyrene and glass contact surfaces. Food Microbiol. 2018, 76, 513–517. [Google Scholar] [CrossRef]
- Piercey, M.J.; Hingston, P.A.; Hansen, L.T. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef]
- González-Machado, C.; Capita, R.; Riesco-Peláez, F.; Alonso-Calleja, C. Visualization and quantification of the cellular and extracellular components of Salmonella Agona biofilms at different stages of development. PLoS ONE 2018, 13, e0200011. [Google Scholar] [CrossRef]
- Shi, W.; Quingping, W.; Jumei, Z.; Moutong, C.; Zéan, Y. Prevalence, antibiotic resistance and genetic diversity of Listeria monocytogenes isolated from retail ready-to-eat foods in China. Food Control 2015, 47, 340–347. [Google Scholar] [CrossRef]
- Matloob, M.; Griffiths, M. Ribotyping and automated ribotyping of Listeria monocytogenes. Methods Mol. Biol. 2014, 1157, 85–93. [Google Scholar] [PubMed]
- International Organization for Standardization. Microbiology of the Food Chain-Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and Listeria spp. Part 1: Detection Method. Reference Number ISO 11290-1:2017(E); International Organization for Standarization: Geneva, Switzerland, 2017. [Google Scholar]
- Capita, R.; Alonso-Calleja, C.; Moreno, B.; García-Fernández, M.C. Occurrence of Listeria species in retail poultry meat and comparison of a cultural/immunoassay for their detection. Int. J. Food Microbiol. 2001, 65, 75–82. [Google Scholar] [CrossRef]
- Orsi, R.H.; Bakker, H.C.D.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.L.; Ricke, S.C.; Donalson, J.R. Establishment of Listeria monocytogenes in the gastrointestinal tract. Microorganisms 2019, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Murga, R.; Stewart, P.S.; Daly, D. Quantitative analysis of biofilm thickness variability. Biotechnol. Bioeng. 1995, 45, 503–510. [Google Scholar] [CrossRef]
- Alonso-Hernando, A.; Capita, R.; Alonso-Calleja, C. Behaviour of co-inoculated pathogenic and spoilage bacteria on poultry following several decontamination treatments. Int. J. Food Microbiol. 2012, 159, 152–159. [Google Scholar] [CrossRef]
- Ristori, C.A.; Rowlands, R.E.; Martins, C.G.; Barbosa, M.L.; Yoshida, J.T.; Franco, B.D. Prevalence and populations of Listeria monocytogenes in meat products retailed in Sao Paulo, Brazil. Foodborne Pathog. Dis. 2014, 11, 969–973. [Google Scholar] [CrossRef]
- Elmali, M.; Can, H.J.; Yaman, H. Prevalence of Listeria monocytogenes in poultry meat. Food Sci. Technol. 2015, 35, 662–665. [Google Scholar] [CrossRef]
- Oliveira, T.S.; Varjão, L.M.; Da Silva, L.N.N.; Pereira, R.C.L.; Hofer, P.; Vallim, D.C.; Almeida, R.C.C. Listeria monocytogenes at chicken slaughterhouse: Occurrence, genetic relationship among isolates and evaluation of antimicrobial susceptibility. Food Control 2018, 88, 131–138. [Google Scholar] [CrossRef]
- Alonso-Hernando, A.; Prieto, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control 2012, 23, 37–41. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Nadon, C.A.; Woodward, D.L.; Young, C.; Rodgers, F.G.; Wiedmann, M. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 2001, 39, 2704–2707. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Louie, M.; Jayaratne, P.; Luchsinger, I.; Devenish, J.; Yao, J.; Schlech, W.; Simor, A. Comparison of ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for molecular typing of Listeria monocytogenes. J. Clin. Microbiol. 1996, 34, 15–19. [Google Scholar]
- Swaminathan, B.; Hunter, S.B.; Desmarchelier, P.M.; Gerner-Smidt, P.; Graves, L.M.; Harlander, S.; Hubner, R.; Jacquet, C.; Pedersen, B.; Reineccius, K.; et al. WHO-sponsored international collaborative study to evaluate methods for subtyping Listeria monocytogenes: Restriction fragment length polymorphism (RFLP) analysis using ribotyping and Southern hybridization with two probes derived from L. monocytogenes chromosome. Int. J. Food Microbiol. 1996, 32, 263–278. [Google Scholar]
- Kerouanton, A.; Brisabois, A.; Denoyer, E.; Dilasser, F.; Grout, J.; Salvat, G.; Picard, B. Comparison of five typing methods for the epidemiological study of Listeria monocytogenes. Int. J. Food Microbiol. 1998, 43, 61–71. [Google Scholar] [CrossRef]
- De Cesare, A.; Bruce, J.L.; Dambaugh, T.R.; Guerzoni, M.E.; Wiedmann, M. Automated ribotyping using different enzymes to improve discrimination of Listeria monocytogenes isolates, with a particular focus on serotype 4b strains. J. Clin. Micobiol. 2001, 39, 3002–3005. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C.; Prieto, M. Prevalence of Salmonella enterica serovars and genovars from chicken carcasses in slaughterhouses in Spain. J. Appl. Microbiol. 2007, 103, 1366–1375. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Molina-González, D.; Blanco-Morán, S.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Prevalence, antimicrobial resistance, and genotypic characterization of vancomycin-resistant enterococci in meat preparations. J. Food Prot. 2016, 79, 748–756. [Google Scholar] [CrossRef]
- Norwood, D.E.; Gilmour, A. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 1999, 86, 576–582. [Google Scholar] [CrossRef]
- Marsh, E.J.; Luo, H.; Wang, H. A three-tiered approach to differentiate Listeria monocytogenes biofilm-forming abilities. FEMS Microbiol. Lett. 2003, 228, 203–210. [Google Scholar] [CrossRef]
- Mosquera-Fernández, M.; Rodríguez-López, P.; Cabo, M.L.; Balsa-Canto, E. Numerical spatio-temporal characterization of Listeria monocytogenes biofilms. Int. J. Food Microbiol. 2014, 182, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Guilbaud, M.; Piveteau, P.; Desvaux, M.; Brisse, S.; Briandet, R. Exploring the diversity of Listeria monocytogenes biofilm architecture by high-throughput confocal laser scanning microscopy and the predominance of the honeycomb-like morphotype. Appl. Environ. Microbiol. 2015, 81, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Melcón, C.; Capita, R.; Rodriguez-Jerez, J.J.; Martínez-Suárez, J.V.; Alonso-Calleja, C. Effect of low doses of disinfectants on the biofilm-forming ability of Listeria monocytogenes. Foodborne Pathog. Dis. 2019, 16, 262–268. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Riesco-Peláez, F.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Susceptibility of Listeria monocytogenes planktonic cultures and biofilms to sodium hypochlorite and benzalkonium chloride. Food Microbiol. 2019, 82, 533–540. [Google Scholar] [CrossRef]
- Orsi, R.H.; Bowen, B.M.; Wiedmann, M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genom. 2010, 11, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, W.B.; Cutter, C.N. An ecological perspective of Listeria monocytogenes biofilms in food processing facilities. Crit. Rev. Food Sci. Nutr. 2013, 53, 801–817. [Google Scholar] [CrossRef]
- Sauders, B.D.; Wiedmann, M. Ecology of Listeria species and L. monocytogenes in the natural environment. In Listeria, Listeriosis, and Food Safety, 3rd ed.; Ryser, E.T., Marth, E.H., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 21–53. [Google Scholar]
- Jeffers, G.T.; Bruce, J.L.; McDonough, P.L.; Scarlett, J.; Boor, K.J.; Wiedmann, M. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 2001, 147, 1095–1104. [Google Scholar] [CrossRef]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef]
- Harvey, J.; Keenan, K.P.; Gilmour, A. Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol. 2007, 24, 380–392. [Google Scholar] [CrossRef]
- Pan, Y.; Breidt, F.; Gorski, L. Synergistic effects of sodium chloride, glucose, and temperature on biofilm formation by Listeria monocytogenes serotype 1/2a and 4b strains. Appl. Environ. Microbiol. 2010, 76, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Combrouse, T.; Sadovskaya, I.; Faille, C.; Kol, O.; Guérardel, Y.; Midelet-Bourdin, G. Quantification of the extracellular matrix of the Listeria monocytogenes biofilms of different phylogenic lineages with optimization of culture conditions. J. Appl. Microbiol. 2013, 114, 1120–1131. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, W.B.; Ostiguy, N.; Cutter, C.N. Multivariate analysis reveals differences in biofilm formation capacity among Listeria monocytogenes lineages. Biofouling 2014, 30, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef]
- Takahashi, H.; Miya, S.; Igarashi, K.; Suda, T.; Kuramoto, S.; Kimura, B. Biofilm formation ability of Listeria monocytogenes isolates from raw ready-to-eat seafood. J. Food Prot. 2009, 72, 1476–1480. [Google Scholar] [CrossRef]
- Notification Details—2019.2989. Foodborne Outbreak Caused by Listeria monocytogenes (>1.5x10E4 CFU/g) in Chilled Pork Products from Spain. Available online: https://webgate.ec.europa.eu/rasff-window/portal/?event=notificationDetail&NOTIF_REFERENCE=2019.2989 (accessed on 20 September 2019).
- Junta de Andalucía. Consejería de Salud y Familias. Available online: https://www.juntadeandalucia.es/organismos/saludyfamilias/actualidad/noticias/detalle/220344.html (accessed on 20 September 2019).
- Kadam, S.R.; den Besten, H.M.W.; van der Veen, S.; Zwietering, M.H.; Moezelaar, R.; Abee, T. Diversity assessment of Listeria monocytogenes biofilm formation: Impact of growth condition, serotype and strain origin. Int. J. Food Microbiol. 2013, 165, 259–264. [Google Scholar] [CrossRef]
- Folsom, J.P.; Siragusa, G.R.; Frank, J.F. Formation of biofilm at different nutrient levels by various genotypes of Listeria monocytogenes. J. Food Prot. 2006, 69, 826–834. [Google Scholar] [CrossRef]
- Takahashi, H.; Suda, T.; Tanaka, Y.; Kimura, B. Cellular hydrophobicity of Listeria monocytogenes involves initial attachment and biofilm formation on the surface of polyvinyl chloride. Lett. Appl. Microbiol. 2010, 50, 618–625. [Google Scholar] [CrossRef]
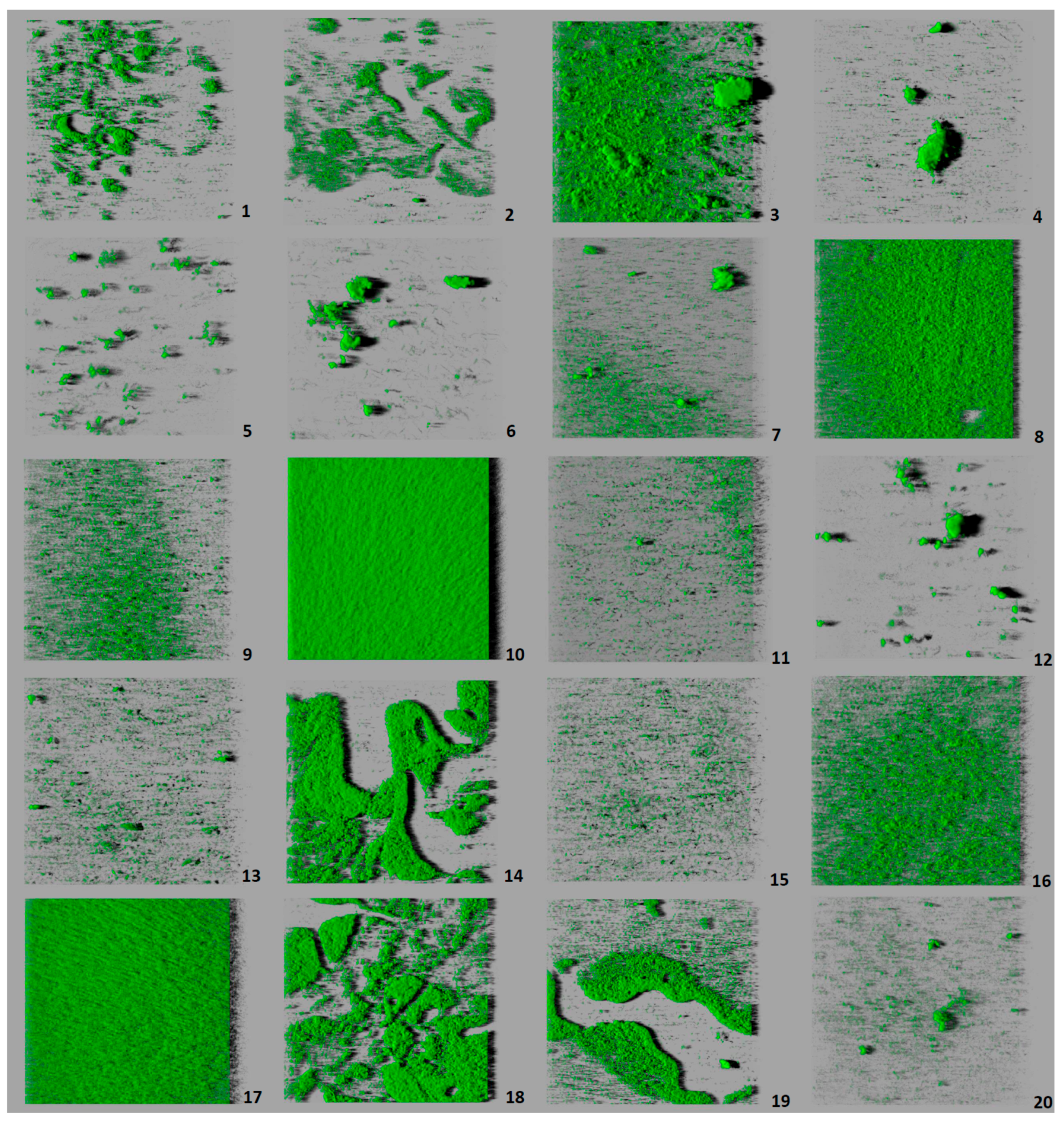
| Strain (Serotype) | Biovolume (µm3) | % Surface Coverage | Roughness | Maximum Thickness (µm) |
|---|---|---|---|---|
| 1 (1/2a) | 12,099.1 ± 12,156.7 a | 20.09 ± 15.25 ab | 0.434 ± 0.048 a | 11.33 ± 2.31 ab |
| 2 (1/2a) | 2877.1 ± 827.2 a | 7.79 ± 2.16 a | 0.485 ± 0.029 ab | 10.33 ± 0.58 ab |
| 3 (1/2b) | 28,184.0 ± 21,540.5 a | 50.50 ± 31.20 ab | 0.548 ± 0.131 ab | 19.00 ± 7.00 ab |
| 4 (1/2b) | 2208.5 ± 2014.4 a | 4.05 ± 0.57 a | 0.661 ± 0.168 ab | 16.33 ± 8.74 ab |
| 5 (1/2c) | 2600.3 ± 3367.7 a | 3.18 ± 1.82 a | 0.739 ± 0.153 ab | 24.00 ± 14.93 a |
| 6 (1/2c) | 1225.4 ± 1097.4 a | 2.17 ± 0.84 a | 0.714 ± 0.031 ab | 17.00 ± 2.65 ab |
| 7 (3a) | 4504.2 ± 2780.8 a | 10.38 ± 0.81 a | 0.606 ± 0.054 ab | 19.33 ± 2.31 ab |
| 8 (3b) | 18,568.1 ± 4367.1 a | 53.85 ± 3.96 ab | 0.455 ± 0.033 ab | 10.67 ± 2.08 ab |
| 9 (3b) | 24,234.6 ± 34,507.0 a | 37.64 ± 46.81 ab | 0.417 ± 0.079 a | 11.00 ± 1.73 ab |
| 10 (3c) | 62,984.0 ± 14,888.2 b | 94.43 ± 3.97 b | 0.399 ± 0.052 a | 14.33 ± 1.53 ab |
| 11 (3c) | 2054.5 ± 780.5 a | 9.97 ± 3.29 a | 0.433 ± 0.048 a | 10.33 ± 1.53 ab |
| 12 (4a) | 1725.9 ± 991.6 a | 2.52 ± 0.82 a | 0.830 ± 0.022 b | 24.00 ± 5.00 a |
| 13 (4a) | 5032.7 ± 2535.7 a | 18.39 ± 9.10 a | 0.592 ± 0.172 ab | 15.67 ± 4.16 ab |
| 14 (4b) | 18,870.8 ± 8290.3 a | 35.92 ± 13.52 ab | 0.401 ± 0.013 a | 12.00 ± 2.00 ab |
| 15 (4b) | 1901.6 ± 517.9 a | 9.68 ± 2.58 a | 0.415 ± 0.026 a | 9.00 ± 0.00 b |
| 16 (4b) | 8059.7 ± 3897.1 a | 28.71 ± 11.62 ab | 0.452 ± 0.035 ab | 10.00 ± 1.00 ab |
| 17 (4b) | 23,727.1 ± 15,800.7 a | 66.49 ± 30.47 ab | 0.443 ± 0.039 a | 10.67 ± 2.52 ab |
| 18 (4c) | 19,969.7 ± 1398.8 a | 35.42 ± 4.74 ab | 0.417 ± 0.005 a | 14.00 ± 1.00 ab |
| 19 (4d) | 13,498.3 ± 1635.5 a | 29.30 ± 5.38 ab | 0.417 ± 0.006 a | 11.67 ± 1.15 ab |
| 20 (4d) | 621.7 ± 36.0 a | 2.27 ± 0.28 a | 0.578 ± 0.057 ab | 12.00 ± 1.00 ab |
| Serotype | Biovolume (µm3) | % Surface Coverage | Roughness | Maximum Thickness (µm) |
|---|---|---|---|---|
| 1/2a (n = 2) | 7488.1 ± 9214.2 a | 13.94 ± 11.85 ab | 0.460 ± 0.045 ab | 10.83 ± 1.60 a |
| 1/2b (n = 2) | 15,196.3 ± 19,739.2 ab | 27.27 ± 32.20 ac | 0.605 ± 0.148 ac | 17.67 ± 7.23 ab |
| 1/2c (n = 2) | 1912.9 ± 2363.4 a | 2.67 ± 1.39 a | 0.727 ± 0.099 c | 20.50 ± 10.33 b |
| 3a (n = 1) | 4504.2 ± 2780.8 a | 10.38 ± 0.81 ab | 0.606 ± 0.054 bc | 19.33 ± 2.31 ab |
| 3b (n = 2) | 21,401.4 ± 22,216.1 ab | 45.74 ± 31.01 bc | 0.436 ± 0.058 ab | 10.83 ± 1.72 a |
| 3c (n = 2) | 32,519.3 ± 34,678.9 b | 52.20 ± 46.37 c | 0.416 ± 0.049 b | 12.33 ± 2.58 ab |
| 4a (n = 2) | 3379.3 ± 2499,1 a | 10.46 ± 10.44 a | 0.711 ± 0.170 c | 19.83 ± 6.15 b |
| 4b (n = 4) | 13,139.8 ± 11,904.2 ab | 35.20 ± 26.15 ac | 0.428 ± 0.033 b | 10.42 ± 1.83 a |
| 4c (n = 1) | 19,969.7 ± 1398.8 ab | 35.42 ± 4.74 ac | 0.417 ± 0.005 ab | 14.00 ± 1.00 ab |
| 4d (n = 2) | 7060.0 ± 7128.3 a | 15.78 ± 15.19 ab | 0.497 ± 0.095 ab | 11.83 ± 0.98 ab |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alonso-Calleja, C.; Gómez-Fernández, S.; Carballo, J.; Capita, R. Prevalence, Molecular Typing, and Determination of the Biofilm-Forming Ability of Listeria monocytogenes Serotypes from Poultry Meat and Poultry Preparations in Spain. Microorganisms 2019, 7, 529. https://doi.org/10.3390/microorganisms7110529
Alonso-Calleja C, Gómez-Fernández S, Carballo J, Capita R. Prevalence, Molecular Typing, and Determination of the Biofilm-Forming Ability of Listeria monocytogenes Serotypes from Poultry Meat and Poultry Preparations in Spain. Microorganisms. 2019; 7(11):529. https://doi.org/10.3390/microorganisms7110529
Chicago/Turabian StyleAlonso-Calleja, Carlos, Sara Gómez-Fernández, Javier Carballo, and Rosa Capita. 2019. "Prevalence, Molecular Typing, and Determination of the Biofilm-Forming Ability of Listeria monocytogenes Serotypes from Poultry Meat and Poultry Preparations in Spain" Microorganisms 7, no. 11: 529. https://doi.org/10.3390/microorganisms7110529
APA StyleAlonso-Calleja, C., Gómez-Fernández, S., Carballo, J., & Capita, R. (2019). Prevalence, Molecular Typing, and Determination of the Biofilm-Forming Ability of Listeria monocytogenes Serotypes from Poultry Meat and Poultry Preparations in Spain. Microorganisms, 7(11), 529. https://doi.org/10.3390/microorganisms7110529