The Endophytic Fungus Chaetomium cupreum Regulates Expression of Genes Involved in the Tolerance to Metals and Plant Growth Promotion in Eucalyptus globulus Roots
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Experiment Design
2.2. Fungus Detection in Roots by Scanning Electron Microscopy (SEM)
2.3. RNA Extraction, Sequencing and Illumina Reads Processing
2.4. De novo Transcriptome Assembly
2.5. Functional Annotation of the Transcriptome
2.6. Differential Expression Analysis
3. Results
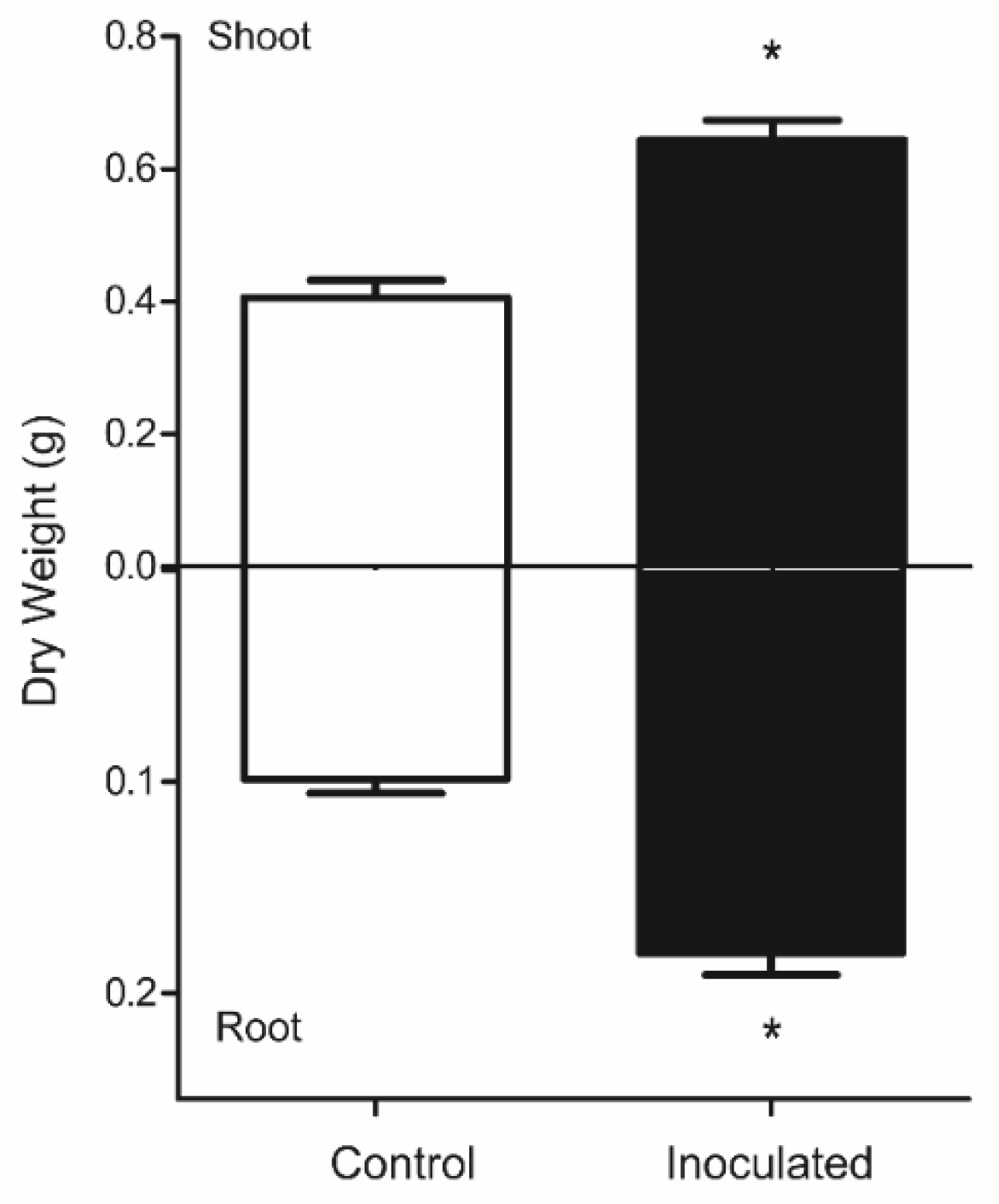
3.1. Plant Biomass and Fungus Detection in Roots
3.2. Sequence Data, de novo Transcriptome Assembly and Annotation
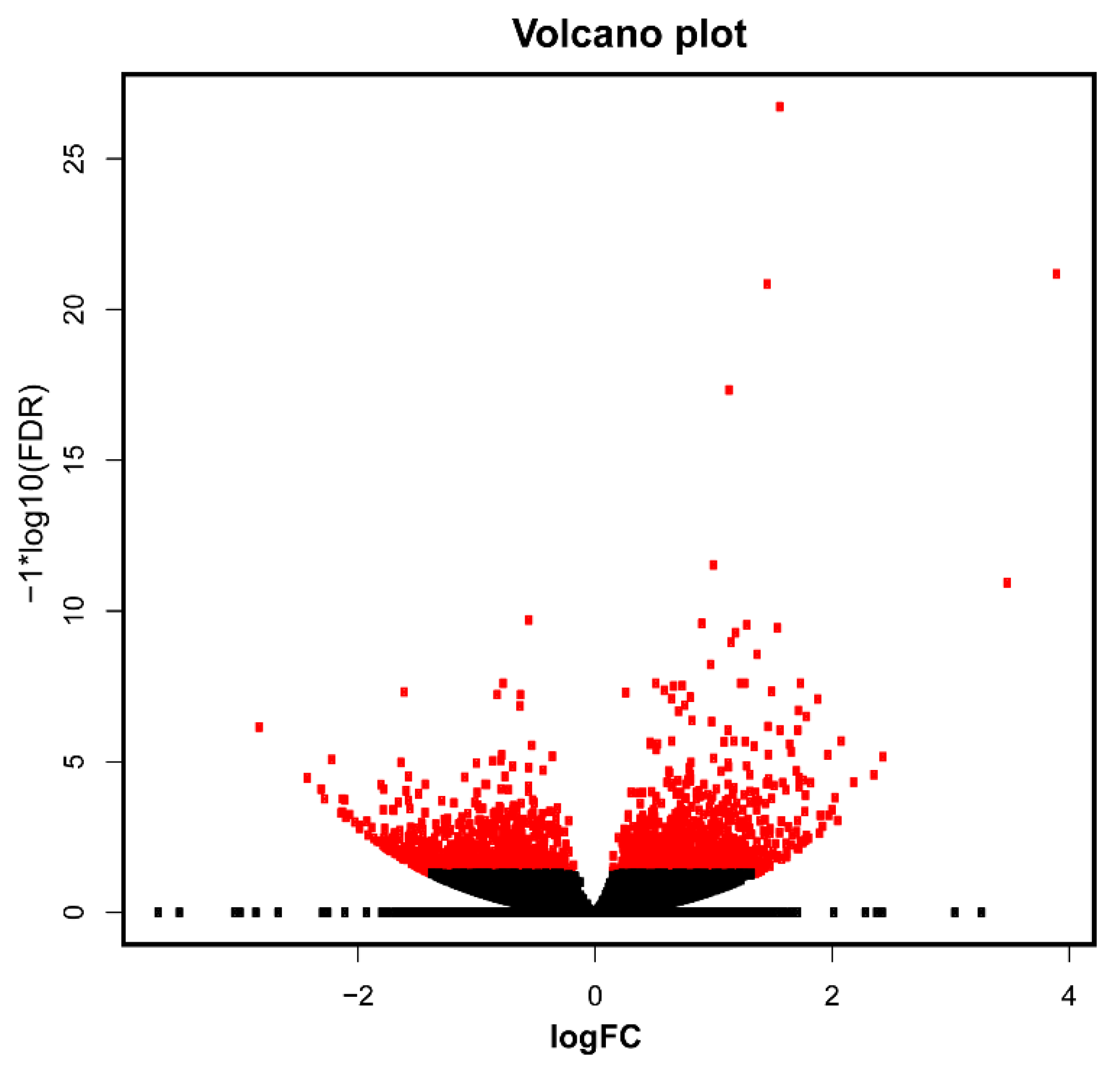
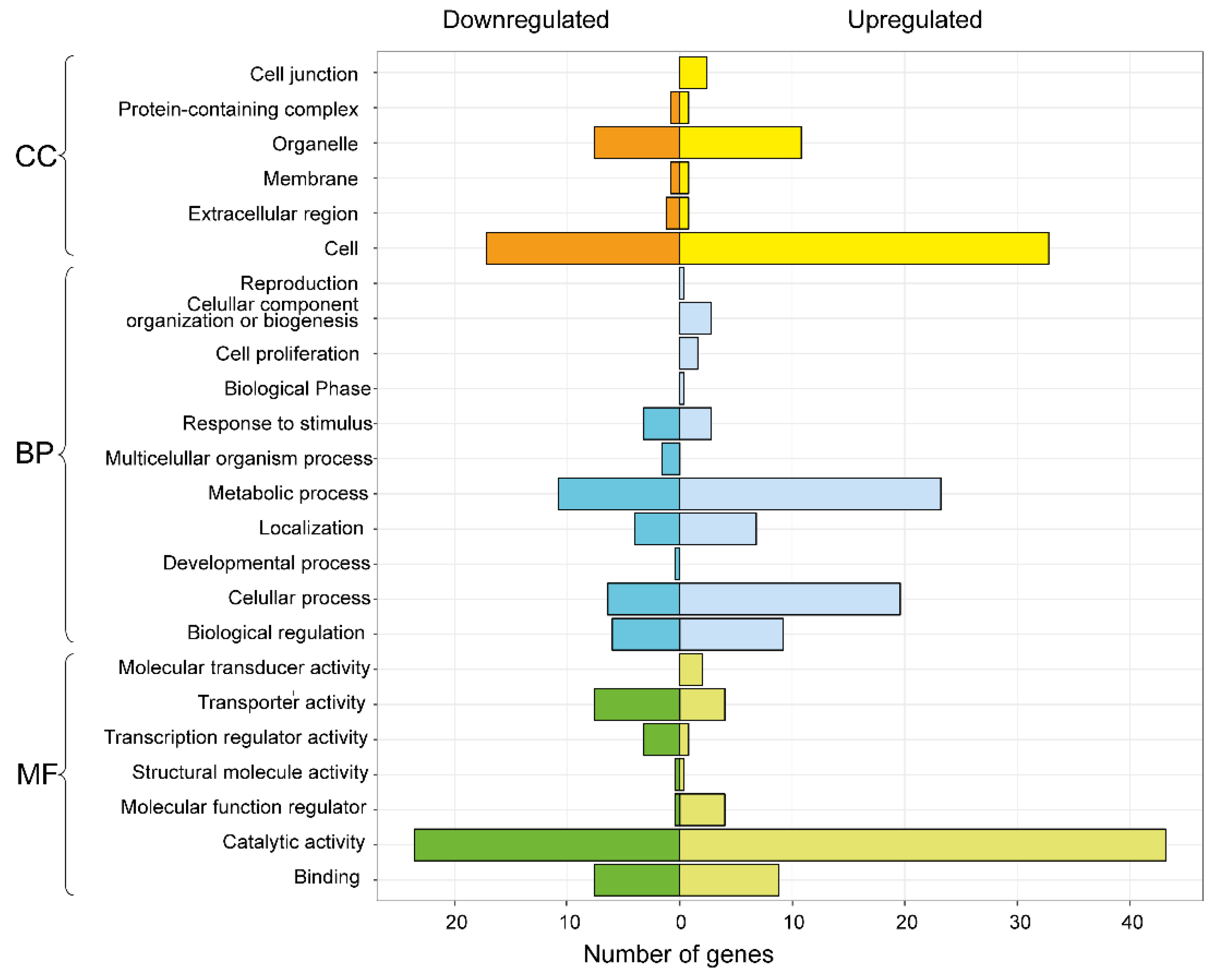
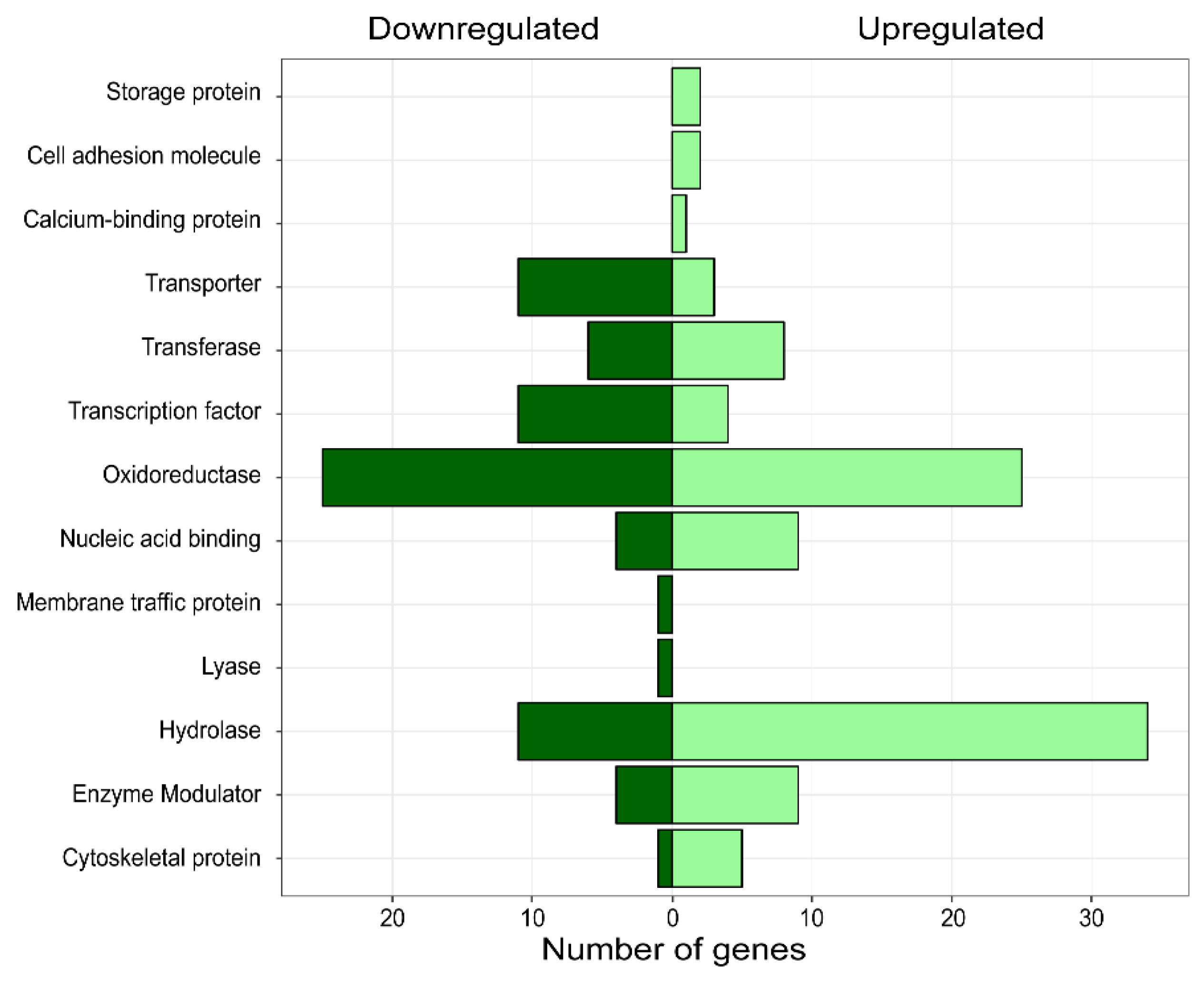
3.3. Differential Expression Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ginocchio, R. Effects of a copper smelter on a grassland community in the Puchuncaví Valley, Chile. Chemosphere 2000, 41, 15–23. [Google Scholar] [CrossRef]
- Gisbert, C.; Clemente, R.; Navarro-Aviñó, J.; Baixauli, C.; Ginér, A.; Serrano, R.; Walker, D.J.; Bernal, M.P. Tolerance and accumulation of heavy metals by Brassicaceae species grown in contaminated soils from Mediterranean regions of Spain. Environ. Exp. Bot. 2006, 56, 19–27. [Google Scholar] [CrossRef]
- Arriagada, C.; Aranda, E.; Sampedro, I.; García-Romera, I.; Ocampo, J. Contribution of the saprobic fungi Trametes versicolor and Trichoderma harzianum and the arbuscular mycorrhizal fungi Glomus deserticola and G. claroideum to arsenic tolerance of Eucalyptus globulus. Bioresour. Technol. 2009, 100, 6250–6257. [Google Scholar] [CrossRef]
- Rajkumar, M.; Sandhya, S.; Prasad, M.; Freitas, H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 2012, 30, 1562–1574. [Google Scholar] [CrossRef]
- Kane, K.H. Effects of endophyte infection on drought stress tolerance of Lolium perenne accessions from the Mediterranean region. Environ. Exp. Bot. 2011, 71, 337–344. [Google Scholar] [CrossRef]
- Deng, Z.; Cao, L. Fungal endophytes and their interactions with plants in phytoremediation: A review. Chemosphere 2017, 168, 1100–1106. [Google Scholar] [CrossRef]
- Ortiz, J.; Soto, J.; Almonacid, L.; Fuentes, A.; Campos-Vargas, R.; Arriagada, C. Alleviation of metal stress by Pseudomonas orientalis and Chaetomium cupreum strains and their effects on Eucalyptus globulus growth promotion. Plant Soil 2019, 436, 449–461. [Google Scholar] [CrossRef]
- Bilal, S.; Shahzad, R.; Khan, A.L.; Al-Harrasi, A.; Kim, C.K.; Lee, I.-J. Phytohormones enabled endophytic Penicillium funiculosum LHL06 protects Glycine max L. from synergistic toxicity of heavy metals by hormonal and stress-responsive proteins modulation. J. Hazard. Mater. 2019, 379, 120824. [Google Scholar] [CrossRef]
- Ban, Y.; Xu, Z.; Yang, Y.; Zhang, H.; Chen, H.; Tang, M. Effect of Dark Septate Endophytic Fungus Gaeumannomyces cylindrosporus on Plant Growth, Photosynthesis and Pb Tolerance of Maize (Zea mays L.). Pedosphere 2017, 27, 283–292. [Google Scholar] [CrossRef]
- Peng, H.; He, X.; Gao, J.; Ma, H.; Zhang, Z.; Shen, Y.; Pan, G.; Lin, H. Transcriptomic changes during maize roots development responsive to Cadmium (Cd) pollution using comparative RNAseq-based approach. Biochem. Biophys. Res. Commun. 2015, 464, 1040–1047. [Google Scholar] [CrossRef]
- Guan, Q.; Yu, J.; Zhu, W.; Yang, B.; Li, Y.; Zhang, L.; Tian, J. RNA-Seq transcriptomic analysis of the Morus alba L. leaves exposed to high-level UVB with or without dark treatment. Gene 2018, 645, 60–68. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, C.; Xue, Z.; Jin, Q.; Xu, Y. De novo transcriptome sequencing and discovery of genes related to copper tolerance in Paeonia ostii. Gene 2016, 576, 126–135. [Google Scholar] [CrossRef]
- Gu, C.; Xu, S.; Wang, Z.; Liu, L.; Zhang, Y.; Deng, Y.; Huang, S. De novo sequencing, assembly, and analysis of Iris lactea var. chinensis roots’ transcriptome in response to salt stress. Plant Physiol. Biochem. 2018, 125, 1–12. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.; Liu, W.; Wang, J.; Zhu, X.; Zhang, K.; Yu, R.; Wang, R.; Xie, Y.; Zhang, W.; et al. De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.). Plant Sci. 2015, 236, 313–323. [Google Scholar] [CrossRef]
- Avis, T.J.; Gravel, V.; Antoun, H.; Tweddell, R.J. Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol. Biochem. 2008, 40, 1733–1740. [Google Scholar] [CrossRef]
- Hossain, M.A.; Piyatida, P.; Da Silva, J.A.T.; Fujita, M. Molecular Mechanism of Heavy Metal Toxicity and Tolerance in Plants: Central Role of Glutathione in Detoxification of Reactive Oxygen Species and Methylglyoxal and in Heavy Metal Chelation. J. Bot. 2012, 2012, 1–37. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef]
- Fuentes, A.; Almonacid, L.; Ocampo, J.A.; Arriagada, C. Synergistic interactions between a saprophytic fungal consortium and Rhizophagus irregularis alleviate oxidative stress in plants grown in heavy metal contaminated soil. Plant Soil 2016, 407, 355–366. [Google Scholar] [CrossRef]
- Xie, X.; Fu, J.; Wang, H.; Liu, J. Heavy metal resistance by two bacteria strains isolated from a copper mine tailing in China. Afr. J. Biotechnol. 2010, 9, 4056–4066. [Google Scholar]
- Almonacid, L.; Fuentes, A.; Ortiz, J.; Salas, C.; García-Romera, I.; Ocampo, J.; Arriagada, C.; Garcia-Romera, I. Effect of mixing soil saprophytic fungi with organic residues on the response of Solanum lycopersicum to arbuscular mycorrhizal fungi. Soil Use Manag. 2015, 31, 155–164. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLOS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinform. 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinform. 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic acids research 2012, 40, D290–D301. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Love, I.M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Boil. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mehmood, A.; Hussain, A.; Irshad, M.; Hamayun, M.; Iqbal, A.; Rahman, H.; Tawab, A.; Ahmad, A.; Ayaz, S. Cinnamic acid as an inhibitor of growth, flavonoids exudation and endophytic fungus colonization in maize root. Plant Physiol. Biochem. 2019, 135, 61–68. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Li, H.; Sivasithamparam, K.; Jones, M.G.K.; Wylie, S.J. A simple and rapid in vitro test for large-scale screening of fungal endophytes from drought-adapted Australian wild plants for conferring water deprivation tolerance and growth promotion in Nicotiana benthamiana seedlings. Arch. Microbiol. 2017, 199, 1357–1370. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals—Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.U.; Rono, J.K.; Zhang, B.Q.; Liu, X.S.; Wang, M.Q.; Wang, L.L.; Wu, X.C.; Chen, X.; Cao, H.W.; Yang, Z.M. Identification of novel rice (Oryza sativa) HPP and HIPP genes tolerant to heavy metal toxicity. Ecotoxicol. Environ. Saf. 2019, 175, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.P.; Awasthi, M.; Singh, S.; Tiwari, S.; Dwivedi, U.N. A Comprehensive Review on Function and Application of Plant Peroxidases. Biochem. Anal. Biochem. 2017, 06. [Google Scholar] [CrossRef]
- Wang, C.; Chen, X.; Yao, Q.; Long, D.; Fan, X.; Kang, H.; Zeng, J.; Sha, L.; Zhang, H.; Zhou, Y.; et al. Overexpression of TtNRAMP6 enhances the accumulation of Cd in Arabidopsis. Gene 2019, 696, 225–232. [Google Scholar] [CrossRef]
- Migocka, M.; Kosieradzka, A.; Papierniak, A.; Maciaszczyk-Dziubinska, E.; Posyniak, E.; Garbiec, A.; Filleur, S. Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. J. Exp. Bot. 2015, 66, 1001–1015. [Google Scholar] [CrossRef]
- Klein, M.; Burla, B.; Martinoia, E. The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett. 2006, 580, 1112–1122. [Google Scholar] [CrossRef]
- Rozpądek, P.; Domka, A.; Ważny, R.; Nosek, M.; Jędrzejczyk, R.; Tokarz, K.; Turnau, K. How does the endophytic fungus Mucor sp. improve Arabidopsis arenosa vegetation in the degraded environment of a mine dump? Environ. Exp. Bot. 2018, 147, 31–42. [Google Scholar] [CrossRef]
- Haruma, T.; Yamaji, K.; Masuya, H.; Hanyu, K. Root endophytic Chaetomium cupreum promotes plant growth and detoxifies aluminum in Miscanthus sinensis Andersson growing at the acidic mine site. Plant Species Biol. 2018, 33, 109–122. [Google Scholar] [CrossRef]
- Wu, C.; Li, B.; Wei, Q.; Pan, R.; Zhang, W. Endophytic fungus Serendipita indica increased nutrition absorption and biomass accumulation in Cunninghamia lanceolata seedlings under low phosphate. Acta Ecol. Sin. 2019, 39, 21–29. [Google Scholar] [CrossRef]
- Sherameti, I.; Shahollari, B.; Venus, Y.; Altschmied, L.; Varma, A.; Oelmüller, R. The Endophytic Fungus Piriformospora indica Stimulates the Expression of Nitrate Reductase and the Starch-degrading Enzyme Glucan-water Dikinase in Tobacco and Arabidopsis Roots through a Homeodomain Transcription Factor That Binds to a Conserved Motif in Their Promoters. J. Boil. Chem. 2005, 280, 26241–26247. [Google Scholar]
- Rodrigues, M.I.; Bravo, J.P.; Sassaki, F.T.; Severino, F.E.; Maia, I.G. The tonoplast intrinsic aquaporin (TIP) subfamily of Eucalyptus grandis: Characterization of EgTIP2, a root-specific and osmotic stress-responsive gene. Plant Sci. 2013, 213, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Bolouri Moghaddam, M.R.; Van den Ende, W. Sugars and plant innate immunity. J. Exp. Bot. 2012, 63, 3989–3998. [Google Scholar] [CrossRef] [PubMed]
- Dastogeer, K.M.; Li, H.; Sivasithamparam, K.; Jones, M.G.; Du, X.; Ren, Y.; Wylie, S.J. Metabolic responses of endophytic Nicotiana benthamiana plants experiencing water stress. Environ. Exp. Bot. 2017, 143, 59–71. [Google Scholar] [CrossRef]
- Molina-Montenegro, M.A.; Oses, R.; Torres-Diaz, C.; Atala, C.; Zurita-Silva, A.; Ruiz-Lara, S. Root-endophytes improve the ecophysiological performance and production of an agricultural species under drought condition. AoB Plants 2016, 8. [Google Scholar] [CrossRef]
- Chen, N.; Yang, Q.; Pan, L.; Chi, X.; Chen, M.; Hu, D.; Yang, Z.; Wang, T.; Wang, M.; Yu, S. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.). Gene 2014, 533, 332–345. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Viana, V.E.; Busanello, C.; Da Maia, L.C.; Pegoraro, C.; De Oliveira, A.C. Activation of rice WRKY transcription factors: an army of stress fighting soldiers? Curr. Opin. Plant Biol. 2018, 45, 268–275. [Google Scholar] [CrossRef]
- Herbel, V.; Sieber-Frank, J.; Wink, M. The antimicrobial peptide snakin-2 is upregulated in the defense response of tomatoes (Solanum lycopersicum) as part of the jasmonate-dependent signaling pathway. J. Plant Physiol. 2017, 208, 1–6. [Google Scholar] [CrossRef]
- Matthes, M.S.; Best, N.B.; Robil, J.M.; Malcomber, S.; Gallavotti, A.; McSteen, P. Auxin EvoDevo: Conservation and Diversification of Genes Regulating Auxin Biosynthesis, Transport, and Signaling. Mol. Plant 2019, 12, 298–320. [Google Scholar] [CrossRef]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Dev. 2013, 140, 943–950. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, Q.; Hu, Z.; Sun, X.; Fan, S.; Zhang, H. Function of the auxin-responsive gene TaSAUR75 under salt and drought stress. Crop. J. 2018, 6, 181–190. [Google Scholar] [CrossRef]
- Gibson, S.W.; Todd, C.D. Arabidopsis AIR12 influences root development. Physiol. Mol. Boil. Plants 2015, 21, 479–489. [Google Scholar] [CrossRef] [PubMed]

| Eucalyptus globulus | |
|---|---|
| Number of transcripts | 131.155 |
| Number of components | 81.210 |
| Size | 147.15 Mbp |
| N10 | 4469 bp |
| N20 | 3580 bp |
| N30 | 3002 bp |
| N40 | 2540 bp |
| N50 | 2128 bp |
| Average length | 1121.97 bp |
| Median | 580 bp |
| Complete BUSCOs | 1133 | 78.7% |
|---|---|---|
| -Single-copy BUSCOs | 574 | 39.9% |
| -Duplicated BUSCOs | 559 | 38.8% |
| Fragmented BUSCOs | 108 | 7.5% |
| Missing BUSCOs | 199 | 13.8% |
| Total BUSCO genes | 1.440 | 100.0% |
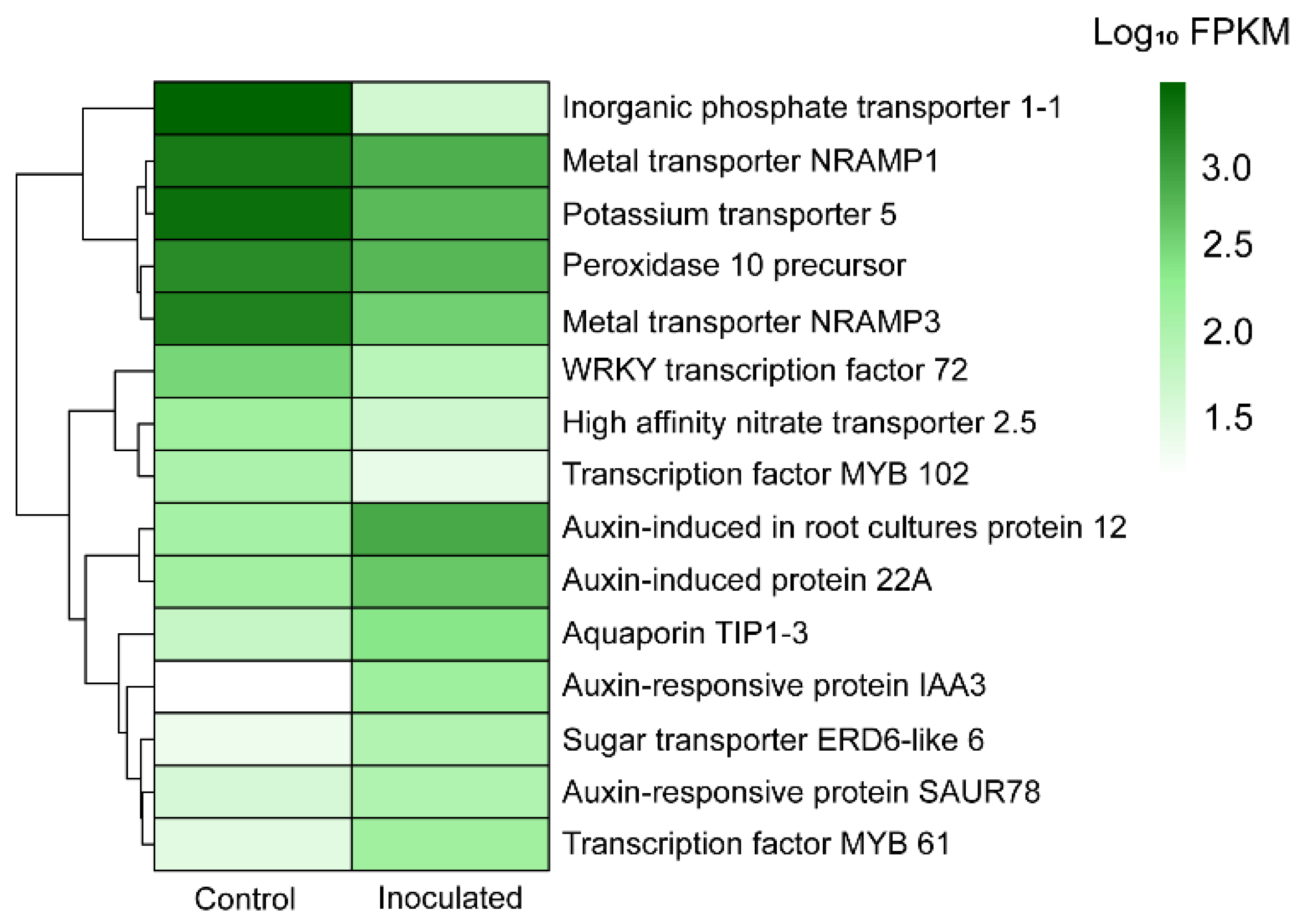
| Gene Description | Ontology | FPKM Control | FPKM Treatment | p-Value | Up/Down |
|---|---|---|---|---|---|
| Stress responses | |||||
| Metallothionein-like protein 1 | MF: Metal ion binding (GO:0046872) | 69932.98 | 32933.82 | 8.65 × 10−7 | Down |
| Peroxidase 10 precursor | MF: Metal ion binding (GO:0046872), BP: Response to oxidative stress (GO:0006979) | 1370.02 | 615.98 | 1.61 × 10−135 | Down |
| Probable 2-oxoglutarate-dependent dioxygenase | BP: Metal ion binding (GO:0046872), Cellular response to toxic substances (GO:0097237) | 1049.91 | 451.091 | 1.63 × 10−6 | Down |
| Protein Cobra precursor | BP: Response to salt stress (GO:0006950) | 85.18 | 1617.25 | 0 | Up |
| Protein High arsenic content 1, mitochondrial | BP: Detoxification of arsenic-containing substance (GO:0071722) | 1046.34 | 318.95 | 5.68 × 10−34 | Down |
| Protein sensitive to proton rhizotoxicity 2 | MF: Metal ion binding (GO:0046872) | 234.41 | 103.58 | 4.83 × 10−103 | Down |
| Putative Multidrug resistance protein | MF: ATPase activity, coupled to transmembrane movement of substances (GO:0042626) | 161.09 | 71.71 | 3.79 × 10−41 | Down |
| Metal tolerance protein 4 | MF: Cation transmembrane transporter activity (GO:0008324) | 723.47 | 280.76 | 0 | Down |
| Transcription factor MYB102 | BP: Response to osmotic stress (GO:0006970), response to salt stress (GO:0009651) | 120.87 | 31.99 | 4.63 × 10−68 | Down |
| Transcription factor MYB74 | BP: Response to salt stress (GO:0009651) | 181.21 | 66 | 2.09 × 10−9 | Down |
| Transcription factor MYB86 | BP: Regulation of stomatal movement (GO:0010119) | 35.69 | 153.43 | 3.96 × 10−23 | Up |
| Heat stress transcription factor A-2 | BP: Cellular response to heat (GO:0034605) | 298.08 | 124.75 | 7.8 × 10−30 | Down |
| Universal stress protein A-like protein | MF: AMP binding (GO:0016208) | 62.21 | 24.3 | 2.78 × 10−10 | Down |
| Probable WRKY transcription factor 72 | BP: Defense response (GO:0006952) | 344.15 | 89.65 | 4.22 × 10−62 | Down |
| Disease resistance protein RPS4B | BP: Defense response signaling pathway (GO:0009870) | 269.97 | 960.33 | 0.003 | Up |
| Disease resistance-like protein DSC1 | MF: Defense response to bacterium (GO:0042742) | 269.97 | 960.33 | 1.48 × 10−5 | Up |
| Snakin-2 precursor | BP: Defense response (GO:0006952) | 70.25 | 323.33 | 5.25 × 10−21 | Up |
| Disease resistance response protein 206 | BP: Defense response (GO:0006952), response to biotic stimulus (GO:0009607) | 75.24 | 308.47 | 5.89 × 10−76 | Up |
| 3,9-dihydroxypterocarpan 6A-monooxygenase | BP: Defense response (GO:0006952) | 234.72 | 1139.56 | 1.69 × 10−25 | Up |
| Cysteine protease X CP2 precursor | MF: Defense response to bacterium (GO:0042742) | 403.5 | 1270.23 | 0 | Up |
| Disease resistance protein RPP4 | BP: Defense response (GO:0006952) | 239.78 | 628.32 | 7.88 × 10−22 | Up |
| Ethylene-responsive transcription factor ERF043 | BP: Ethylene-activated signaling pathway (GO:0009873) | 42.47 | 147.73 | 1.04 × 10−40 | Up |
| Cell transport | |||||
| Metal transporter Nramp1 | BP: Iron ion homeostasis (GO:0055072), iron ion transmembrane transport (GO:0034755) | 1849.04 | 706.19 | 1.68 × 10−23 | Down |
| Metal transporter Nramp3 | BP: Iron ion transmembrane transport (GO:0034755) | 1613.53 | 378.21 | 3.64 × 10−34 | Down |
| Metal transporter Nramp5 | BP: Iron ion homeostasis (GO:0055072) | 1849.04 | 706.19 | 0 | Down |
| Metal transporter Nramp6 | BP: Iron ion transmembrane transport (GO:0034755) | 1613.53 | 378.21 | 7.81 × 10–6 | Down |
| Heavy metal-associated isoprenylated plant protein 28 | MF: Metal ion binding (GO:0046872), BP: Metal ion transport (GO:0030001) | 2010.9 | 911.05 | 8.38 × 10–25 | Down |
| High affinity nitrate transporter 2.5 | BP: Cellular response to nitrate (GO:0071249), nitrate transport (GO:0015706) | 156.4 | 53.16 | 4.63 × 10−22 | Down |
| High-affinity nitrate transporter 3.2 | BP: Nitrate assimilation (GO:0042128), nitrate transport (GO:0015706) | 7323.21 | 3085.33 | 8.34 × 10−49 | Down |
| Inorganic phosphate transporter 1-1 | BP: Phosphate ion transmembrane transporter activity (GO:0015114) | 2858.46 | 50.92 | 8.77 × 10−27 | Down |
| Potassium transporter 5 | BP: Potassium ion transport (GO:0006813) | 2330.51 | 570.91 | 4.86 × 10−34 | Down |
| Sodium transporter HKT1 | BP: Response to osmotic stress (GO:0006970), sodium ion transport (GO:0006814) | 116.31 | 50.95 | 1.17 × 10−23 | Down |
| Magnesium transporter MRS2-3 | BP: Magnesium ion transport (GO:0015693) | 924.34 | 365.83 | 6.95 × 10−139 | Down |
| Ammonium transporter 1 member 1 | BP: Ammonium transmembrane transport (GO:0072488) | 3059.06 | 1050.29 | 0 | Down |
| High affinity sulfate transporter 1 | MF: Secondary activate sulfate transmembrane transporter activity (GO:0008271) | 433.26 | 100.61 | 1.59 × 10−10 | Down |
| Aluminum-activated malate transporter 10 | BP: Malate transport (GO:0015743) | 379.74 | 126.8 | 3.78 × 10−22 | Down |
| Sugar transporter ERD6-like 6 | BP: Glucose homeostasis (GO:0042593) | 28.58 | 102.73 | 3.24 × 10−36 | Up |
| Probable Aquaporin PIP2-2 | MF: Water channel activity (GO:0015250), BP: Response to water deprivation (GO:0009414) | 78.13 | 1168.64 | 1.39 × 10−55 | Up |
| Aquaporin TIP1-3 | MF: Water channel activity (GO:0015250), BP: Water transport (GO:0006833) | 64.88 | 252.7 | 8.49 × 10−32 | Up |
| Plant growth | |||||
| Auxin-induced protein AUX22 | MF: Auxin-activated signaling pathway (GO:0009734) | 106.53 | 349.04 | 2.86 × 10−20 | Up |
| Auxin-responsive protein IAA3 | BP: Auxin-activated signaling pathway (GO:0009734), response to auxin (GO:0009733) | 18.84 | 167.76 | 5.82 × 10−40 | Up |
| Auxin-responsive protein IAA4 | BP: Auxin-activated signaling pathway (GO:0009734) | 18.84 | 167.76 | 3.26 × 10−45 | Up |
| Auxin-induced protein 22D | BP: Auxin-activated signaling pathway (GO:0009734) | 18.84 | 167.76 | 1.33 × 10−57 | Up |
| Auxin efflux carrier component 2 | BP: Auxin-activated signaling pathway (GO:0009734), auxin efflux (GO:0010315) | 10.46 | 115.46 | 0 | Up |
| Auxin-induced in root cultures protein 12 | BP: Auxin-activated signaling pathway (GO:0009734) | 142.02 | 772.68 | 4.19 × 10−48 | Up |
| Auxin-responsive protein SAUR50 | BP: Auxin-activated signaling pathway (GO:0009734), regulation of growth (GO:0040008) | 18.44 | 108.18 | 1.11 × 10−15 | Up |
| Auxin-responsive protein SAUR78 | BP: Positive regulation of cell growth (GO:0030307), responsive to auxin (GO:0009733) | 46.66 | 107.16 | 2.42 × 10−22 | Up |
| Auxin- induced protein 22A | BP: Auxin-activated signaling pathway (GO:0009734) | 147.72 | 441.66 | 1.15 × 10−60 | Up |
| Indole-3-acetic acid-amino synthetase GH3.17 | BP: Auxin homeostasis (GO:0010252) | 41.6 | 137.52 | 0 | Up |
| Transcription factor MYB61 | BP: Response to auxin (GO:0009733), root development (GO:0048364) | 35.69 | 153.43 | 1.29 × 10−88 | Up |
| Protein RALF-like 24 precursor | MF: Hormone activity (GO:0005179), BP: Cell-cell signaling (GO:0007267) | 182.13 | 487.29 | 3.06 × 10−23 | Up |
| Metal-binding | |||||
| Laccase-15 precursor | MF: Copper ion binding (GO:0005507), oxidoreductase activity (GO:0016491) | 590.37 | 1885 | 6.34 × 10−14 | Up |
| Laccase-14 precursor | MF: Copper ion binding (GO:0005507), oxidoreductase activity (GO:0016491) | 675.65 | 2738.33 | 9.02 × 10−51 | Up |
| Laccase-16 precursor | MF: Copper ion binding (GO:0005507), oxidoreductase activity (GO:0016491) | 675.65 | 2738.33 | 1.01 × 10−110 | Up |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz, J.; Soto, J.; Fuentes, A.; Herrera, H.; Meneses, C.; Arriagada, C. The Endophytic Fungus Chaetomium cupreum Regulates Expression of Genes Involved in the Tolerance to Metals and Plant Growth Promotion in Eucalyptus globulus Roots. Microorganisms 2019, 7, 490. https://doi.org/10.3390/microorganisms7110490
Ortiz J, Soto J, Fuentes A, Herrera H, Meneses C, Arriagada C. The Endophytic Fungus Chaetomium cupreum Regulates Expression of Genes Involved in the Tolerance to Metals and Plant Growth Promotion in Eucalyptus globulus Roots. Microorganisms. 2019; 7(11):490. https://doi.org/10.3390/microorganisms7110490
Chicago/Turabian StyleOrtiz, Javier, Javiera Soto, Alejandra Fuentes, Héctor Herrera, Claudio Meneses, and César Arriagada. 2019. "The Endophytic Fungus Chaetomium cupreum Regulates Expression of Genes Involved in the Tolerance to Metals and Plant Growth Promotion in Eucalyptus globulus Roots" Microorganisms 7, no. 11: 490. https://doi.org/10.3390/microorganisms7110490
APA StyleOrtiz, J., Soto, J., Fuentes, A., Herrera, H., Meneses, C., & Arriagada, C. (2019). The Endophytic Fungus Chaetomium cupreum Regulates Expression of Genes Involved in the Tolerance to Metals and Plant Growth Promotion in Eucalyptus globulus Roots. Microorganisms, 7(11), 490. https://doi.org/10.3390/microorganisms7110490






