Emerging Species and Genome Editing Tools: Future Prospects in Cyanobacterial Synthetic Biology
Abstract
1. Introduction
2. “Non-Model” Species: Requirements for Uptake and Genetic Manipulation
2.1. The Emergence of Fast-Growing and Stress-Tolerant Synechococcus Strains
2.2. Nostoc punctiforme ATCC 29133
2.3. Cyanothece sp.
2.4. Arthrospira sp.
2.5. Leptolyngbya sp.
2.6. Fremyella diplosiphon
2.7. Marine Synechococcus sp. and Prochlorococcus sp.
2.8. Thermosynechococcus elongatus
2.9. Chlorogloeopsis fritschii and Fischerella muscicola
2.10. Chroococcidiopsis thermalis
2.11. Gloeobacter violaceus PCC 7421
| Species | Strain | Desirable Features from Table 1 | Transformation Method | Reported Selection Markers | Agar Medium | References |
|---|---|---|---|---|---|---|
| Synechococcus sp. | UTEX 2973 | 1), 2), 3), 4), 5), 6) | Conjugation CRISPR/Cas pilN mutants are naturally transformable | Apramycin Chloramphenicol Kanamycin Spectinomycin Streptomycin | BG-11 | [20,33,35,58] |
| PCC 11801 | 1), 2), 3), 5), 6) | Naturally transformable | Spectinomycin | BG-11 | [23] | |
| PCC 11901 | 1), 2), 3), 5), 6) | Naturally transformable | Acrylic acid Spectinomycin | AD7 | [24] | |
| Nostoc punctiforme | ATCC 29133 | 1), 2), 3), 4), 5), 6) | Conjugation | Chloramphenicol Neomycin sacB markers Streptomycin | Allen and Arnon | [62,63,90] |
| Cyanothece sp. | PCC 7822 | 1), 2), 3), 5) | Electroporation | Spectinomycin | BG-11 | [66] |
| ATCC 51142 | 1), 2), 3), 4), 6) | Conjugation | Kanamycin | BG-11 ASP2 | [67] | |
| Arthrospira platensis | C1 | 1), 2), 3), 4), 5) | Electroporation | Spectinomycin | Zarrouk | [53] |
| Leptolyngbya sp. | BL0902 | 1), 2), 3), 4), 5), 6) | Conjugation | Chloramphenicol Erythromycin Neomycin Spectinomycin Streptomycin | BG-11 | [72,73] |
| Fremyella diplosiphon | SF33 | 1), 2), 3), 4), 6) | Conjugation | Kanamycin Neomycin sacB markers | BG-11/HEPES | [74,75,91] |
| Marine Synechococcus sp. | WH7803 WH8102 WH8103 | 1), 2), 3), 6) | Conjugation Electroporation | Kanamycin | SN | [76,77,92] |
| Marine Prochlorococcus sp. | 1), 3) | - | Spectinomycin | Pro99 | [76,93] | |
| Thermosynechococcus elongatus | BP-1 | 1), 2), 3), 4), 5), 6) | Naturally transformable | Chloramphenicol Kanamycin | BG-11 | [81] |
| PKUAC-SCTE542 | 1), 2), 3), 6) | Naturally transformable | Spectinomycn | BG-11 | [82] | |
| Chlorogloeopsis fritschii | PCC 6912 | 1), 2), 3), 4), 5) | Biolistic Conjugation | Kanamycin Neomycin | Allen and Arnon | [83,90] |
| Fischerella muscicola | PCC 7414 | 1), 2), 3), 4), 5) | Biolistic Conjugation | Kanamycin Neomycin | Allen and Arnon | [83,90] |
| Chroococcidiopsis thermalis | 1), 2), 3), 4), 5) | Conjugation | Neomycin | BG-11 | [84] | |
| Gloeobacter violaceus | PCC 7421 | 1), 2), 3), 4), 5) | Conjugation | Streptomycin | BG-11 | [89] |
3. Current and Future Strategies for Genome Engineering in Cyanobacteria
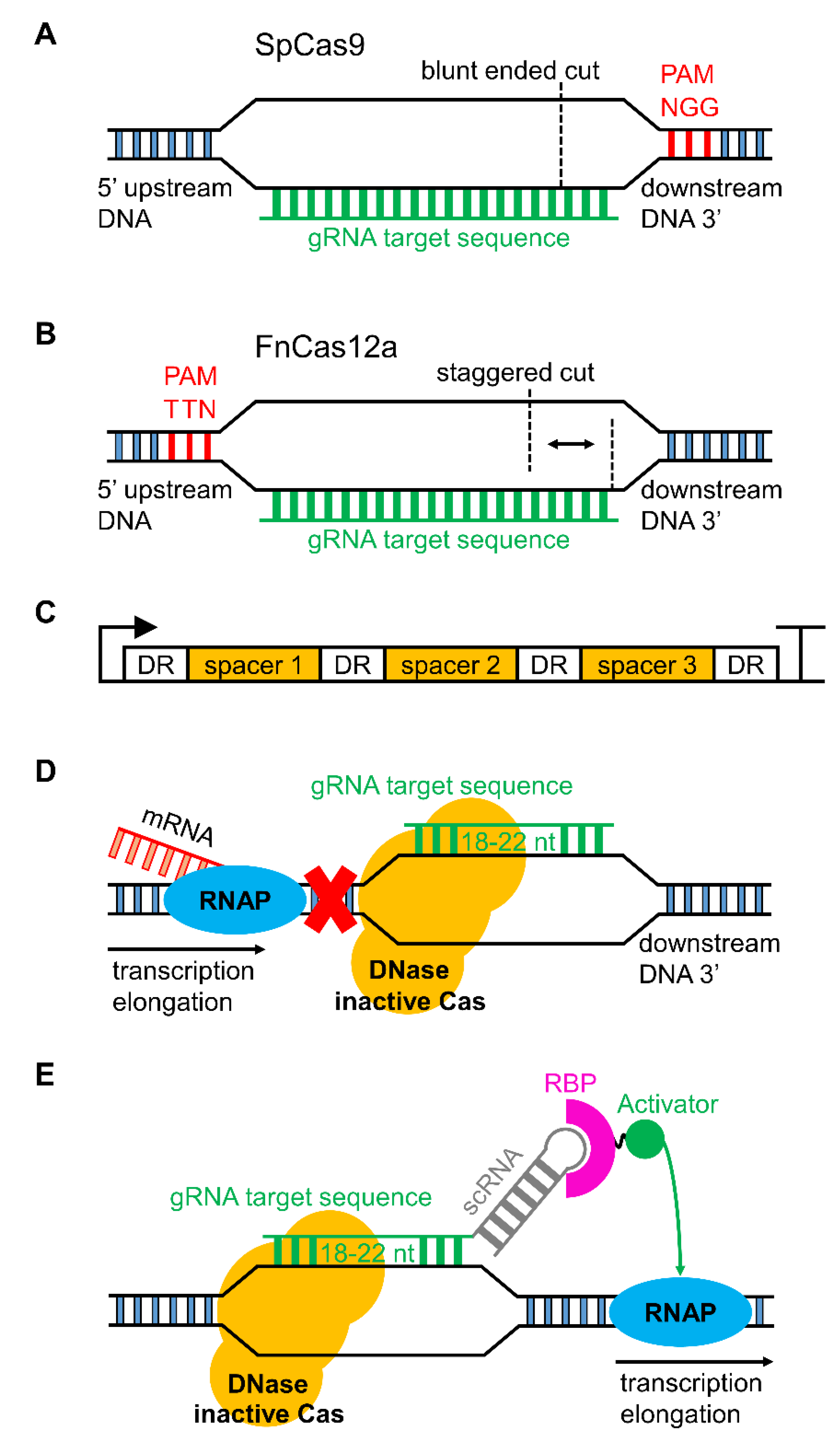
3.1. CRISPR/Cas Genome Editing in Cyanobacteria
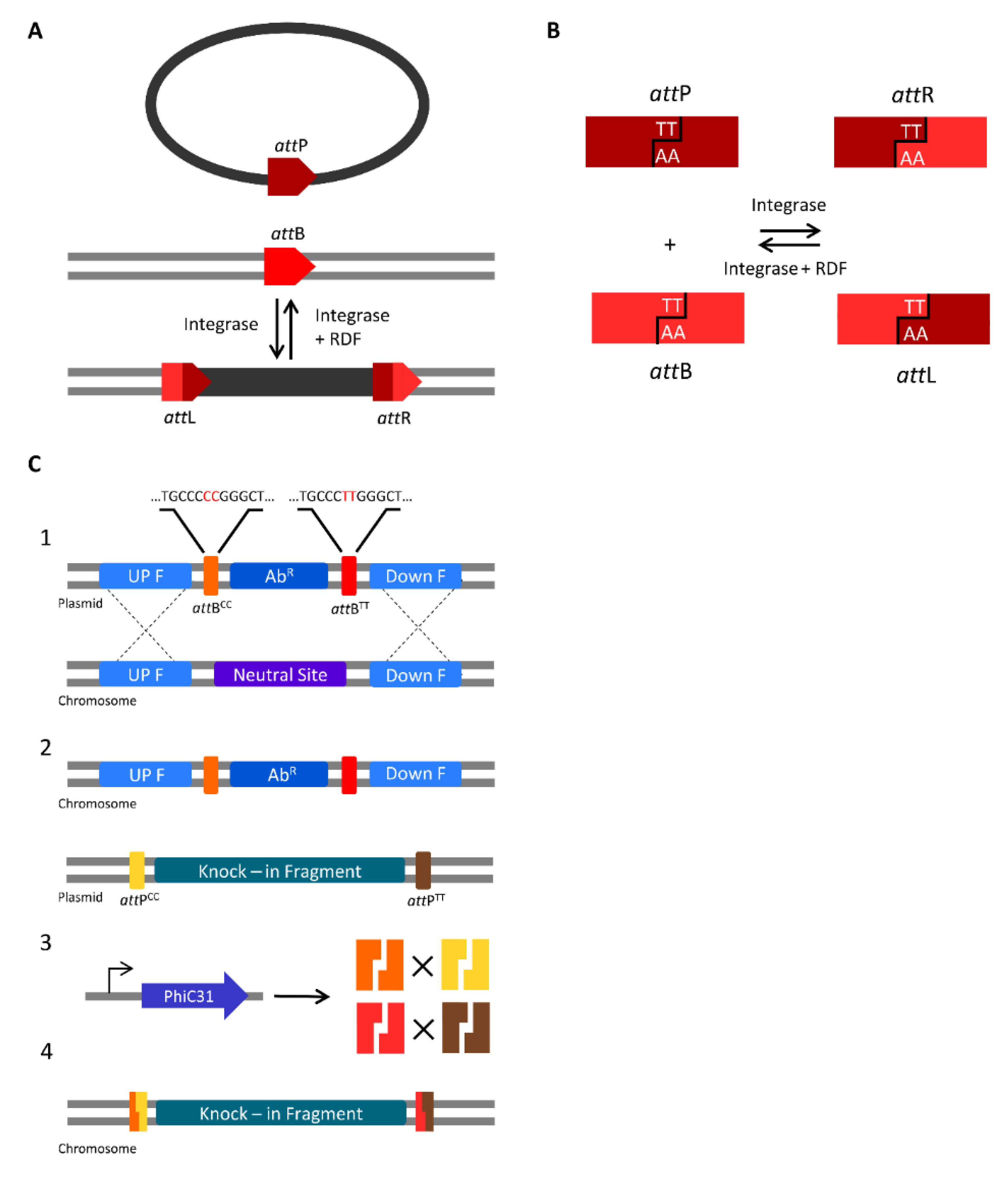
3.2. Serine Integrases for Generating Multiple Knock-ins
4. Known and Novel Tools for Regulating Gene Expression in Cyanobacteria
4.1. Gene Regulation with CRISPRi and Synthetic Small Regulatory RNAs
4.2. Sigma Factors and RNA Polymerase as Regulatory Tools for Gene Transcription
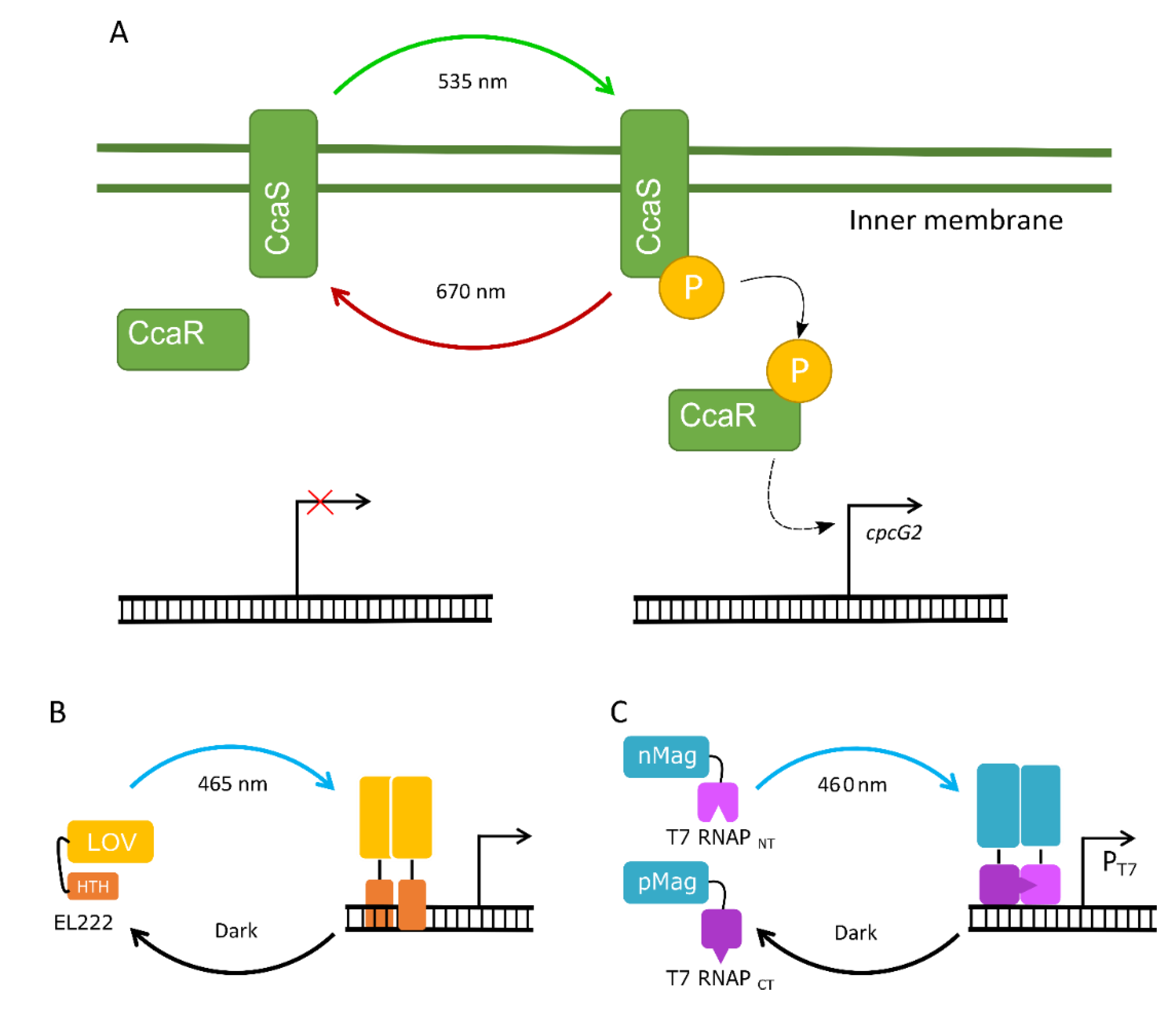
4.3. The Potential of Optogenetic Systems
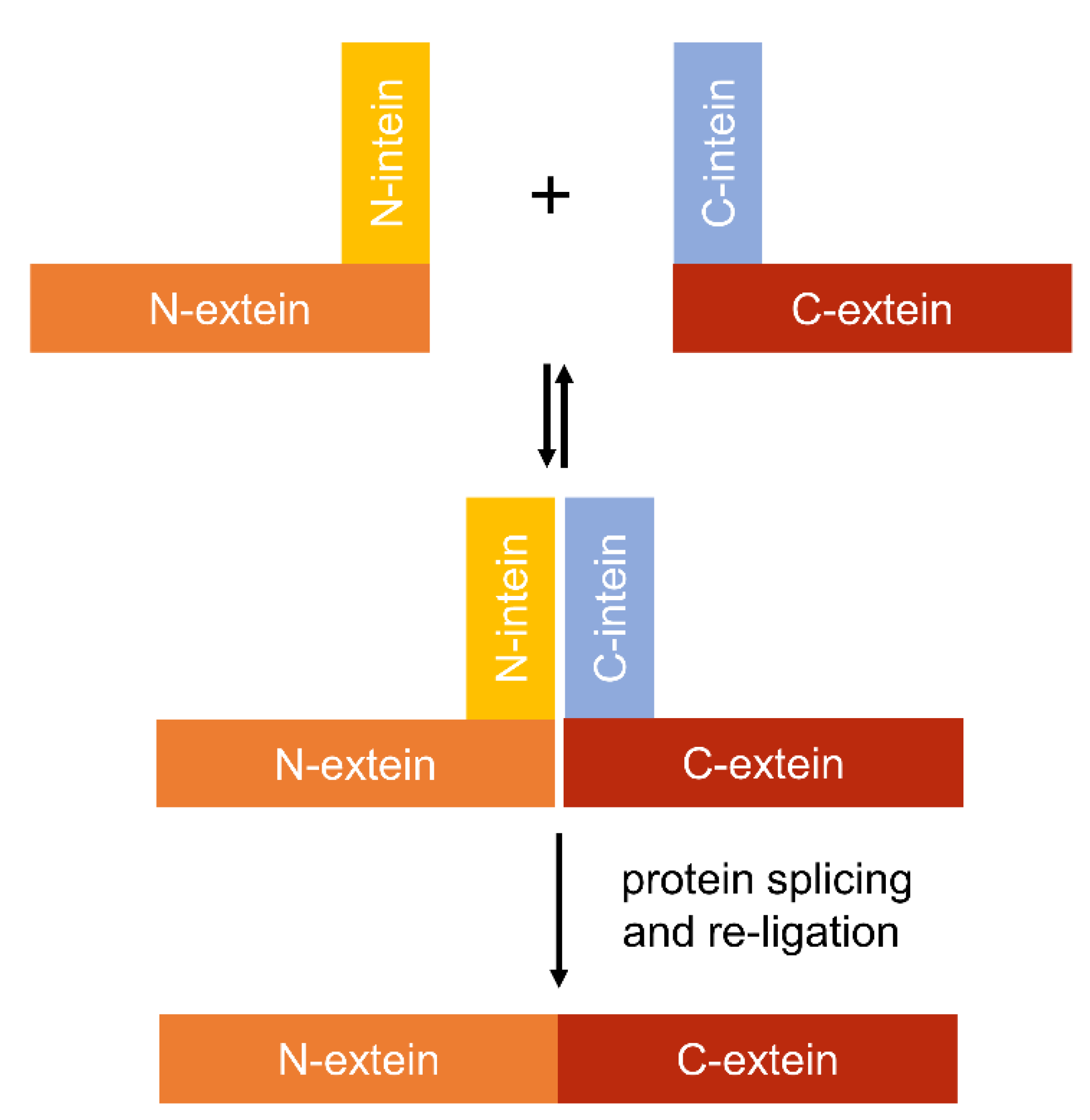
4.4. Using Inteins to Progress Genetic Circuit Research in Cyanobacteria
5. Genome-Scale Models
6. Development of CyanoSource: A Barcoded Mutant Library for Synechocystis sp. PCC 6803
7. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Seckbach, J. Algae and Cyanobacteria in Extreme Environments; Cellular Origin, Life in Extreme Habitats and Astrobiology; Springer: Dordrecht, The Netherlands, 2007; ISBN 978-1-4020-6111-0. [Google Scholar]
- Flombaum, P.; Gallegos, J.L.; Gordillo, R.A.; Rincón, J.; Zabala, L.L.; Jiao, N.; Karl, D.M.; Li, W.K.W.; Lomas, M.W.; Veneziano, D.; et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 2013, 110, 9824–9829. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, D.; Miller, S.R. Photosynthetic temperature adaptation during niche diversification of the thermophilic cyanobacterium Synechococcus A/B clade. ISME J. 2017, 11, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Puente-Sánchez, F.; Arce-Rodríguez, A.; Oggerin, M.; García-Villadangos, M.; Moreno-Paz, M.; Blanco, Y.; Rodríguez, N.; Bird, L.; Lincoln, S.A.; Tornos, F.; et al. Viable cyanobacteria in the deep continental subsurface. Proc. Natl. Acad. Sci. USA 2018, 115, 10702–10707. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeister, B.E.; Gugger, M.; Donoghue, P.C.J. Cyanobacteria and the great oxidation event: Evidence from genes and fossils. Palaeontology 2015, 58, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Knoll, A.H. Paleobiological perspectives on early microbial evolution. Cold Spring Harb. Perspect. Biol. 2015, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nutman, A.P.; Bennett, V.C.; Friend, C.R.L.; Van Kranendonk, M.J.; Chivas, A.R. Rapid emergence of life shown by discovery of 3700-million-year-old microbial structures. Nature 2016, 537, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.J.; Grim, S.L.; Klatt, J.M. Controls on O2 production in cyanobacterial mats and implications for Earth’s oxygenation. Annu. Rev. Earth Planet. Sci. 2018, 46, 123–147. [Google Scholar] [CrossRef]
- Pisciotta, J.M.; Zou, Y.; Baskakov, I.V. Light-dependent electrogenic activity of cyanobacteria. PLoS ONE 2010, 5, e10821. [Google Scholar] [CrossRef]
- Lea-Smith, D.J.; Howe, C.J. The use of cyanobacteria for biofuel production. In Biofuels and Bioenergy; Love, J., Bryant, J.A., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2017; pp. 143–155. ISBN 9781118350553. [Google Scholar]
- Singh, R.; Parihar, P.; Singh, M.; Bajguz, A.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Uncovering potential applications of cyanobacteria and algal metabolites in biology, agriculture and medicine: Current status and future prospects. Front. Microbiol. 2017, 8, 1–37. [Google Scholar] [CrossRef]
- Carroll, A.L.; Case, A.E.; Zhang, A.; Atsumi, S. Metabolic engineering tools in model cyanobacteria. Metab. Eng. 2018, 50, 47–56. [Google Scholar] [CrossRef]
- Dismukes, G.C.; Carrieri, D.; Bennette, N.; Ananyev, G.M.; Posewitz, M.C. Aquatic phototrophs: Efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 2008, 19, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Long, B.M.; Hee, W.Y.; Sharwood, R.E.; Rae, B.D.; Kaines, S.; Lim, Y.L.; Nguyen, N.D.; Massey, B.; Bala, S.; von Caemmerer, S.; et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 2018, 9, 3570. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.S.; Matsui, M.; Abdullah, A.A.A. Cyanobacteria: Photoautotrophic microbial factories for the sustainable synthesis of industrial products. BioMed Res. Int. 2015, 2015, 754934. [Google Scholar] [CrossRef] [PubMed]
- Lea-Smith, D.J.; Vasudevan, R.; Howe, C.J. Generation of marked and markerless mutants in model cyanobacterial species. J. Vis. Exp. 2016, 2016, e54001. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J. Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 2004, 91, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.Z.; Mellor, S.B.; Vavitsas, K.; Wlodarczyk, A.J.; Gnanasekaran, T.; Perestrello Ramos H de Jesus, M.; King, B.C.; Bakowski, K.; Jensen, P.E. Extending the biosynthetic repertoires of cyanobacteria and chloroplasts. Plant J. 2016, 87, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Pakrasi, H.B. Engineering cyanobacteria for production of terpenoids. Planta 2019, 249, 145–154. [Google Scholar] [CrossRef]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep. 2015, 5, 8132. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Snoep, J.L.; Mrwebi, M.; Schuurmans, J.M.; Rohwer, J.M.; Teixeira de Mattos, M.J. Control of specific growth rate in Saccharomyces cerevisiae. Microbiology 2009, 155, 1699–1707. [Google Scholar] [CrossRef]
- Jaiswal, D.; Sengupta, A.; Sohoni, S.; Sengupta, S.; Phadnavis, A.G.; Pakrasi, H.B.; Wangikar, P.P. Genome features and biochemical characteristics of a robust, fast growing and naturally transformable cyanobacterium Synechococcus elongatus PCC 11801 isolated from India. Sci. Rep. 2018, 8, 16632. [Google Scholar] [CrossRef] [PubMed]
- Włodarczyk, A.; Selão, T.T.; Norling, B.; Nixon, P.J. Unprecedented biomass and fatty acid production by the newly discovered cyanobacterium Synechococcus sp. PCC 11901. bioRxiv 2019, 684944. [Google Scholar] [CrossRef]
- Behler, J.; Vijay, D.; Hess, W.R.; Akhtar, M.K. CRISPR-based technologies for metabolic engineering in cyanobacteria. Trends Biotechnol. 2018, 36, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, S.; Song, X.; Diao, J.; Chen, L.; Zhang, W. Toolboxes for cyanobacteria: Recent advances and future direction. Biotechnol. Adv. 2018, 36, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Santos-Merino, M.; Singh, A.K.; Ducat, D.C. New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front. Bioeng. Biotechnol. 2019, 7, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Vijay, D.; Akhtar, M.K.; Hess, W.R. Genetic and metabolic advances in the engineering of cyanobacteria. Curr. Opin. Biotechnol. 2019, 59, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Markley, A.L.; Begemann, M.B.; Clarke, R.E.; Gordon, G.C.; Pfleger, B.F. Synthetic biology toolbox for controlling gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. ACS Synth. Biol. 2015, 4, 595–603. [Google Scholar] [CrossRef]
- Nielsen, J.; Keasling, J.D. Engineering cellular metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef]
- Immethun, C.M.; DeLorenzo, D.M.; Focht, C.M.; Gupta, D.; Johnson, C.B.; Moon, T.S. Physical, chemical, and metabolic state sensors expand the synthetic biology toolbox for Synechocystis sp. PCC 6803. Biotechnol. Bioeng. 2017, 114, 1561–1569. [Google Scholar] [CrossRef]
- Ferreira, E.A.; Pacheco, C.C.; Pinto, F.; Pereira, J.; Lamosa, P.; Oliveira, P.; Kirov, B.; Jaramillo, A.; Tamagnini, P. Expanding the toolbox for Synechocystis sp. PCC 6803: Validation of replicative vectors and characterization of a novel set of promoters. Synth. Biol. 2018, 3, 1–15. [Google Scholar] [CrossRef]
- Li, S.; Sun, T.; Xu, C.; Chen, L.; Zhang, W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab. Eng. 2018, 48, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Roulet, J.; Taton, A.; Golden, J.W.; Arabolaza, A.; Burkart, M.D.; Gramajo, H. Development of a cyanobacterial heterologous polyketide production platform. Metab. Eng. 2018, 49, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, R.; Gale, G.A.R.; Schiavon, A.A.; Puzorjov, A.; Malin, J.; Gillespie, M.D.; Vavitsas, K.; Zulkower, V.; Wang, B.; Howe, C.J.; et al. CyanoGate: A modular cloning suite for engineering cyanobacteria based on the plant MoClo syntax. Plant Physiol. 2019, 180, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Eckert, C.; Maness, P.C.; Yu, J. A Genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synth. Biol. 2018, 7, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.L.; Taylor, G.M.; Hitchcock, A.; Torres-Méndez, A.; Heap, J.T. A rhamnose-inducible system for precise and temporal control of gene expression in cyanobacteria. ACS Synth. Biol. 2018, 7, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Heidorn, T.; Camsund, D.; Huang, H.H.; Lindberg, P.; Oliveira, P.; Stensjö, K.; Lindblad, P. Synthetic biology in cyanobacteria. In Methods in Enzymology; Voigt, C., Ed.; Elsevier: Boston, MA, USA, 2011; Volume 497, pp. 539–579. ISBN 9780123850751. [Google Scholar]
- Fujisawa, T.; Narikawa, R.; Maeda, S.I.; Watanabe, S.; Kanesaki, Y.; Kobayashi, K.; Nomata, J.; Hanaoka, M.; Watanabe, M.; Ehira, S.; et al. CyanoBase: A large-scale update on its 20th anniversary. Nucleic Acids Res. 2017, 45, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Aikawa, S.; Kojima, Y.; Toya, Y.; Furusawa, C.; Kondo, A.; Shimizu, H. Construction of a genome-scale metabolic model of Arthrospira platensis NIES-39 and metabolic design for cyanobacterial bioproduction. PLoS ONE 2015, 10, e0144430. [Google Scholar] [CrossRef]
- Broddrick, J.T.; Rubin, B.E.; Welkie, D.G.; Du, N.; Mih, N.; Diamond, S.; Lee, J.J.; Golden, S.S.; Palsson, B.O. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc. Natl. Acad. Sci. USA 2016, 113, E8344–E8353. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Pakrasi, H.B.; Maranas, C.D. Elucidation of photoautotrophic carbon flux topology in Synechocystis PCC 6803 using genome-scale carbon mapping models. Metab. Eng. 2018, 47, 190–199. [Google Scholar] [CrossRef]
- Hendry, J.I.; Gopalakrishnan, S.; Ungerer, J.; Pakrasi, H.B.; Tang, Y.J.; Maranas, C.D. Genome-scale fluxome of Synechococcus elongatus UTEX 2973 using transient 13 C-labeling data. Plant Physiol. 2019, 179, 761–769. [Google Scholar] [CrossRef]
- Monk, J.M.; Lloyd, C.J.; Brunk, E.; Mih, N.; Sastry, A.; King, Z.; Takeuchi, R.; Nomura, W.; Zhang, Z.; Mori, H.; et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 2017, 35, 904–908. [Google Scholar] [CrossRef] [PubMed]
- Shestakov, S.V.; Khyen, N.T. Evidence for genetic transformation in blue-green alga Anacystis nidulans. MGG Mol. Gen. Genet. 1970, 107, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Stevens, S.E.; Porter, R.D. Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. USA 1980, 77, 6052–6056. [Google Scholar] [CrossRef] [PubMed]
- Elhai, J. Strong and regulated promoters in the cyanobacterium Anabaena PCC 7120. FEMS Microbiol. Lett. 1993, 114, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Wendt, K.E.; Pakrasi, H.B. Genomics approaches to deciphering natural transformation in cyanobacteria. Front. Microbiol. 2019, 10, 1259. [Google Scholar] [CrossRef] [PubMed]
- Taton, A.; Unglaub, F.; Wright, N.E.; Zeng, W.Y.; Paz-Yepes, J.; Brahamsha, B.; Palenik, B.; Peterson, T.C.; Haerizadeh, F.; Golden, S.S.; et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 2014, 42, e136. [Google Scholar] [CrossRef] [PubMed]
- Elhai, J.; Wolk, C.P. Conjugal transfer of DNA to cyanobacteria. In Methods in Enzymology; Packer, L., Glazer, A.N., Eds.; Academic Press: Cambridge, MA, USA, 1988; Volume 167, pp. 747–754. ISBN 9780121820688. [Google Scholar]
- Masukawa, H.; Inoue, K.; Sakurai, H.; Wolk, C.P.; Hausinger, R.P. Site-directed mutagenesis of the Anabaena sp. strain PCC 7120 nitrogenase active site to increase photobiological hydrogen production. Appl. Environ. Microbiol. 2010, 76, 6741–6750. [Google Scholar] [CrossRef]
- Mandakovic, D.; Trigo, C.; Andrade, D.; Riquelme, B.; Gómez-Lillo, G.; Soto-Liebe, K.; Díez, B.; Vásquez, M. CyDiv, a conserved and novel filamentous cyanobacterial cell division protein involved in septum localization. Front. Microbiol. 2016, 7, 94. [Google Scholar] [CrossRef]
- Jeamton, W.; Dulsawat, S.; Tanticharoen, M.; Vonshak, A.; Cheevadhanarak, S. Overcoming intrinsic restriction enzyme barriers enhances transformation efficiency in Arthrospira platensis C1. Plant Cell Physiol. 2017, 58, 822–830. [Google Scholar] [CrossRef]
- Ried, J.L.; Collmer, A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 1987, 57, 239–246. [Google Scholar] [CrossRef]
- Kratz, W.A.; Myers, J. Nutrition and growth of several blue-green algae. Am. J. Bot. 1955, 42, 282–287. [Google Scholar] [CrossRef]
- Ungerer, J.; Lin, P.C.; Chen, H.Y.; Pakrasi, H.B. Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. MBio 2018, 9, e02327-17. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, J.; Wendt, K.E.; Hendry, J.I.; Maranas, C.D.; Pakrasi, H.B. Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc. Natl. Acad. Sci. USA 2018, 115, E11761–E11770. [Google Scholar] [CrossRef] [PubMed]
- Wendt, K.E.; Ungerer, J.; Cobb, R.E.; Zhao, H.; Pakrasi, H.B. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb. Cell Fact. 2016, 15, 115. [Google Scholar] [CrossRef]
- Begemann, M.B.; Zess, E.K.; Walters, E.M.; Schmitt, E.F.; Markley, A.L.; Pfleger, B.F. An organic acid based counter selection system for cyanobacteria. PLoS ONE 2013, 8, e76594. [Google Scholar] [CrossRef] [PubMed]
- Meeks, J.C. Symbiosis between nitrogen-fixing cyanobacteria and plants. Bioscience 1998, 48, 266–276. [Google Scholar] [CrossRef]
- Gao, Q.; Garcia-Pichel, F. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 2011, 9, 791–802. [Google Scholar] [CrossRef]
- Cohen, M.F.; Wallis, J.G.; Campbell, E.L.; Meeks, J.C. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple cellular differentiation alternatives. Microbiology 1994, 140, 3233–3240. [Google Scholar] [CrossRef] [PubMed]
- Soule, T.; Stout, V.; Swingley, W.D.; Meeks, J.C.; Garcia-Pichel, F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2007, 189, 4465–4472. [Google Scholar] [CrossRef]
- Reddy, K.J.; Haskell, J.B.; Sherman, D.M.; Sherman, L.A. Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J. Bacteriol. 1993, 175, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.; Elvitigala, T.; Welsh, E.; Stöckel, J.; Liberton, M.; Min, H.; Sherman, L.A.; Pakrasi, H.B. Novel metabolic attributes of the genus Cyanothece, comprising a group of unicellular nitrogen-fixing cyanobacteria. MBio 2011, 2, e00214-11. [Google Scholar] [CrossRef] [PubMed]
- Min, H.; Sherman, L.A. Genetic transformation and mutagenesis via single-stranded DNA in the unicellular, diazotrophic cyanobacteria of the genus Cyanothece. Appl. Environ. Microbiol. 2010, 76, 7641–7645. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liberton, M.; Bandyopadhyay, A.; Pakrasi, H.B. Enhanced nitrogen fixation in a glgX -deficient strain of Cyanothece sp. strain ATCC 51142, a unicellular nitrogen-fixing cyanobacterium. Appl. Environ. Microbiol. 2019, 85, e02887-18. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, M.; Ehira, S. Spirulina: An example of cyanobacteria as nutraceuticals. In Cyanobacteria; Sharma, N.K., Rai, A.K., Stal, L.J., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2013; pp. 103–118. ISBN 9781118402238. [Google Scholar]
- Adir, N.; Bar-Zvi, S.; Harris, D. The amazing phycobilisome. Biochim. Biophys. Acta Bioenerg. 2019. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Narikawa, R.; Okamoto, S.; Ehira, S.; Yoshimura, H.; Suzuki, I.; Masuda, T.; Mochimaru, M.; Takaichi, S.; Awai, K.; et al. Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39. DNA Res. 2010, 17, 85–103. [Google Scholar] [CrossRef]
- Shimura, Y.; Hirose, Y.; Misawa, N.; Osana, Y.; Katoh, H.; Yamaguchi, H.; Kawachi, M. Comparison of the terrestrial cyanobacterium Leptolyngbya sp. NIES-2104 and the freshwater Leptolyngbya boryana PCC 6306 genomes. DNA Res. 2015, 22, 403–412. [Google Scholar] [CrossRef]
- Taton, A.; Lis, E.; Adin, D.M.; Dong, G.; Cookson, S.; Kay, S.A.; Golden, S.S.; Golden, J.W. Gene Transfer in Leptolyngbya sp. strain BL0902, a cyanobacterium suitable for production of biomass and bioproducts. PLoS ONE 2012, 7, e30901. [Google Scholar] [CrossRef]
- Taton, A.; Ma, A.T.; Ota, M.; Golden, S.S.; Golden, J.W. NOT gate genetic circuits to control gene expression in cyanobacteria. ACS Synth. Biol. 2017, 6, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.W.U.; Montgomery, B.L. The Tryptophan-Rich Sensory Protein (TSPO) is involved in stress-related and light-dependent processes in the cyanobacterium Fremyella diplosiphon. Front. Microbiol. 2015, 6, 1393. [Google Scholar] [CrossRef]
- Pattanaik, B.; Montgomery, B.L. FdTonB is involved in the photoregulation of cellular morphology during complementary chromatic adaptation in Fremyella diplosiphon. Microbiology 2010, 156, 731–741. [Google Scholar] [CrossRef]
- Morris, J.J.; Kirkegaard, R.; Szul, M.J.; Johnson, Z.I.; Zinser, E.R. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl. Environ. Microbiol. 2008, 74, 4530–4534. [Google Scholar] [CrossRef] [PubMed]
- Brahamsha, B. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 1996, 62, 1747–1751. [Google Scholar] [PubMed]
- Ostrowski, M.; Mazard, S.; Tetu, S.G.; Phillippy, K.; Johnson, A.; Palenik, B.; Paulsen, I.T.; Scanlan, D.J. PtrA is required for coordinate regulation of gene expression during phosphate stress in a marine Synechococcus. ISME J. 2010, 4, 908–921. [Google Scholar] [CrossRef]
- Yamaoka, T.; Satoh, K.; Katoh, S. Photosynthetic activities of a thermophilic blue-green alga. Plant Cell Physiol. 1978, 19, 943–954. [Google Scholar] [CrossRef]
- Liang, Y.; Kaczmarek, M.B.; Kasprzak, A.K.; Tang, J.; Shah, M.M.R.; Jin, P.; Klepacz-Smółka, A.; Cheng, J.J.; Ledakowicz, S.; Daroch, M. Thermosynechococcaceae as a source of thermostable C-phycocyanins: Properties and molecular insights. Algal Res. 2018, 35, 223–235. [Google Scholar] [CrossRef]
- Onai, K.; Tabata, S.; Ishiura, M.; Kaneko, T.; Morishita, M. Natural transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: A simple and efficient method for gene transfer. Mol. Genet. Genom. 2004, 271, 50–59. [Google Scholar] [CrossRef]
- Liang, Y.; Tang, J.; Luo, Y.; Kaczmarek, M.B.; Li, X.; Daroch, M. Thermosynechococcus as a thermophilic photosynthetic microbial cell factory for CO2 utilisation. Bioresour. Technol. 2019, 278, 255–265. [Google Scholar] [CrossRef]
- Stucken, K.; Ilhan, J.; Roettger, M.; Dagan, T.; Martin, W.F. Transformation and Conjugal Transfer of Foreign Genes into the Filamentous Multicellular Cyanobacteria (Subsection V) Fischerella and Chlorogloeopsis. Curr. Microbiol. 2012, 65, 552–560. [Google Scholar] [CrossRef]
- Billi, D.; Friedmann, E.I.; Helm, R.F.; Potts, M. Gene transfer to the desiccation-tolerant Cyanobacterium Chroococcidiopsis. J. Bacteriol. 2001, 183, 2298–2305. [Google Scholar] [CrossRef]
- Nürnberg, D.J.; Morton, J.; Santabarbara, S.; Telfer, A.; Joliot, P.; Antonaru, L.A.; Ruban, A.V.; Cardona, T.; Krausz, E.; Boussac, A.; et al. Photochemistry beyond the red limit in chlorophyll f–containing photosystems. Science 2018, 360, 1210–1213. [Google Scholar] [CrossRef]
- Verseux, C.; Baqué, M.; Lehto, K.; De Vera, J.P.P.; Rothschild, L.J.; Billi, D. Sustainable life support on Mars—The potential roles of cyanobacteria. Int. J. Astrobiol. 2016, 15, 65–92. [Google Scholar] [CrossRef]
- Rippka, R.; Waterbury, J.; Cohen-Bazire, G. A cyanobacterium which lacks thylakoids. Arch. Microbiol. 1974, 100, 419–436. [Google Scholar] [CrossRef]
- Rexroth, S.; Mullineaux, C.W.; Ellinger, D.; Sendtko, E.; Rögner, M.; Koenig, F. The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell 2011, 23, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Araki, M.; Shimada, Y.; Mimuro, M.; Tsuchiya, T. Establishment of the reporter system for a thylakoid-lacking cyanobacterium, Gloeobacter violaceus PCC 7421. FEBS Open Bio 2013, 3, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.B.; Arnon, D.I. Studies on nitrogen-fixing blue-algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 1955, 27, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Bordowitz, J.R.; Montgomery, B.L. Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon. J. Bacteriol. 2008, 190, 4069–4074. [Google Scholar] [CrossRef] [PubMed]
- Waterbury, J.B.; Willey, J.M. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 1988, 167, 100–105. [Google Scholar]
- Moore, L.R.; Coe, A.; Zinser, E.R.; Saito, M.A.; Sullivan, M.B.; Lindell, D.; Frois-Moniz, K.; Waterbury, J.; Chisholm, S.W. Culturing the marine cyanobacterium Prochlorococcus. Limnol. Oceanogr. Methods 2007, 5, 353–362. [Google Scholar] [CrossRef]
- Naduthodi, M.I.S.; Barbosa, M.J.; van der Oost, J. Progress of CRISPR-Cas Based Genome Editing in Photosynthetic Microbes. Biotechnol. J. 2018, 13, 1700591. [Google Scholar] [CrossRef]
- Khumsupan, P.; Donovan, S.; McCormick, A.J. CRISPR/Cas in Arabidopsis: Overcoming challenges to accelerate improvements in crop photosynthetic efficiencies. Physiol. Plant. 2019, 166, 428–437. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Jiang, J.Y.; Shi, T.Q.; Sun, X.M.; Zhao, Q.Y.; Huang, H.; Ren, L.J. Application of the CRISPR/Cas system for genome editing in microalgae. Appl. Microbiol. Biotechnol. 2019, 103, 3239–3248. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Murovec, J.; Pirc, Ž.; Yang, B. New variants of CRISPR RNA-guided genome editing enzymes. Plant Biotechnol. J. 2017, 15, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Alshareef, S.; Mahfouz, M.M. CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 2018, 475, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Kellner, M.J.; Zhang, F. Nucleic acid detection of plant genes using CRISPR-Cas13. CRISPR J. 2019, 2, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Campa, C.C.; Weisbach, N.R.; Santinha, A.J.; Incarnato, D.; Platt, R.J. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods 2019, 6, 887–893. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef]
- Swarts, D.C.; van der Oost, J.; Jinek, M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell 2017, 66, 221–233.e4. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Fontana, J.; Patel, A.; Carothers, J.M.; Zalatan, J.G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 2018, 9, 2489. [Google Scholar] [CrossRef]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Vento, J.M.; Crook, N.; Beisel, C.L. Barriers to genome editing with CRISPR in bacteria. J. Ind. Microbiol. Biotechnol. 2019, 1–15. [Google Scholar] [CrossRef]
- Li, H.; Shen, C.R.; Huang, C.H.; Sung, L.Y.; Wu, M.Y.; Hu, Y.C. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab. Eng. 2016, 38, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, S.; Rommelfanger, S.; Balassy, A.; Barba-Ostria, C.; Gu, P.; Galazka, J.M.; Zhang, F. Developing a Cas9-based tool to engineer native plasmids in Synechocystis sp. PCC 6803. Biotechnol. Bioeng. 2018, 115, 2305–2314. [Google Scholar] [CrossRef]
- Niu, T.C.; Lin, G.M.; Xie, L.R.; Wang, Z.Q.; Xing, W.Y.; Zhang, J.Y.; Zhang, C.C. Expanding the potential of CRISPR-Cpf1-based genome editing technology in the cyanobacterium Anabaena PCC 7120. ACS Synth. Biol. 2019, 8, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, J.; Pakrasi, H.B. Cpf1 Is A versatile tool for CRISPR genome editing ccross diverse species of cyanobacteria. Sci. Rep. 2016, 6, 39681. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Fonfara, I.; Richter, H.; BratoviÄ, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Cheng, Q.; Zheng, X.; Zhao, G.; Wang, J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017, 3, 17018. [Google Scholar] [CrossRef] [PubMed]
- Nakahira, Y.; Ogawa, A.; Asano, H.; Oyama, T.; Tozawa, Y. Theophylline-dependent riboswitch as a novel genetic tool for strict regulation of protein expression in cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol. 2013, 54, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.T.; Schmidt, C.M.; Golden, J.W. Regulation of gene expression in diverse cyanobacterial species by using theophylline-responsive riboswitches. Appl. Environ. Microbiol. 2014, 80, 6704–6713. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Cox, D.B.T.; Yan, W.X.; Manteiga, J.C.; Schneider, M.W.; Yamano, T.; Nishimasu, H.; Nureki, O.; Crosetto, N.; Zhang, F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017, 35, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef]
- Tsujimoto, R.; Kotani, H.; Yokomizo, K.; Yamakawa, H.; Nonaka, A.; Fujita, Y. Functional expression of an oxygen-labile nitrogenase in an oxygenic photosynthetic organism. Sci. Rep. 2018, 8, 7380. [Google Scholar] [CrossRef]
- Merrick, C.A.; Zhao, J.; Rosser, S.J. Serine integrases: Advancing synthetic biology. ACS Synth. Biol. 2018, 7, 299–310. [Google Scholar] [CrossRef]
- Colloms, S.D.; Merrick, C.A.; Olorunniji, F.J.; Stark, W.M.; Smith, M.C.M.; Osbourn, A.; Keasling, J.D.; Rosser, S.J. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 2014, 42, e23. [Google Scholar] [CrossRef]
- Snoeck, N.; De Mol, M.L.; Van Herpe, D.; Goormans, A.; Maryns, I.; Coussement, P.; Peters, G.; Beauprez, J.; De Maeseneire, S.L.; Soetaert, W. Serine integrase recombinational engineering (SIRE): A versatile toolbox for genome editing. Biotechnol. Bioeng. 2019, 116, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Guiziou, S.; Mayonove, P.; Bonnet, J. Hierarchical composition of reliable recombinase logic devices. Nat. Commun. 2019, 10, 456. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chai, C.; Yang, S.; Jiang, W.; Gu, Y. Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii. Metab. Eng. 2019, 52, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Andreas, S.; Schwenk, F.; Küter-Luks, B.; Faust, N.; Kühn, R. Enhanced efficiency through nuclear localization signal fusion on phage phiC31-integrase: Activity comparison with Cre and FLPe recombinase in mammalian cells. Nucleic Acids Res. 2002, 30, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Groth, A.C.; Fish, M.; Nusse, R.; Calos, M.P. Construction of transgenic Drosophila by using the site-specific integrase from phage φC31. Genetics 2004, 166, 1775–1782. [Google Scholar] [CrossRef] [PubMed]
- Na, D.; Yoo, S.M.; Chung, H.; Park, H.; Park, J.H.; Lee, S.Y. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 2013, 31, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, S.; Song, X.; Pei, G.; Diao, J.; Cui, J.; Shi, M.; Chen, L.; Zhang, W. Re-direction of carbon flux to key precursor malonyl-CoA via artificial small RNAs in photosynthetic Synechocystis sp. PCC 6803. Biotechnol. Biofuels 2018, 11, 26. [Google Scholar] [CrossRef]
- Higo, A.; Isu, A.; Fukaya, Y.; Ehira, S.; Hisabori, T. Application of CRISPR interference for metabolic engineering of the heterocyst-forming multicellular cyanobacterium Anabaena sp. PCC 7120. Plant Cell Physiol. 2018, 59, 119–127. [Google Scholar] [CrossRef]
- Zess, E.K.; Begemann, M.B.; Pfleger, B.F. Construction of new synthetic biology tools for the control of gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. Biotechnol. Bioeng. 2016, 113, 424–432. [Google Scholar] [CrossRef]
- Gordon, G.C.; Korosh, T.C.; Cameron, J.C.; Markley, A.L.; Begemann, M.B.; Pfleger, B.F. CRISPR interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. strain PCC 7002. Metab. Eng. 2016, 38, 170–179. [Google Scholar] [CrossRef]
- Huang, C.H.; Shen, C.R.; Li, H.; Sung, L.Y.; Wu, M.Y.; Hu, Y.C. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb. Cell Fact. 2016, 15, 196. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarzyk, D.; Cengic, I.; Yao, L.; Hudson, E.P. Diversion of the long-chain acyl-ACP pool in Synechocystis to fatty alcohols through CRISPRi repression of the essential phosphate acyltransferase PlsX. Metab. Eng. 2018, 45, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Shabestary, K.; Anfelt, J.; Ljungqvist, E.; Jahn, M.; Yao, L.; Hudson, E.P. Targeted repression of essential genes to arrest growth and increase carbon partitioning and biofuel titers in cyanobacteria. ACS Synth. Biol. 2018, 7, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Choe, D.; Lee, E.; Kim, S.C.; Palsson, B.; Cho, B.K. High-Level dCas9 Expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 2018, 7, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Cengic, I.; Anfelt, J.; Hudson, E.P. Multiple Gene Repression in Cyanobacteria Using CRISPRi. ACS Synth. Biol. 2016, 5, 207–212. [Google Scholar] [CrossRef]
- Miao, C.; Zhao, H.; Qian, L.; Lou, C. Systematically investigating the key features of the DNase deactivated Cpf1 for tunable transcription regulation in prokaryotic cells. Synth. Syst. Biotechnol. 2019, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Martella, A.; Firth, M.A.; Taylor, B.J.M.; Goeppert, A.U.; Cuomo, E.M.; Roth, R.G.; Dickson, A.J.; Fisher, D.I. Systematic evaluation of CRISPRa and CRISPRi modalities enables development of a multiplexed, orthogonal gene activation and repression system. ACS Synth. Biol. 2019, 8, 1998–2006. [Google Scholar] [CrossRef]
- Fontana, J.; Dong, C.; Kiattisewee, C.; Chavali, V.P.; Tickman, B.I.; Carothers, J.M.; Zalatan, J.G. Effective CRISPRa-mediated control of gene expression in bacteria must overcome strict target site requirements. bioRxiv 2019, 770891. [Google Scholar] [CrossRef]
- Bradley, R.W.; Buck, M.; Wang, B. Tools and principles for microbial gene circuit engineering. J. Mol. Biol. 2016, 428, 862–888. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.A.K.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychalski, E.A.; Ross, D.; Densmore, D.; Voigt, C.A. Genetic circuit design automation. Science 2016, 352, aac7341. [Google Scholar] [CrossRef] [PubMed]
- Lalwani, M.A.; Zhao, E.M.; Avalos, J.L. Current and future modalities of dynamic control in metabolic engineering. Curr. Opin. Biotechnol. 2018, 52, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.; Cannon, W. Specific binding of the transcription factor sigma-54 to promoter DNA. Nature 1992, 358, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Feklístov, A.; Sharon, B.D.; Darst, S.A.; Gross, C.A. Bacterial sigma factors: A historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014, 68, 357–376. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.C.; Kesthely, C.A.; Franklin, E.A.; MacLellan, S.R. The essential activities of the bacterial sigma factor. Can. J. Microbiol. 2017, 63, 89–99. [Google Scholar] [CrossRef]
- Stensjö, K.; Vavitsas, K.; Tyystjärvi, T. Harnessing transcription for bioproduction in cyanobacteria. Physiol. Plant. 2018, 162, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Helmann, J.D. Where to begin? Sigma factors and the selectivity of transcription initiation in bacteria. Mol. Microbiol. 2019, 112, 335–347. [Google Scholar] [CrossRef]
- Imamura, S.; Asayama, M. Sigma factors for cyanobacterial transcription. Gene Regul. Syst. Biol. 2009, 3, 65–87. [Google Scholar] [CrossRef]
- Wells, K.N.; Videau, P.; Nelson, D.; Eiting, J.E.; Philmus, B. The influence of sigma factors and ribosomal recognition elements on heterologous expression of cyanobacterial gene clusters in Escherichia coli. FEMS Microbiol. Lett. 2018, 365, fny164. [Google Scholar] [CrossRef]
- Bervoets, I.; Van Brempt, M.; Van Nerom, K.; Van Hove, B.; Maertens, J.; De Mey, M.; Charlier, D. A sigma factor toolbox for orthogonal gene expression in Escherichia coli. Nucleic Acids Res. 2018, 46, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.; Zhang, Y.; Lin, Z. Bacterial sigma factors as targets for engineered or synthetic transcriptional control. Front. Bioeng. Biotechnol. 2014, 2, 33. [Google Scholar] [CrossRef]
- Iyer, L.M.; Koonin, E.V.; Aravind, L. Evolution of bacterial RNA polymerase: Implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene 2004, 335, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Liu, M.; Burgess, R.R. Promoter and regulon analysis of nitrogen assimilation factor, σ54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 2010, 38, 1273–1283. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, X.; Wang, B. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria. Nat. Commun. 2019, 10, 3693. [Google Scholar] [CrossRef]
- Meyer, A.J.; Ellefson, J.W.; Ellington, A.D. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry. ACS Synth. Biol. 2015, 4, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Hussey, B.J.; McMillen, D.R. Programmable T7-based synthetic transcription factors. Nucleic Acids Res. 2018, 46, 9842–9854. [Google Scholar] [CrossRef]
- Temme, K.; Hill, R.; Segall-Shapiro, T.H.; Moser, F.; Voigt, C.A. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012, 40, 8773–8781. [Google Scholar] [CrossRef]
- Kim, W.J.; Lee, S.M.; Um, Y.; Sim, S.J.; Woo, H.M. Development of SyneBrick vectors as a synthetic biology platform for gene expression in Synechococcus elongatus PCC 7942. Front. Plant Sci. 2017, 8, 293. [Google Scholar] [CrossRef]
- Jin, H.; Lindblad, P.; Bhaya, D. Building an inducible T7 RNA polymerase/T7 promoter circuit in Synechocystis sp. PCC6803. ACS Synth. Biol. 2019, 8, 655–660. [Google Scholar] [CrossRef]
- Pu, J.; Zinkus-Boltz, J.; Dickinson, B.C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 2017, 13, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Komura, R.; Aoki, W.; Motone, K.; Satomura, A.; Ueda, M. High-throughput evaluation of T7 promoter variants using biased randomization and DNA barcoding. PLoS ONE 2018, 13, e0196905. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Chen, Q.; Liu, H. Engineered photoactivatable genetic switches based on the bacterium phage T7 RNA Polymerase. ACS Synth. Biol. 2017, 6, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J.; Segall-Shapiro, T.H.; Glassey, E.; Zhang, J.; Voigt, C.A. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat. Chem. Biol. 2019, 15, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K. Molecular engineering of the salicylate-inducible transcription factor Sal7AR for orthogonal and high gene expression in Escherichia coli. PLoS ONE 2018, 13, e0194090. [Google Scholar] [CrossRef]
- Kelly, C.L.; Liu, Z.; Yoshihara, A.; Jenkinson, S.F.; Wormald, M.R.; Otero, J.; Estévez, A.; Kato, A.; Marqvorsen, M.H.S.; Fleet, G.W.J.; et al. Synthetic chemical inducers and genetic decoupling enable orthogonal control of the rhaBAD Promoter. ACS Synth. Biol. 2016, 5, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.H.; Lindblad, P. Wide-dynamic-range promoters engineered for cyanobacteria. J. Biol. Eng. 2013, 7, 10. [Google Scholar] [CrossRef]
- Albers, S.C.; Gallegos, V.A.; Peebles, C.A.M. Engineering of genetic control tools in Synechocystis sp. PCC 6803 using rational design techniques. J. Biotechnol. 2015, 216, 36–46. [Google Scholar] [CrossRef]
- Cao, Y.Q.; Li, Q.; Xia, P.F.; Wei, L.J.; Guo, N.; Li, J.W.; Wang, S.G. AraBAD based toolkit for gene expression and metabolic robustness improvement in Synechococcus elongatus. Sci. Rep. 2017, 7, 18059. [Google Scholar] [CrossRef]
- Huang, H.H.; Camsund, D.; Lindblad, P.; Heidorn, T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010, 38, 2577–2593. [Google Scholar] [CrossRef]
- Camsund, D.; Heidorn, T.; Lindblad, P. Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. J. Biol. Eng. 2014, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Blasi, B.; Peca, L.; Vass, I.; Kós, P.B. Characterization of stress responses of heavy metal and metalloid inducible promoters in Synechocystis PCC6803. J. Microbiol. Biotechnol. 2012, 22, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.A.; Gajewski, J.P.; Ferlez, B.H.; Ludwig, M.; Baker, C.S.; Golbeck, J.H.; Bryant, D.A. Zn2+ -inducible expression platform for Synechococcus sp. strain PCC 7002 based on the smtA promoter/operator and smtB repressor. Appl. Environ. Microbiol. 2017, 83, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Englund, E.; Liang, F.; Lindberg, P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2016, 6, 36640. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Gao, C.; Hou, J.; Xu, P.; Guo, L.; Chen, X.; Hu, G.; Ye, C.; Edwards, H.; Chen, J.; Chen, W.; et al. Programmable biomolecular switches for rewiring flux in Escherichia coli. Nat. Commun. 2019, 10, 3751. [Google Scholar] [CrossRef]
- Polstein, L.R.; Gersbach, C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015, 11, 198–200. [Google Scholar] [CrossRef]
- Lee, D.; Hyun, J.H.; Jung, K.; Hannan, P.; Kwon, H.B. A calcium- and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol. 2017, 35, 858–863. [Google Scholar] [CrossRef]
- Mansouri, M.; Strittmatter, T.; Fussenegger, M. Light-controlled mammalian cells and their therapeutic applications in synthetic biology. Adv. Sci. 2019, 6, 1800952. [Google Scholar] [CrossRef]
- Tandar, S.T.; Senoo, S.; Toya, Y.; Shimizu, H. Optogenetic switch for controlling the central metabolic flux of Escherichia coli. Metab. Eng. 2019, 55, 68–75. [Google Scholar] [CrossRef]
- Zhao, E.M.; Suek, N.; Wilson, M.Z.; Dine, E.; Pannucci, N.L.; Gitai, Z.; Avalos, J.L.; Toettcher, J.E. Light-based control of metabolic flux through assembly of synthetic organelles. Nat. Chem. Biol. 2019, 15, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, P.; Devarajan, K.; Chua, T.K.; Zhang, H.; Gunawan, E.; Poh, C.L. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Res. 2016, 44, 6994–7005. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Rodriguez, J.; Moser, F.; Song, M.; Voigt, C.A. Engineering RGB color vision into Escherichia coli. Nat. Chem. Biol. 2017, 13, 706–708. [Google Scholar] [CrossRef] [PubMed]
- Rost, B.R.; Schneider-Warme, F.; Schmitz, D.; Hegemann, P. Optogenetic tools for subcellular applications in neuroscience. Neuron 2017, 96, 572–603. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Wang, Y.; Zhao, S.; Ren, Y. Photocontrolled reversible self-assembly of dodecamer nitrilase. Bioresour. Bioprocess. 2017, 4, 36. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirose, Y.; Shimada, T.; Narikawa, R.; Katayama, M.; Ikeuchi, M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. USA 2008, 105, 9528–9533. [Google Scholar] [CrossRef] [PubMed]
- Tilbrook, K.; Arongaus, A.B.; Binkert, M.; Heijde, M.; Yin, R.; Ulm, R. The UVR8 UV-B Photoreceptor: Perception, signaling and response. Arab. Book 2013, 11, e0164. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, P.; Tabor, J.J. Repurposing synechocystis PCC6803 UirS-UirR as a UV-violet/green photoreversible transcriptional regulatory tool in E. coli. ACS Synth. Biol. 2016, 5, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Möglich, A.; Ayers, R.A.; Moffat, K. Design and signaling mechanism of light-regulated histidine kinases. J. Mol. Biol. 2009, 385, 1433–1444. [Google Scholar] [CrossRef]
- Jin, X.; Riedel-Kruse, I.H. Biofilm Lithography enables high-resolution cell patterning via optogenetic adhesin expression. Proc. Natl. Acad. Sci. USA 2018, 115, 3698–3703. [Google Scholar] [CrossRef]
- Abe, K.; Miyake, K.; Nakamura, M.; Kojima, K.; Ferri, S.; Ikebukuro, K.; Sode, K. Engineering of a green-light inducible gene expression system in Synechocystis sp. PCC6803. Microb. Biotechnol. 2014, 7, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Abe, K.; Ferri, S.; Nakajima, M.; Nakamura, M.; Yoshida, W.; Kojima, K.; Ikebukuro, K.; Sode, K. A green-light inducible lytic system for cyanobacterial cells. Biotechnol. Biofuels 2014, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Ong, N.T.; Tabor, J.J. A miniaturized Escherichia coli green light sensor with high dynamic range. ChemBioChem 2018, 19, 1255–1258. [Google Scholar] [CrossRef] [PubMed]
- Tabor, J.J.; Levskaya, A.; Voigt, C.A. Multichromatic control of gene expression in Escherichia coli. J. Mol. Biol. 2011, 405, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Ong, N.T.; Olson, E.J.; Tabor, J.J. Engineering an E. coli near-infrared light sensor. ACS Synth. Biol. 2018, 7, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Takakado, A.; Nakasone, Y.; Terazima, M. Sequential DNA binding and dimerization processes of the photosensory protein EL222. Biochemistry 2018, 57, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Du, Z.; Liu, R.; Li, T.; Zhao, Y.; Chen, X.; Yang, Y. A single-component optogenetic system allows stringent switch of gene expression in yeast cells. ACS Synth. Biol. 2018, 7, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Papanatsiou, M.; Petersen, J.; Henderson, L.; Wang, Y.; Christie, J.M.; Blatt, M.R. Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 2019, 363, 1456–1459. [Google Scholar] [CrossRef]
- Baumschlager, A.; Aoki, S.K.; Khammash, M. Dynamic blue light-inducible T7 RNA Polymerases (Opto-T7RNAPs) for precise spatiotemporal gene expression control. ACS Synth. Biol. 2017, 6, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Cho, H.S.; Cho, J.I.; Jeon, J.S.; Lagarias, J.C.; Park, Y.I. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2011, 108, 10780–10785. [Google Scholar] [CrossRef]
- Badary, A.; Abe, K.; Ferri, S.; Kojima, K.; Sode, K. The Development and characterization of an exogenous green-light-regulated gene expression system in marine cyanobacteria. Mar. Biotechnol. 2015, 17, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Wiltbank, L.B.; Kehoe, D.M. Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 2019, 17, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Luimstra, V.M.; Schuurmans, J.M.; Verschoor, A.M.; Hellingwerf, K.J.; Huisman, J.; Matthijs, H.C.P. Blue light reduces photosynthetic efficiency of cyanobacteria through an imbalance between photosystems I and II. Photosynth. Res. 2018, 138, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Volpetti, F.; Petrova, E.; French, C.; Maerkl, S.J.; Wang, B. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol. 2019, 15, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Buck, M. Rapid engineering of versatile molecular logic gates using heterologous genetic transcriptional modules. Chem. Commun. 2014, 50, 11642–11644. [Google Scholar] [CrossRef] [PubMed]
- Brophy, J.A.N.; Voigt, C.A. Principles of genetic circuit design. Nat. Methods 2014, 11, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Dalchau, N.; Wang, B. Scaling up genetic circuit design for cellular computing: Advances and prospects. Nat. Comput. 2018, 17, 833–853. [Google Scholar] [CrossRef]
- Xia, P.; Ling, H.; Foo, J.L.; Chang, M.W. Synthetic biology toolkits for metabolic engineering of cyanobacteria. Biotechnol. J. 2019, 14, 1800496. [Google Scholar] [CrossRef]
- Immethun, C.M.; Ng, K.M.; Delorenzo, D.M.; Waldron-Feinstein, B.; Lee, Y.C.; Moon, T.S. Oxygen-responsive genetic circuits constructed in Synechocystis sp. PCC 6803. Biotechnol. Bioeng. 2016, 113, 433–442. [Google Scholar] [CrossRef]
- Wang, B.; Barahona, M.; Buck, M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens. Bioelectron. 2013, 40, 368–376. [Google Scholar] [CrossRef]
- Noren, C.J.; Wang, J.; Perler, F.B. Dissecting the chemistry of protein splicing and its applications. Angew. Chem. Int. Ed. 2000, 39, 450–466. [Google Scholar] [CrossRef]
- Caspi, J.; Amitai, G.; Belenkiy, O.; Pietrokovski, S. Distribution of split DnaE inteins in cyanobacteria. Mol. Microbiol. 2003, 50, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Perler, F.B. InBase: The intein database. Nucleic Acids Res. 2002, 30, 383–384. [Google Scholar] [CrossRef] [PubMed]
- Novikova, O.; Topilina, N.; Belfort, M. Enigmatic distribution, evolution, and function of inteins. J. Biol. Chem. 2014, 289, 14490–14497. [Google Scholar] [CrossRef] [PubMed]
- Saleh, L.; Perler, F.B. Protein splicing in cis and in trans. Chem. Rec. 2006, 6, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Lockless, S.W.; Muir, T.W. Traceless protein splicing utilizing evolved split inteins. Proc. Natl. Acad. Sci. USA 2009, 106, 10999–11004. [Google Scholar] [CrossRef] [PubMed]
- Schaerli, Y.; Gili, M.; Isalan, M. A split intein T7 RNA polymerase for transcriptional AND-logic. Nucleic Acids Res. 2014, 42, 12322–12328. [Google Scholar] [CrossRef]
- Zeng, Y.; Jones, A.M.; Thomas, E.E.; Nassif, B.; Silberg, J.J.; Segatori, L. A split transcriptional repressor that links protein solubility to an orthogonal genetic circuit. ACS Synth. Biol. 2018, 7, 2126–2138. [Google Scholar] [CrossRef]
- Carvajal-Vallejos, P.; Pallissé, R.; Mootz, H.D.; Schmidt, S.R. Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources. J. Biol. Chem. 2012, 287, 28686–28696. [Google Scholar] [CrossRef]
- Huang, H.H.; Seeger, C.; Helena Danielson, U.; Lindblad, P. Analysis of the leakage of gene repression by an artificial TetR-regulated promoter in cyanobacteria. BMC Res. Notes 2015, 8, 459. [Google Scholar] [CrossRef]
- Goodall, E.C.A.; Robinson, A.; Johnston, I.G.; Jabbari, S.; Turner, K.A.; Cunningham, A.F.; Lund, P.A.; Cole, J.A.; Henderson, I.R. The Essential Genome of Escherichia coli K-12. MBio 2018, 9, e02096-17. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, H.U. Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 2015, 33, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Baroukh, C.; Muñoz-Tamayo, R.; Bernard, O.; Steyer, J.P. Mathematical modeling of unicellular microalgae and cyanobacteria metabolism for biofuel production. Curr. Opin. Biotechnol. 2015, 33, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.J.; Peebles, C.A.M.; Prasad, A. Modeling and analysis of flux distribution and bioproduct formation in Synechocystis sp. PCC 6803 using a new genome-scale metabolic reconstruction. Algal Res. 2017, 27, 295–310. [Google Scholar] [CrossRef]
- Triana, J.; Montagud, A.; Siurana, M.; Fuente, D.; Urchueguía, A.; Gamermann, D.; Torres, J.; Tena, J.; de Córdoba, P.; Urchueguía, J. Generation and evaluation of a genome-scale metabolic network model of Synechococcus elongatus PCC7942. Metabolites 2014, 4, 680–698. [Google Scholar] [CrossRef] [PubMed]
- Broddrick, J.T.; Welkie, D.G.; Jallet, D.; Golden, S.S.; Peers, G.; Palsson, B.O. Predicting the metabolic capabilities of Synechococcus elongatus PCC 7942 adapted to different light regimes. Metab. Eng. 2019, 52, 42–56. [Google Scholar] [CrossRef]
- Mueller, T.J.; Ungerer, J.L.; Pakrasi, H.B.; Maranas, C.D. Identifying the metabolic differences of a fast-growth phenotype in Synechococcus UTEX 2973. Sci. Rep. 2017, 7, 41569. [Google Scholar] [CrossRef]
- Vu, T.T.; Hill, E.A.; Kucek, L.A.; Konopka, A.E.; Beliaev, A.S.; Reed, J.L. Computational evaluation of Synechococcus sp. PCC 7002 metabolism for chemical production. Biotechnol. J. 2013, 8, 619–630. [Google Scholar] [CrossRef]
- Qian, X.; Kim, M.K.; Kumaraswamy, G.K.; Agarwal, A.; Lun, D.S.; Dismukes, G.C. Flux balance analysis of photoautotrophic metabolism: Uncovering new biological details of subsystems involved in cyanobacterial photosynthesis. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 276–287. [Google Scholar] [CrossRef]
- Malatinszky, D.; Steuer, R.; Jones, P.R. A comprehensively curated genome-scale two-cell model for the heterocystous cyanobacterium Anabaena sp. PCC 7120. Plant Physiol. 2017, 173, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; Gudmundsson, S.; Knight, E.M.; Palsson, B.O.; Thiele, I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc. Natl. Acad. Sci. USA 2012, 109, 2678–2683. [Google Scholar] [CrossRef] [PubMed]
- Knoop, H.; Gründel, M.; Zilliges, Y.; Lehmann, R.; Hoffmann, S.; Lockau, W.; Steuer, R. Flux balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 2013, 9, e1003081. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.E.; Wetmore, K.M.; Price, M.N.; Diamond, S.; Shultzaberger, R.K.; Lowe, L.C.; Curtin, G.; Arkin, A.P.; Deutschbauer, A.; Golden, S.S. The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. USA 2015, 112, E6634–E6643. [Google Scholar] [CrossRef] [PubMed]
- Abernathy, M.H.; Yu, J.; Ma, F.; Liberton, M.; Ungerer, J.; Hollinshead, W.D.; Gopalakrishnan, S.; He, L.; Maranas, C.D.; Pakrasi, H.B.; et al. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol. Biofuels 2017, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Shabestary, K.; Hudson, E.P. Computational metabolic engineering strategies for growth-coupled biofuel production by Synechocystis. Metab. Eng. Commun. 2016, 3, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Suthers, P.F.; Maranas, C.D. OptForce: An optimization procedure for identifying All genetic manipulations leading to targeted overproductions. PLoS Comput. Biol. 2010, 6, e1000744. [Google Scholar] [CrossRef] [PubMed]
- Rocha, I.; Maia, P.; Evangelista, P.; Vilaça, P.; Soares, S.; Pinto, J.P.; Nielsen, J.; Patil, K.R.; Ferreira, E.C.; Rocha, M. OptFlux: An open-source software platform for in silico metabolic engineering. BMC Syst. Biol. 2010, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Saha, R.; Zhang, F.; Pakrasi, H.B. Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2017, 7, 17503. [Google Scholar] [CrossRef]
- Lv, Q.; Ma, W.; Liu, H.; Li, J.; Wang, H.; Lu, F.; Zhao, C.; Shi, T. Genome-wide protein-protein interactions and protein function exploration in cyanobacteria. Sci. Rep. 2015, 5, 15519. [Google Scholar] [CrossRef]
- Vivek-Ananth, R.P.; Samal, A. Advances in the integration of transcriptional regulatory information into genome-scale metabolic models. Biosystems 2016, 147, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zavřel, T.; Očenášová, P.; Červený, J. Phenotypic characterization of Synechocystis sp. PCC 6803 substrains reveals differences in sensitivity to abiotic stress. PLoS ONE 2017, 12, e0189130. [Google Scholar] [CrossRef] [PubMed]
- Soppa, J.; Ludt, K.; Zerulla, K. The ploidy level of Synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology 2016, 162, 730–739. [Google Scholar]
- Morris, J.N.; Eaton-Rye, J.J.; Summerfield, T.C. Phenotypic variation in wild-type substrains of the model cyanobacterium Synechocystis sp. PCC 6803. N. Z. J. Bot. 2017, 55, 25–35. [Google Scholar] [CrossRef]
- Li, X.; Patena, W.; Fauser, F.; Jinkerson, R.E.; Saroussi, S.; Meyer, M.T.; Ivanova, N.; Robertson, J.M.; Yue, R.; Zhang, R.; et al. A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 2019, 51, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; André, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Watabe, K.; Mimuro, M.; Tsuchiya, T. Development of a high-frequency in vivo transposon mutagenesis system for Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942. Plant Cell Physiol. 2014, 55, 2017–2026. [Google Scholar] [CrossRef]
- Chambers, S.; Kitney, R.; Freemont, P. The Foundry: The DNA synthesis and construction Foundry at Imperial College. Biochem. Soc. Trans. 2016, 44, 687–688. [Google Scholar] [CrossRef]
- Kuchmina, E.; Wallner, T.; Kryazhov, S.; Zinchenko, V.V.; Wilde, A. An expression system for regulated protein production in Synechocystis sp. PCC 6803 and its application for construction of a conditional knockout of the ferrochelatase enzyme. J. Biotechnol. 2012, 162, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Krynická, V.; Tichý, M.; Krafl, J.; Yu, J.; Kaňa, R.; Boehm, M.; Nixon, P.J.; Komenda, J. Two essential FtsH proteases control the level of the Fur repressor during iron deficiency in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 2014, 94, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Schuergers, N.; Nürnberg, D.J.; Wallner, T.; Mullineaux, C.W.; Wilde, A. PilB localization correlates with the direction of twitching motility in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 2015, 161, 960–966. [Google Scholar] [CrossRef] [PubMed]

|
| Species and Strain | Cas Type | Expression System | Reference |
|---|---|---|---|
| Synechococcus elongatus PCC 7942 | SpCas9 | episomal | [111] |
| Synechocystis sp. PCC 6803 | FnCas12a | episomal | [114] |
| Synechococcus elongatus UTEX 2973 | |||
| Nostoc sp. PCC 7120 | |||
| Synechococcus elongatus UTEX 2973 | SpCas9 | episomal | [58] |
| Synechocystis sp. PCC 6803 | SpCas9 | chromosomal | [112] |
| Synechococcus elongatus PCC 7942 | FnCas12a | episomal | [57] |
| Synechococcus elongatus UTEX 2973 | |||
| Nostoc sp. PCC 7120 | FnCas12a | episomal | [113] |
| Cyanobacteria | Genome Size (bp) | SpCas9 (NGG) | FnCas12a (TTN) | AsCas12a and LbCas12a (TTTV) | AsCas12a-RR (TYCV) | ASCas12a-RVR (TATV) | CasX (TTCN) |
|---|---|---|---|---|---|---|---|
| Arthrospira platensis C1 | 6,089,210 | 134 | 168 | 24 | 46 | 21 | 33 |
| Arthospira plantensis NIES 39 | 6,788,435 | 114 | 171 | 37 | 46 | 22 | 33 |
| Chroococcidiopsis thermalis PCC 7203 | 6,315,792 | 89 | 175 | 38 | 41 | 17 | 34 |
| Cyanothece sp. ATCC 51142 | 4,934,271 | 118 | 221 | 35 | 40 | 23 | 39 |
| Cyanothece sp. PCC 7822 | 6,091,620 | 114 | 210 | 33 | 39 | 21 | 36 |
| Gleobacter violaceus PCC 7421 | 4,659,019 | 170 | 89 | 17 | 42 | 6 | 26 |
| Nostoc punctiforme strain ATCC 29133 | 8,234,322 | 114 | 194 | 43 | 40 | 20 | 35 |
| Nostoc sp. PCC 7120 | 6,413,771 | 119 | 191 | 27 | 39 | 39 | 34 |
| Synechococcus elongatus PCC 6301 | 2,696,255 | 142 | 113 | 10 | 41 | 7 | 28 |
| Synechococcus elongatus PCC 7942 | 2,695,903 | 141 | 113 | 10 | 41 | 6 | 28 |
| Synechococcus elongatus PCC 11801 | 2,691,022 | 139 | 115 | 10 | 41 | 7 | 29 |
| Synechococcus sp. PCC 7002 | 3,008,047 | 153 | 163 | 22 | 48 | 11 | 33 |
| Synechococcus sp. PCC 11901 | 3,081,514 | 152 | 163 | 22 | 47 | 11 | 33 |
| Synechococcus sp. UTEX 2973 | 2,690,418 | 142 | 113 | 10 | 41 | 6 | 28 |
| Synechococcus sp. WH 8102 | 2,434,428 | 173 | 86 | 7 | 49 | 4 | 28 |
| Synechocystis sp. PCC 6803 | 3,569,561 | 161 | 174 | 23 | 51 | 12 | 32 |
| Thermosynechococcus elongatus BP-1 | 2,593,857 | 176 | 126 | 13 | 42 | 10 | 27 |
| Cyanobacteria | GSM Name | Reference |
|---|---|---|
| Synechocystis sp. PCC 6803 | iSynCJ816 | [230] |
| imSyn716 | [42] | |
| Synechococcus sp. PCC 7942 | iSyf715 | [231] |
| iJB785 | [41] | |
| iJB792 | [232] | |
| Synechococcus sp. UTEX 2973 | iSyu683 | [233] |
| imSyu593 | [43] | |
| Synechococcus sp. PCC 7002 | iSpy708 | [234] |
| iSpy821 | [235] | |
| Arthrospira platensis NIES-39 | n/a | [40] |
| Nostoc sp. PCC 7120 | n/a | [236] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gale, G.A.R.; Schiavon Osorio, A.A.; Mills, L.A.; Wang, B.; Lea-Smith, D.J.; McCormick, A.J. Emerging Species and Genome Editing Tools: Future Prospects in Cyanobacterial Synthetic Biology. Microorganisms 2019, 7, 409. https://doi.org/10.3390/microorganisms7100409
Gale GAR, Schiavon Osorio AA, Mills LA, Wang B, Lea-Smith DJ, McCormick AJ. Emerging Species and Genome Editing Tools: Future Prospects in Cyanobacterial Synthetic Biology. Microorganisms. 2019; 7(10):409. https://doi.org/10.3390/microorganisms7100409
Chicago/Turabian StyleGale, Grant A. R., Alejandra A. Schiavon Osorio, Lauren A. Mills, Baojun Wang, David J. Lea-Smith, and Alistair J. McCormick. 2019. "Emerging Species and Genome Editing Tools: Future Prospects in Cyanobacterial Synthetic Biology" Microorganisms 7, no. 10: 409. https://doi.org/10.3390/microorganisms7100409
APA StyleGale, G. A. R., Schiavon Osorio, A. A., Mills, L. A., Wang, B., Lea-Smith, D. J., & McCormick, A. J. (2019). Emerging Species and Genome Editing Tools: Future Prospects in Cyanobacterial Synthetic Biology. Microorganisms, 7(10), 409. https://doi.org/10.3390/microorganisms7100409








