Abstract
Acremonium is known to be regularly isolated from food and also to be a cause of human disease. Herein, we resolve some sources of confusion that have strongly hampered the accurate interpretation of these and other isolations. The recently designated type species of the genus Acremonium, A. alternatum, is known only from a single isolate, but it is the closest known relative of what may be one of the planet’s most successful organisms, Acremonium sclerotigenum/egyptianum, shown herein to be best called by its earliest valid name, A. egyptiacum. The sequencing of ribosomal internal transcribed spacer (ITS) regions, actin genes, or both for 72 study isolates within this group allowed the full range of morphotypes and ITS barcode types to be elucidated, along with information on temperature tolerance and habitat. The results showed that nomenclatural confusion and frequent misidentifications facilitated by morphotaxonomy, along with misidentified early sequence deposits, have obscured the reality that this species is, in many ways, the definitive match of the historical concept of Acremonium: a pale orange or dull greenish-coloured monophialidic hyphomycete, forming cylindrical, ellipsoidal, or obovoid conidia in sticky heads or obovoid conidia in dry chains, and acting ecologically as a soil organism, marine organism, plant pathogen, plant endophyte, probable insect pathogen, human opportunistic pathogen, food contaminant, probable dermatological communicable disease agent, and heat-tolerant spoilage organism. Industrially, it is already in exploratory use as a producer of the antibiotic ascofuranone, active against trypanosomes, cryptosporidia, and microsporidia, and additional applications are in development. The genus-level clarification of the phylogeny of A. egyptiacum shows other historic acremonia belong to separate genera, and two are here described, Parasarocladium for the Acremonium radiatum complex and Kiflimonium for the Acremonium curvulum complex.
1. Introduction
Acremonium is one of the genera selected for this Special Issue, as it is well established as a potentially food-borne organism that also plays a role in infectious diseases of humans and animals [1]. The study of overlapping biotypes commonly isolated from food and cases of human disease soon led to a focus on one particular group of Acremonium species as significant in these areas. The sequence analysis of DNA loci was needed to clarify the circumscription of the group of interest.
Molecular phylogeny is particularly revelatory in the case of simply structured organisms, which produce few morphological clues to their natural affinities. Phylogenetic revisions [2,3,4] of the species grouped in the morphogenus Acremonium by Gams [5] contained at least two major surprises. One surprise was that multiple families and orders of fungi were encompassed in the concept. Indeed, species of radically different phylogenetic affinity could look almost indistinguishable; an example is the convergence of the Hypocrealean Acremonium sclerotigenum (particularly mucoid-conidial isolates not forming sclerotia) and its distant relative Acremonium (now Sarocladium) strictum, as well as the far-removed Plectosphaerellaceous Acremonium cucurbitacearum, now Plectosphaerella melonis. The other surprise was that conidiogenesis was strongly pleomorphic within some species. Different morphs with identical or near-identical sequences at standard biosystematically used loci had sometimes been described as separate species. In the especially dramatic case of Acremonium sclerotigenum/egyptiacum, conidiogenesis, though always phialidic, differed markedly among isolates. Some mainly produced cylindrical to long-ellipsoidal conidia in mucoid heads, while others produced broadly ellipsoidal conidia in heads. There were also many isolates producing obclavate to obovoid conidia in dry chains. The different conidial morphs had distinct colours: the mucoid morphs were pale to medium salmonaceous, while the catenate morphs matured as dull greenish grey. This unexpected plasticity helped to inform a revised view of the taxonomic informativeness of fungal conidiogenesis [6].
The long-standing practical consequence of this sort of plasticity was that very few historic identifications of the Acremonium species from substrates of practical interest, like food, could be trusted. The exceptions were a few cases where voucher isolates were retained and could later be sequence-identified. The small number of such isolates in collections indicated that the name Acremonium sclerotigenum was sometimes isolated from contaminated food [7]. The species concepts had to be redefined, however, in order to be certain which isolates, if any, should truly bear this name, particularly as early sequencing had deposited many reference sequences under names known to be incorrect. The phylogenetic analysis used to clarify species definitions also frequently brought the names of genera into question, and further biosystematic decisions had to be made at that level.
The morphological convergence among Acremonium types was so extensive that, prior to 2011, there were at least five major, phylogenetically distinct contenders among sequence types for the role of epitype of Acremonium alternatum, the type species. This typification was only substantiated by a scant dried type specimen and illustrations. The choice of epitype would root the future phylogenetic concept of Acremonium. Even when the contenders were vetted by exact morphological comparison with Gams’ illustrations of dried type material [8], as well as habitat considerations (leaf litter, near Rostock, Germany), the name could not be narrowed down to a single phylogenetic species. At the time, the most economically, medically, and industrially important species in Acremonium were (a) those related to A. kiliense and A. strictum (two closely related species), (b) the antibiotic producers A. chrysogenum and Emericellopsis salmosynnemata (which had an Acremonium asexual mvorph), and (c) Acremonium sclerotigenum. The importance of A. sclerotigenum was masked by circumstance: its morphological diversity meant that most isolates had been identified under other names, usually A. strictum or A. potronii for isolates with mucoid conidia and A. egyptiacum or A. alternatum for isolates with catenate conidia. Sequence evidence revealed that the importance of A. strictum as an opportunistic human pathogen was based on misidentification: all such identifications that could be definitively traced were A. sclerotigenum or A. kiliense [7]. A. kiliense was closely related to the type species of a valid genus, Sarocladium, originally distinguished by a minor apomorphy, slightly penicillate conidiophore branching. It was not especially closely related to any candidate for A. alternatum, nor were A. chrysogenum or E. salmosynnemata.
Meanwhile, sequenced isolates revealed that A. sclerotigenum was going unrecognised in many important roles and microhabitats that were typical of existing concepts of Acremonium. Though not infrequently identified as A. alternatum itself when catenate, it had a distinct grey-green tone in chained conidial masses that was not mentioned in the description of the type. It was, however, closely related to an isolate that was an excellent candidate for ‘the real’ A. alternatum in every way, CBS 407.66 (CBS = Westerdijk Institute, Utrecht, the Netherlands). For all these reasons (and others), A. kiliense and A. strictum were recombined into Sarocladium, and A. alternatum was epitypified with CBS 407.66 with the express aim of including A. sclerotigenum in the core group of the revised genus Acremonium.
The object of the present paper is to document the intrinsic phylogenetic biodiversity, protean ecology, multifaceted anthropic significance, and complex nomenclature of this archetypical Acremonium species. For practical purposes, in reference to the theme of this Special Issue, this clarification is expected to provide an essential underpinning for interpreting Acremonium isolations, as well as nucleic acid and secondary metabolite data, from food-related and biomedical sources. Without such a study, given the prevalence of misidentified reference sequences in public databases and the many known reports already marred by use of these inappropriate names [7], authors remain at risk of inadvertently generating data that are misleading or arduous to interpret.
2. Materials and Methods
Isolates were obtained from the collections of the Westerdijk Institute (previously called the CBS Fungal Biodiversity Centre; herbarium designation still CBS) and the FMR collection (Facultad de Medicina, Reus, Tarragona, Spain). Many of the latter isolates had originally been received from the University of Texas Health Science Center at San Antonio, TX, USA, and had UTHSC (University of Texas Health Science Center) numbers. Several isolates from the Ontario Ministry of Health (OMH) were tested, and while most were deposited into CBS and listed under those numbers, a few have only OMH numbers.
The isolates were grown for microscopic observation and temperature growth tests on malt extract agar (MEA; 3% malt extract, Oxoid, Basingstoke, Hampshire, UK) and oatmeal agar (OA; [9]). Growth at human body temperature (37 °C) was monitored at 7 and 14 days for selected isolates considered to represent the range of morphological diversity seen in the isolates.
The DNA for the isolates handled at the Westerdijk Institute was extracted with a FastDNA kit (Qbiogene, Heidelberg, Germany) from mycelium grown for 5–14 days in liquid complete medium [10]. One mL of each DNA sample (containing 50–100 ng DNA mL–1) was subjected to PCR amplification. The nuclear ribosomal internal transcribed spacer (ITS) region was amplified (ITS1 spacer, 5.8S ribosomal region, ITS2 spacer) using primers ITS1 and ITS4 as described by White et al. [11].
The amplicons were cleaned using the GFX PCR DNA Purification Kit (Amersham Pharmacia, Little Chalfont, UK) as per the kit’s instructions. The cleaned DNA was subjected to cycle sequencing using the same primers used for PCR. The cycle sequencing program was as follows: 25 cycles of 95 °C (20 s), 50 °C (15 s), and 60 °C (60 s). The cycle sequencing PCR products were purified using Sephadex G-50 (Amersham Pharmacia) and sequenced at the Westerdijk Institute using BigDye chemistry and an ABI 3730xl sequencer (Applied Biosystems, Carlsbad, CA, USA).
At FMR, total genomic DNA was extracted from colonies grown on potato dextrose agar (PDA; 4 g of potato infusion, 20 g dextrose, 15 g of agar-agar, 1 L tap water) after 7 days of incubation at 20 ± 1 °C, using the FastDNA kit protocol (per Bio101, Vista, CA, USA), with a FastPrep FP120 instrument (Thermo Savant, Holbrook, NY, USA) following the manufacturer’s protocol. The DNA was quantified using the Nanodrop 2000 (Thermo Scientific, Madrid, Spain). The ITS region and a fragment of the actin gene were amplified with the primer pairs ITS1/ITS4 [11] and ACT1/ACT4R [12], respectively. The amplicons were sequenced in both directions with the same primer pair used for amplification at Macrogen Europe (Macrogen Inc., Amsterdam, The Netherlands). The consensus sequences were obtained using the SeqMan software version 7.0.0 (DNAStar Lasergene, Madison, WI, USA).
At the Australian National Herbarium, the Lasergene SeqMan and EditSeq software modules (DNAstar Lasergene) were used to assemble and edit the sequence files. The sequences were then aligned manually in Mesquite version 3.40 [13]. The two gene regions were analysed separately. The ITS dataset included a total of 72 isolates, and six of these (four species of Emericellopsis, one species of Stilbella, and the ex-type strain of Acremonium tubakii) were used to root the tree. The actin dataset included a total of 49 taxa, with Acremonium curvulum, Acremonium spinosum, and Emericellopsis glabra as outgroup taxa. A GTRCAT model was applied to the two markers, and the two datasets were analysed using maximum likelihood (ML) with the software RAxML VI-HPC v.8.2.9; [14,15]), as implemented on the CIPRES Web Portal (http://www.phylo.org; [16]). Support values were obtained using a fast bootstrap analysis of 1000 pseudoreplicates. The trees were visualised in PAUP* [17] and edited with Illustrator (Adobe Systems, San Jose, CA, USA). The datasets were deposited in TreeBase (accession 22890), sequences in GenBank, and nomenclature in MycoBank.
3. Results
DNA sequencing at both the ITS and actin loci showed that isolates with a range of morphologies were grouped into a cohesive sequence group. Within this group, ITS sequences were consistently subdivided into three minimally delimited genetic variants. The sequence differences for these ITS barcode variants are shown in Table 1. A small number of sequences within each barcode type had other minor divergences, not consistent from strain to strain.

Table 1.
Major variants in internal transcribed spacer (ITS) sequences among isolates identified as conspecific with the type of Acremonium sclerotigenum.
A select group of isolates at CBS and at FMR were studied to analyse morphological characters. A smaller group of isolates held at CBS were tested for growth at 37 °C. Figure 1A–C,E,F show the range of morphologies seen: catenate conidia of CBS 545.89 (Figure 1A) consistent with Gams’ concept of A. alternatum, ellipsoidal, mucoid conidia of CBS 251.95 (Figure 1C) consistent with Gams’ concept of A. potronii, and cylindrical, mucoid conidia of CBS 113276 (Figure 1E) consistent with the original description of A. sclerotigenum. Sclerotia are shown both for the catenate-conidial 545.89 (Figure 1B) and at lower magnification for the mucoid-conidial Zare 60-#3. The original 1910 line-drawing of Acremonium potronii (Figure 1D), reproduced here for comparison, is described in the Discussion.
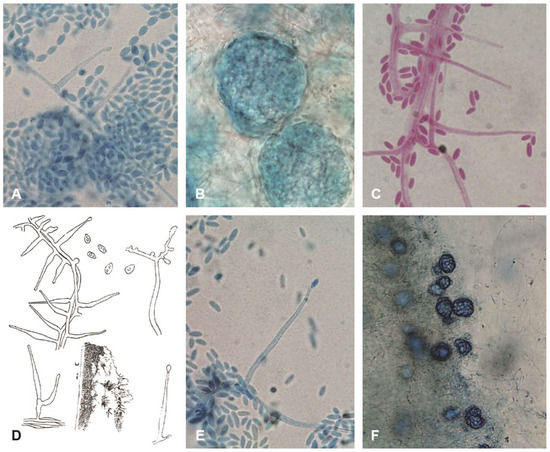
Figure 1.
Morphological diversity of Acremonium sclerotigenum/egyptiacum plus a comparison with an illustration of the original Acremonium potronii. (A–E) ×1000, (F) ×400. Blue structures stained in lactic-acid-cotton-blue stain, reddish in lactofuchsin: (A) CBS 545.89, catenate-conidial isolate originally identified as A. alternatum; (B) CBS 545.89 sclerotia; (C) CBS 251.95 mucoid-conidial isolate with ellipsoidal conidia, originally identified as A. potronii; (D) original Vuillemin drawing of A. potronii; (E) CBS 113276, typical mucoid-conidial A. sclerotigenum isolate with cylindrical conidia; and (F) heavily sclerotial Iranian isolate Zare 60-#3.
In Table 2, the characteristics of the isolates studied in detail are listed with the identification and isolation details of all the other isolates in this study for which ITS sequences were obtained. Table 3 lists isolation details for isolates included in actin sequencing but not elsewhere in the study. It also lists details for outgroup isolates used in analysis in the ITS and actin studies.

Table 2.
Isolates in the Acremonium sclerotigenum/egyptiacum complex typed by ITS barcode type.

Table 3.
Additional isolates included in actin sequencing only or as outgroups.
Table 2 shows that among the three common ITS barcode types, only the first one, 33C/106C/388C, was predominantly seen in isolates obtained outside clinical (including medical and veterinary) environments. Of 32 such isolates, 23 were from environmental sources, eight from clinical sources, and one of unrecorded origin. In contrast, for ITS barcode 2 (33T/106C/388C), six of 11 isolates were from clinical sources. For barcode type 3, (33T/106T/388A), only two of 19 isolates were known to be from environmental sources, while just one was from an unrecorded source, leaving the remaining 16 clinical. Each barcode type contained an isolate involved in a published medical or veterinary case: lethal infection of an ostrich egg in barcode 1 [18], dialysis-related peritonitis in barcode 2 [19], and systemic infection of an immunocompromised patient in barcode 3 [20]. Most isolates obtained from humans were from infected toenails. Many were from repetition-confirmed infections as described by Gupta et al. [21]; the 32 “Acremonium sp.” isolates listed in that publication as confirmed causes of onychomycosis were all later identified as A. sclerotigenum.
The isolates from plant sources were diverse and mostly not ecologically interpretable, but barcode types 1 and 2, particularly the former, were regularly isolated as endophytes of grapevines, including numerous isolates from California vine arms obtained by one of us (A.E.).
The isolates derived from food-related sources were all of barcode type 1 and were obtained from fishmeal, cucumber, muskmelon, and a grapefruit juice can. Whether the many isolates from grapevines have any connection to the quality of grapes as food was not investigated.
Studies of growth at 37 °C revealed that many of the isolates in barcode types 1 and 2 could survive 14 days at body temperature but could not grow. In each barcode type, at least one isolate was seen that could grow minimally off the inoculum plug into the surrounding MEA during 14 days at 37 °C. All three test isolates of barcode group 3 grew at least minimally at 37 °C, and isolate 287.70O from Egyptian soil grew 9 mm in diameter in 14 days.
The majority of the catenate-conidial isolates conforming to the prototype Acremonium egyptiacum morphology belonged to barcode type 1, but one such isolate, CBS 545.89, transpired to belong to barcode type 2. Perdomo et al. [4] reported that an isolate consistent with barcode type 3, UTHSC 05-1172, formed conidia in chains, but that isolate was subsequently lost, and no similar isolate has been detected so far.
Alignments of the ITS and actin regions showed that ITS barcode types were minimally resolved, although in the ITS dendrogram itself (Figure 2), the barcode type 2 isolates tended to cluster together, and the barcode type 3 isolates made up a distinct branch. In the actin dendrogram (Figure 3), the ITS barcode type 3 isolates again mostly clustered together on a unique branch, but there were two anomalies. The branch containing the type 3 isolates also consistently contained, on a short side-branch, CBS 734.69, an ITS type 1 strain with catenate conidiation isolated from tomato roots in Turkey. Also, CBS 287.70O, an ITS type 3 isolate from Egyptian soil, did not cluster with other type 3 isolates in actin analysis but instead clustered among the type 1 and 2 isolates.

Figure 2.
Phylogenetic disposition of the Acremonium sclerotigenum/egyptiacum complex as shown by complete ribosomal internal transcribed spacer (ITS) sequences analysed using maximum likelihood (ML) with RAxML VI-HPC. Support values were obtained using a fast bootstrap analysis of 1000 pseudoreplicates. Members of the Emericellopsis-Stilbella complex are used as an outgroup.
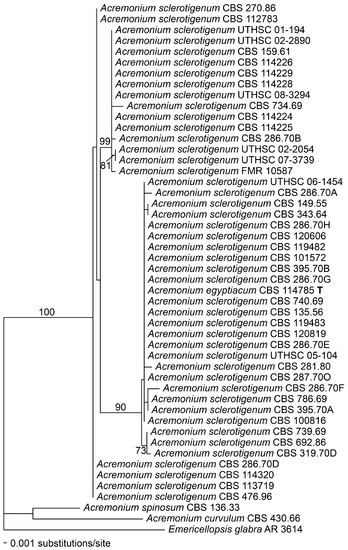
Figure 3.
Phylogenetic disposition of the Acremonium sclerotigenum/egyptiacum complex as shown by a fragment of the actin gene amplified using the primers ACT1 and ACT4R, and analysed using maximum likelihood (ML) with RAxML VI-HPC. Support values were obtained using a fast bootstrap analysis of 1000 pseudoreplicates. Acremonium spinosum, A. curvulum, and Emericellopsis glabra appear as outgroups.
Figure 2 and Figure 3 also show that the genus Acremonium sensu stricto (s. str.), as represented by its apparently rare type species, A. alternatum, and the common A. sclerotigenum/egyptiacum group, is well demarcated from the most closely situated genus in ITS analysis, the recently described Ovicillium, composed of isolates conforming to the outdated Verticillium sect. Albo-Erecta. Also distinctly delimited is the cluster containing Stilbella s. str. and Emericellopsis (along with the closely associated Stanjemonium, not shown). Other related genera, Geosmithia and Clonostachys, are too distant to align reliably in the ITS analyses used here, and few compatible actin sequences exist for related fungi.
4. Discussion
4.1. Taxonomy and Nomenclature of the Acremonium sclerotigenum/egyptiacum Cluster
As Perdomo et al. [4] noted, there is a need to settle the matter of the correct naming of the unexpectedly phenotypically diverse group of isolates encompassed in the A. sclerotigenum/egyptiacum cluster. The balance of the Discussion will be most comprehensible if this matter is settled before other matters are addressed.
We conclude that the ITS and actin sequences used do not support the naming of additional species or infraspecific taxa in this group. Apparent contradictions between ITS and actin placements of a few strains suggest that a sexual or hybridisation process may be shuffling markers within the isolates studied and their conspecifics. Although this re-assortment might well indicate linkage disequilibrium within a sexual species, the differences are small enough that chance alone may also be a factor.
Despite their morphological differences, the isolates ex-type of the original species names Cephalosporium sclerotigenum Moreau and R. Moreau ex Valenta 1948 (validation of an invalid name from 1941) and Oospora egyptiaca J.F.H. Beyma 1933 are very closely related, both belonging to ITS barcode type 1. The earlier valid name, in its modern generic combination, is thus Acremonium egyptiacum (J.F.H. Beyma) W. Gams. Both names are underutilised in the literature, because isolates are so frequently misidentified as Sarocladium (Acremonium) strictum or A. alternatum, and there is no eminent economic or cultural momentum that would argue for the junior name being formally conserved against the senior.
The main outstanding question, then, is whether another of the names frequently found in use for this group, Acremonium potronii Vuill. 1910, might be the true earliest name. Although the isolate ex-type, illustrated by Vuillemin [22] and later in more detail by Pollacci and Nannizzi [23], has not survived to be analysed genetically, the illustrations show a superficially plausible, Acremonium sclerotigenum-like form with ellipsoidal to obovate conidia in mucoid heads (Figure 1D). The isolation is from a medical case possibly involving systemic dissemination (see detailed synopsis by Summerbell [24]). Very few Acremonium-like species are confirmed as causing such cases or growing at 37 °C, usually a prerequisite for human infection, but members of the A. sclerotigenum group have been shown to do both. Close examination of Vuillemin’s illustrations and descriptions, however, reveal that nearly all of the phialides he drew for A. potronii were adelophialides—that is, they lacked a basal septum. Some of them were foreshortened to minute phialidic necks protruding laterally along hyphae, while others were fully developed aculeate structures, frequently somewhat swollen between the basal region and the mid-region. The drawings were of impeccable accuracy; Vuillemin, in the same publication [22], coined and defined the term ‘phialide’ for the first time, giving diverse well-scrutinised examples, and he specifically excluded the structures seen in A. potronii from that term because they were not delimited from the subtending hypha [25]. Gams [5] accepted A. potronii as a name for Acremonium types whose phialides all had basal septa, and he identified several A. sclerotigenum-group isolates with this name (e.g., CBS 251.95), although none of the three A. potronii isolates illustrated in his 1971 monograph is a member of this group. Based on current research findings, it appears likely that A. potronii was a Phialemonium-, Phialemoniopsis-, or Coniochaeta-like fungus [26,27]. Its exact identity is unclear, and we consider it a nomen dubium.
The correct name for the species studied by us, then, is Acremonium egyptiacum (J.F.H. Beyma) W. Gams.
4.2. Ecology and Medical and Economic Significance of A. egyptiacum
As detailed by Summerbell and Scott [7], most published medical cases historically attributed to “Acremonium strictum” turned out to be caused by A. egyptiacum in cases where sequence identification or re-identification became available. Members of ITS barcode group 3 appear to be the most important, causing, to begin with, two well-documented cases of disseminated infection in immunocompromised patients, one attributed by Novicki et al. [20] to ‘Acremonium strictum genogroup 2’, and a more recent case attributed by Guitard et al. [28] to ‘Acremonium sclerotigenum/egyptiacum’. Numerous onychomycosis cases studied by Gupta et al. [21] also can now be attributed to this subclade [6], and there are many isolates obtained from infected toenails in other regions (Table 2 and Table 3), though not all are repetition-confirmed as true etiologic agents [29]. This subclade is isolated so regularly from toenail onychomycosis, mostly in the elderly, that it may be hypothesised to have developed an ongoing niche as a communicable disease of the nails. If it were environmentally common, as are other nail infecting non-dermatophyte fungi like Scopulariopsis brevicaulis, it could be interpreted as a regular invader of nails from other environmental substrates; but, its relatively rare environmental isolation suggests that it may instead be transmitted from person to person via transient environmental inoculum as are dermatophytes. This would make its ecology comparable to that of Neoscytalidium dimidiatum, which has wild-type subtypes that are seen both in woody plant infections and in human skin and nail infections, as well as divergent subtypes, featuring slow growth or loss of melanisation, which are only known from dermatological infections [30].
The presence of A. egyptiacum in the nails may become a systemic hazard if the patient becomes immunocompromised: the case of blood-borne disseminated infection studied by Guitard et al. [28] appeared to arise from a toenail with a prior A. egyptiacum infection.
A case of prosthetic mitral heart valve infestation documented by Guarro et al. [31] can now be attributed to A. egyptiacum ITS barcode type 3, based on its sequence, in GenBank as AM990178.
The relative virulence of two ITS barcode type 3 isolates, UTHSC 01-194 and 05-2270, both originally from blood, was studied in an immunocompromised mouse model by Fernández-Silva et al. [32]. The isolates proved only to infect the immunosuppressed mice, but not immunocompetent controls, and killed 70% of the susceptible animals within 16 days even when low inoculum levels were used.
Barcode type 2 can be credited with a case of kidney dialysis-related peritonitis in a Greek patient [19]. The isolate was kindly forwarded to CBS and is included in this study as CBS 112783.
Barcode type 1 isolates have been isolated from toenails, and there is an isolate, UTHSC 04-3176, listed as derived from cerebrospinal fluid. This is a normally sterile bodily material, but even though microorganisms are normally absent in it, medical significance cannot automatically be attributed to common environmental fungi isolated from it. Process contamination, such as microscopic-scale exposure of syringe needle interiors to contaminant fungal spores from room air or alcohol-swabbed skin, cannot be entirely excluded in the making of clinical isolations from normally sterile bodily materials. Confirmation of pathogenicity requires not just isolation alone, but also supportive case information, such as pathology findings or repeated isolations. Thus, UTHSC 04-3176 is of uncertain medical significance. The one well-confirmed case of clinical significance caused by an isolate of this barcode type (specifically, CBS 100816) is an infection of a farm-produced ostrich egg investigated in Italy because it failed to hatch [18].
A. egyptiacum, as seen in Table 2 and Table 3, occurs regularly in many areas as an endophyte of grapevines; one of us (R.C.S.) has identified many isolates from this habitat sent in to the CBS identification service but not retained in the collection. The species was also found, as A. sclerotigenum, to be a year-round endophyte of Quercus ilex in north-central Spain [33]. Barcode type 1 isolates, listed as Acremonium sp., were obtained in Korea as an endophyte of the medicinal Chinese boxwood, Lycium chinense [34]. A. egyptiacum was a moderately prevalent isolate from diseased or dead larvae of the rose stem girdler beetle Agrilus aurichalcenus in the Kerman Province of south-eastern Iran [35]. Here, the identifications were not stated as having been confirmed by sequencing, but a photograph shows the recognisable pale greyish green of catenulate isolates of A. egyptiacum, and the authors were aware of our as yet unpublished findings about this name. Most isolations from plants are of unknown significance: for example, scattered isolations from the phylloplane of barley (CBS 114320; [36]) may reflect growth in situ or sedimentation from air spora.
A phytopathogenic role for A. egyptiacum (as A. sclerotigenum) was documented by Li et al. [37], who fulfilled Koch’s postulates after discovering the fungus caused a spot disease on bagged apples. The sequences obtained were likely of barcode type 1, because CBS 384.65 (GenBank HQ232129) was cited as the reference used in identification. Both a morph with mucoid heads and a morph with long conidial chains were involved; they had identical sequences at three loci. This finding may connect with the isolation of CBS 343.64 from lenticel of an apple tree.
Koch’s postulates were again fulfilled for A. egyptiacum as a causal agent of rose dieback in Iran’s Fars province [38]. Both isolates sequenced in the study (GenBank KU532330, KU532331) conformed to ITS barcode type 1 and had conidia in mucoid heads. Evidence of significant aerial transmission of inoculum was obtained by spore trapping, possibly indicating that some chains were also formed in situ.
Sequences obtained via denaturing gradient gel electrophoresis (DGGE) from the root zone of the Scrophulariaceous medicinal plant Rehmannia glutinosa (‘sheng di huang’ approximately transcribes the common-name 生地黄) in Henan Province, China, were compatible with ITS barcode type 1, with a GenBank ITS sequence of A. sclerotigenum ex-type CBS 124.42 (HQ232209) cited as a reference comparison [39]. The authors interpreted these sequences as indications of a potential pathogenic contribution from A. egyptiacum (as A. sclerotigenum) in a toxic-soil replant problem with the plant.
The correction via sequencing of a long-standing misidentification of an A. egyptiacum barcode type 1 isolate (GenBank LC063776) as Ascochyta viciae allowed this species (as A. sclerotigenum) to be identified as the producer of the meroterpenoid compound ascofuranone, a cyanide-insensitive alternative-oxidase inhibitor with proven efficacy against rodent infections of African trypanosomiasis and cryptosporidiosis [40], as well as laboratory-demonstrated potential against microsporidia [41]. A. egyptiacum also produces the related compounds ascochlorin and ascofuranol [42]. Other industrial explorations have shown that the species can affect some potentially useful transformations of pharmaceutically produced steroids [43]. A recent study has stated that an A. egyptiacum (identified as A. sclerotigenum) isolate obtained from Flabellia petiolata marine alga produces amyloidogenic hydrophobin proteins that may be of interest in the manufacture of ‘green’ bio-coatings [44]. However, a search in GenBank for sequences from this study yielded only accession KR014351 for strain MUT-4872; this sequence matches a known undescribed species related to Acremonium hennebertii. It may have BLASTed most closely to A. egyptiacum sequences.
Correctly identified marine isolates with ITS sequences 99% similar to those of CBS 114785, the ex-type of A. egyptiacum, were cited from seawater near the Northern Antarctic Peninsula by Gonçalves et al. [45]. Examination of the sequences showed they were of barcode type 3. “Acremonium alternatum” isolates listed as having 99% similarity to GenBank record U57674.1, the ITS sequence of CBS 223.70, were obtained from marine sediments associated with coal deposits [46]. The exact sequences obtained were not deposited. A. egyptiacum barcode type 1 was found repeatedly in petroleum-contaminated soils of the Khuzestan Province, southern Iran [47]. Note that GenBank accession KY039283, the first of four sequences listed by these authors as A. sclerotigenum, is actually diagnostic of an Emericellopsis species, with a characteristic ACAAAACTTT motif at the beginning of the 5.8S rDNA.
Certain isolations, such as that of CBS 149.55 from a rust fungus lesion, raise the possibility that A. egyptiacum may be a facultative mycoparasite. There is little evidence for this as yet, but because closely related species, especially Ovicillium oosporum Zare and W. Gams, are well-established fungicolous fungi, evidence of this role in A. egyptiacum should be watched for. Plant endophytism, for example, may in some cases include or be based upon parasitism of other fungal endophytes.
It is likely that A. egyptiacum will be found to be one of the major Acremonium-like organisms associated with food spoilage, if not the most important. The species’ association with multiple plant and animal sources, and its ability to grow at relatively high temperatures, in combination with its general morphological compatibility with Acremonium sp. contaminants described at the genus level in food-related textbooks, makes this seem likely. There may be medical implications, because the intestine can be a portal of entry for infection in severely immunocompromised patients. A disseminated infection attributed to an untraceable “Acremonium strictum” isolate by Schell and Perfect [48] appeared to start in the gut, thus probably originating from food. This isolate was probably either A. egyptiacum or A. kiliense but was most likely the former, because the latter could generally be distinguished easily in pre-molecular identification by the formation of chlamydospores, and also was well known to produce distinctive melanoid pigment on Sabouraud agar. A. egyptiacum has elevated heat tolerance: it is able to survive 90 °C heat treatment for 50 min [49]. Its production of sclerotia probably significantly increases its ability to survive under adverse conditions [38]. This sort of heat tolerance may facilitate the contamination of heat-disinfected foods.
Recently, one of us (R.C.S.) curated the alignment of 170 A. egyptiacum sequences assembled in the Plutof Biodiversity Platform (https://plutof.ut.ee/) [50]. Many of these sequences were deposited under incorrect names or as unknown sequences obtained in molecular community studies. Study of the origins of these sequences, and tracking of any published material involved, could shed considerable further light on the ecology of this fungal group. A preliminary survey of the first few items listed yields variously identified barcode type 1 isolates in a wide range of substrata. There are colonisers of Peniophora basidiomycetes in Texan attine ant nests (GenBank HQ607928; sequence misidentified as Peniophora sp.), as well as endophytes of Bletilla ochracea (Chinese butterfly orchid) (HM751796) and Atractylodes lancea (Asteraceae) (KC172079) in China. Other isolates are from Taxus globosa bark in Mexico (JF773645), and from human nails in China (KT878342, KT878343). Barcode 3 isolates appear as colonisers of human nail (KC254088, MITS56|KP132613) and blood (KC254089|MITS52) in Greece. Others appear as endophytes of Gossypium in China (KF999020) and as colonisers of marine macroalgae (HQ914912) and deep-sea materials (KM274118). Already, combined with the results of Gonçalves et al. [45], mentioned above, a possible trend for barcode type 3 to be isolated from marine materials can be perceived.
4.3. Broader Taxonomic Situation of the Genus Acremonium
The compilation of information on the biodiversity within A. egyptiacum shows that this organism’s range of ecological, clinical, and economic roles epitomises most of the generalities assigned to the genus Acremonium in general textbooks, such as the Introduction to Food- and Airborne Fungi [51]. Thus, prior concepts of Acremonium as a genus are preserved in this phylogenetic nomenclature even though the great majority of species historically given this genus name will be excised. There may be some additional species related to A. sordidulum and A. brachypenium that will remain within Acremonium, but the close phylogenetic proximity of a very different genus, Ovicillium, effectively barricades extension of the name beyond the point where Acremonium and Ovicillium diverge. The next proximal major clade, the Emericellopsis-Stanjemonium-Stilbella one, now incorporates a venerable (1900) and famous genus name, Stilbella, and potentially unites this generic name with several heretofore excluded organisms forming synnemata, such as Emericellopsis salmosynnemata and E. synnematicola.
Most of the groups currently containing Acremonium names are in clades that need revision to clarify their unity and extension. There are, however, two highly distinct and isolated genus-level clades, as shown by Summerbell et al. [3], that could be rescued immediately from anachronistic nomenclature. The highly distinct nature of these groups extends to the possession of variant forms of the usual AAACTTT motif that begins the 5.8 ribosomal DNA segment throughout the filamentous Ascomycota in fungi ranging from Acremonium egyptiacum to Fusarium, Chaetomium, Aspergillus, and Ochroconis. The Acremonium radiatum clade has CAACTTT, while the Acremonium curvulum clade has AAACCTT. These groups are sufficiently distinct that their ITS sequences cannot be meaningfully aligned with those of other groups. Their current taxonomic situation has been determined via the 18S ribosomal and 28S LSU sequences published previously [3], after updating showed no recently deposited sequences have altered their positions of isolation. Both groups require revision to deal with undescribed species in their midst, but we believe that a good beginning would be to focus interest on these orphaned Acremonium clades by giving them working generic names.
A. Parasarocladium Summerbell, Scott, Guarro, and Crous, gen. nov. (MYCOBANK 826815).
Etymology: name derived from ancient Greek παρά (beside, juxtaposed to) and the genus name Sarocladium.
Type species: Parasarocladium radiatum (Sukap. and Thirum.) Summerbell, Scott, Guarro, and Crous.
Ascomatal morph not observed. Mycelium consisting of hyaline, septate, branched hyphae. Colonies smooth to thinly floccose, with salmonaceous, orange, yellow, or greenish-black reverse colouration. Conidiophores arising laterally from somatic hyphae, erect, cylindrical to subcylindrical, unbranched or less commonly branched, aseptate or septate, smooth, hyaline. Conidiogenous cells phialidic, arising laterally from hyphae or in terminal pairs, or verticils of three, or small monopodially branched tufts of up to four from conidiophores, monophialidic, hyaline, smooth, mostly aseptate but up to two-septate, elongate-ampulliform to subcylindrical, with inconspicuous collarettes. Conidia aseptate, smooth-walled or with chromophilic roughening, ellipsoidal, ovate, bacilliform or fusiform, sometimes slightly curved, forming slimy heads on the phialides. Chlamydospores absent but hyphal swellings may be present. Characteristic CAACTTT motif at 5′ end of 5.8S rDNA.
1. Parasarocladium radiatum (Sukap. and Thirum.) Summerbell, Scott, Guarro, and Crous, comb. et stat. nov., MYCOBANK 826816.
Basionym: Cephalosporium acremonium Corda var. radiatum Sukapure and Thirum. in Sydowia 19: 172 (1966).
Acremonium radiatum (Sukap. and Thirum.) W. Gams, Cephalosporium-artige Schimmelpilze: 125 (1971).
Type: CBS 142.62, ITS accession MH424699.
2. Parasarocladium breve (W. Gams) Summerbell, Scott, Guarro, and Crous, comb. et stat. nov., MYCOBANK 826817.
Basionym: Cephalosporium roseum var. breve Sukap. and Thirum. in Sydowia 19: 174, 1966.
≡ Acremonium breve W. Gams, Cephalosporium-artige Schimmelpilze: 60 (1971).
Type: CBS 150.62, ITS accession MH424706.
3. Parasarocladium gamsii (Tichelaar) Summerbell, Scott, Guarro, and Crous, comb. nov. MYCOBANK MB 826818.
Basionym: Acremonium gamsii Tichelaar, Acta Botanica Neerlandica 21: 197 (1972).
Type: CBS 726.71, ITS accession MH424707.
Known members of this genus have been almost exclusively isolated from soil or from possible conidial dispersal media such as air. B. Kiflimonium Summerbell, Scott, Guarro, and Crous, gen. nov., MYCOBANK 826819.
Etymology: Named for the resemblance of the curved shapes of the conidia to crescent-shaped pastries known in many areas by the originally Hungarian name kifli (singular noun form), which is adopted as English in some extended dictionaries such as Wiktionary.org; also known by the cognate Serbian, Bosnian, Macedonian, and Bulgarian name kifla and the Austrian kipfel. (The basic Latin and Greek roots for ‘curved,’ ‘crescent-shaped,’and so forth, are overused in other fungal names, and members of this group of simply structured organisms share no distinctive visible feature other than their curved conidia). -Monium, meaning ‘little singularity,’ derives from Acremonium.
Type species: Kiflimonium curvulum (W. Gams) Summerbell, Scott, Guarro, and Crous.
Ascomatal morph not observed. Mycelium consisting of hyaline, septate, branched hyphae. Colonies pale to yellow-orange, sometimes producing brown reverse pigments. Conidiophores mostly short side branches arising laterally from somatic hyphae, subcylindrical, unbranched, or branched and bearing mostly 1–4 conidiogenous cells, smooth, hyaline. Conidiogenous cells phialidic, arising laterally and singly from hyphae or on conidiophores in subverticillate clusters of mostly 2–4, mostly monophialidic, hyaline, smooth, generally aseptate, elongate-subcylindrical to aciculate, gently tapered, with inconspicuous collarettes; short, adelophialidic side-branches may be present. Conidia 0–1 septate, smooth-walled, lunate to falcate with rounded or acute ends, or in some cases ellipsoidal; in all cases, forming slimy heads on the phialides. Chlamydospores absent. Characteristic AAACCTT motif at 5′ end of 5.8S rDNA.
1. Kiflimonium curvulum (W. Gams) Summerbell, Scott, Guarro, and Crous, comb. nov. MYCOBANK MB 826820
Basionym: Acremonium curvulum W. Gams, Cephalosporium-artige Schimmelpilze: 57, 1971.
No Kiflimonium isolate studied so far has been found to grow at 37 °C. Some of the features in the generic description are only known from as yet undescribed organisms stored at CBS.
Members of this genus are mostly isolated from soil; CBS 214.70, distinct by possessing many septate conidia [5], was isolated from rust-infected Lolium ryegrass. Isolates accepted as K. curvulum ss. str. include (with ITS sequence accessions as substantiation) CBS 430.66 (ex-type; MH424698), CBS 229.75 (MH424700), CBS 101442 (MH424701), CBS 333.92 (MH424702), CBS 898.85 (MH424703), CBS 384.70A (MH424704), and CBS 761.69 (MH424705).
There are at least 10 other major clades of Acremonium species that will require new generic names or will need to be recombined into genera originally erected for ascomatal species or for ‘charismatic apomorphs’ (i.e., groups drawing attention with distinctive, evolutionarily derived conidiophore types [3]—cf. Ovicillium, Stilbella, Trichothecium, Sarocladium). Species that will be affected by name changes include some very recently described taxa, such as Acremonium asperulatum and A. variecolor [52], which may belong to Bulbithecium or an as yet unnamed neighbour-genus, and A. pilosum, A. parvum, and A. citrinum, which are situated in the phylogenetically isolated Acremonium fusidioides clade [53]. We would encourage authors not to hesitate to give simply structured, phylogenetically distinct organisms generic names; hesitation to await discovery of distinctive phenetic features would be an error in scientific strategy. The ecological and biochemical significance of the phylogenetically unified, morphologically flexible concept of Acremonium egyptiacum illustrates an important reality: the impact of Acremonium-like taxa can only be obscured if biosystematists place too great a focus on morphological delimitation.
Author Contributions
Conceptualisation, R.C.S., J.G. (Josep Guarro), J.F.C.-L., and C.G.; Methodology, R.C.S., J.G. (Josep Guarro), J.F.C.-L., and C.G.; Software, C.G., J.F.C.-L., R.C.S., and J.G. (Josepa Gené); Validation, R.C.S., J.G. (Josep Guarro), J.F.C.-L., and C.G.; Formal Analysis, C.G.; Investigation R.C.S., C.G., J.G. (Josep Guarro), A.E., A.K.G., J.G. (Josepa Gené), J.F.C.-L., A.v.I., M.S., and J.A.S.; Resources, P.W.C., J.G. (Josep Guarro), A.K.G., C.G., and A.E.; Data Curation, R.C.S., C.G., and J.F.C.-L.; Original Draft Preparation, R.C.S., C.G., J.F.C.-L., and J.A.S.; Review and Editing of Manuscript, R.C.S., C.G., J.G. (Josep Guarro), J.F.C.-L., J.A.S., and P.W.C.; Visualisation, R.C.S., C.G., and J.A.S.; Supervision, R.C.S., J.G. (Josep Guarro), J.G. (Josepa Gené), and J.F.C.-L.; Project Administration, R.C.S. J.A.S., J.G. (Josep Guarro), J.F.C.-L., J.G. (Josepa Gené), A.K.G., A.E., and C.G.; Funding Acquisition, R.C.S., J.G. (Josep Guarro), C.G., J.A.S., A.K.G., and P.W.C.
Funding
This project of 18-years duration was funded by many pockets. The Dutch component was mostly from the core budget of the Westerdijk Institute. The Spanish component was partly from the Spanish Ministerio de Educación y Ciencia, notably grants CGL 2009-08698/BOS and CGL 2008-04226/BOS. In Canada, A.K. Gupta provided funding from Mediprobe Laboratories, and J.A. Scott provided funding from Sporometrics. In Australia, the Australian National Herbarium supported C. Gueidan.
Acknowledgments
The late Walter Gams provided immensely valuable consultation. The Collection staff of the Westerdijk Institute also played an invaluable role, as did Saleh Albreish and his staff at OMH. The late Deanna Sutton and her colleagues at the University of Texas Health Science Center sent many isolates from medical sources that were re-used in this project. Administratively, the late Dirk van der Mei was the key sponsor of this study’s beginning, and he, along with the other departed crew of this deceptively colossal enterprise, is remembered with great affection.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Paterson, R.R.M.; Lima, N. Filamentous fungal human pathogens from food emphasising Aspergillus, Fusarium and Mucor. Microorganisms 2017, 5, E44. [Google Scholar] [CrossRef] [PubMed]
- Glenn, A.E.; Bacon, C.W.; Price, R.; Hanlin, R.T. Molecular phylogeny of Acremonium and its taxonomic implications. Mycologia 1996, 88, 369–383. [Google Scholar] [CrossRef]
- Summerbell, R.C.; Gueidan, C.; Schroers, H.J.; de Hoog, G.S.; Starink, M.; Arocha Rosete, Y.; Guarro, J.; Scott, J.A. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 2011, 68, 139–162. [Google Scholar] [CrossRef]
- Perdomo, H.; Sutton, D.A.; García, D.; Fothergill, A.W.; Cano, J.; Gené, J.; Summerbell, R.C.; Rinaldi, M.G.; Guarro, J. Spectrum of clinically relevant Acremonium species in the United States. J. Clin. Microbiol. 2011, 49, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Gams, W. Cephalosporium-artige Schimmelpilze (Hyphomycetes); G. Fischer: Stuttgart, Germany, 1971. [Google Scholar]
- Summerbell, R.C.; Scott, J.A. Conidiogenesis: Its evolutionary aspects in the context of a philosophy of opportunity (lectics). In Biology of Microfungi; Li, D.W., Ed.; Springer Science: Berlin/Heidelberg, Germany, 2016; pp. 169–195. ISBN 978-3-319-29137-6. [Google Scholar]
- Summerbell, R.C.; Scott, J.A. Acremonium. In Molecular Biology of Food and Water Borne Mycotoxigenic and Mycotic Fungi; Paterson, R.R.M., Lima, N., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 115–128. ISBN 9781466559868. [Google Scholar]
- Gams, W. Typisierung der Gattung Acremonium. Nova Hedwigia 1968, 16, 141–145. [Google Scholar]
- Gams, W.; Hoekstra, E.S.; Aptroot, A. CBS Course of Mycology, 4th ed.; Centraalbureau voor Schimmelcultures: Baarn, The Netherlands; Delft, The Netherlands, 1998. [Google Scholar]
- Raper, J.R.; Raper, C.A. Genetic analysis of the life cycle of Agaricus bisporus. Mycologia 1972, 64, 1088–1117. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, a Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Hoffmann, K.; Discher, S.; Voigt, K. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol. Res. 2007, 111, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. 2017. Available online: http://www.mesquiteproject.org (accessed on 30 May 2018).
- Stamatakis, A.; Ludwig, T.; Meier, H. RAxML-III: A fast program for maximum likelihood—Based inference of large phylogenetic trees. Bioinformatics 2005, 21, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008, 75, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees; Gateway Computing Environments Workshop (GCE): New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4.0 b10Sinauer: Sunderland, MA, USA, 2002. [Google Scholar]
- Isaia, M.C.; Gams, W.; Sola, S.C. Isolation of Acremonium sclerotigenum from an ostrich’s egg. Avian Pathol. 2000, 29, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Bibashi, E.; Kokolina, E.; Sigler, L.; Sofianou, D.; Tsakiris, D.; Visvardis, G.; Papadimitriou, M.; Memmos, D. Three cases of uncommon fungal peritonitis in patients undergoing peritoneal dialysis. Perit. Dial. Int. 2002, 22, 523–525. [Google Scholar] [PubMed]
- Novicki, T.J.; LaFe, K.; Bui, L.; Bui, U.; Geise, R.; Marr, K.; Cookson, B.T. Genetic diversity among clinical isolates of Acremonium strictum determined during an investigation of a fatal mycosis. J. Clin. Microbiol. 2003, 41, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Cooper, E.A.; McDonald, P.; Summerbell, R.C. Inoculum counting (Walshe/English criteria) in the clinical diagnosis of onychomycosis caused by non-dermatophytic filamentous fungi. J. Clin. Microbiol. 2000, 39, 2115–2121. [Google Scholar] [CrossRef] [PubMed]
- Vuillemin, P. Les conidiosporées. Bull. Séances Soc. Sci. Nancy 1910, 11, 129–172. [Google Scholar]
- Pollacci, G.; Nannizzi, A. I Miceti Patogeni DELL’UOMO e Degli Animali, Descritti, Delineati e Preparati per l’osservazione al Microscopio, con Notizie Sopra i Rimedi per Combatterli; L. Cappelli: Bologna, Italy, 1922–1930. [Google Scholar]
- Summerbell, R.C. Aspergillus, Fusarium, Sporothrix, Piedraia and their relatives. In Pathogenic Fungi in Humans and Animals; Howard, D.H., Ed.; Marcel Dekker Press: New York, NY, USA, 2003; pp. 237–498. ISBN 9780824706838. [Google Scholar]
- Minter, D.W.; Sutton, B.C.; Brady, B.L. What are phialides anyway? Trans. Br. Mycol. Soc. 1983, 81, 109–120. [Google Scholar] [CrossRef]
- Perdomo, H.; García, D.; Gené, J.; Cano, J.; Sutton, D.A.; Summerbell, R.C.; Guarro, J. Phialemoniopsis, a new genus of Sordariomycetes, and new species of Phialemonium and Lecythophora. Mycologia 2013, 105, 398–421. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, H.; Sutton, D.A.; García, D.; Fothergill, A.W.; Gené, J.; Cano, J.; Summerbell, R.C.; Rinaldi, M.G.; Guarro, J. Molecular and phenotypic characterization of Phialemonium and Lecythophora isolates from clinical samples. J. Clin. Microbiol. 2011, 49, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Guitard, J.; Degulys, A.; Buot, G.; Aline-Fardin, A.; Dannaoui, E.; Rio, B.; Marie, J.P.; Lapusan, S.; Hennequin, C. Acremonium sclerotigenum-Acremonium egyptiacum: A multi-resistant fungal pathogen complicating the course of aplastic anaemia. Clin. Microbiol. Infect. 2014, 20, O30–O32. [Google Scholar] [CrossRef] [PubMed]
- Summerbell, R.C.; Cooper, E.; Bunn, U.; Jamieson, F.; Gupta, A.K. Onychomycosis: A critical study of techniques and criteria for confirming the etiologic significance of nondermatophytes. Med. Mycol. 2005, 43, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Madrid, H.; Ruíz-Cendoya, M.; Cano, J.; Stchigel, A.; Orofino, R.; Guarro, J. Genotyping and in vitro antifungal susceptibility of Neoscytalidium dimidiatum isolates from different origins. Int. J. Antimicrob. Agents 2009, 34, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Guarro, J.; Del Palacio, A.; Gené, J.; Cano, J.; González, C.G. A case of colonization of a prosthetic mitral valve by Acremonium strictum. Rev. Iberoam. Micol. 2009, 26, 146–148. [Google Scholar] [CrossRef]
- Fernández-Silva, F.; Capilla, J.; Mayayo, E.; Sutton, D.; Guarro, J. Experimental murine acremoniosis: An emerging opportunistic human infection. Med. Mycol. 2014, 52, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Collado, J.; Platas, G.; González, I.; Peláez, F. Geographical and seasonal influences on the distribution of fungal endophytes in Quercus ilex. New Phytol. 1999, 144, 525–532. [Google Scholar] [CrossRef]
- Paul, N.C.; Lee, H.B.; Lee, J.H.; Shin, K.S.; Ryu, T.H.; Kwon, H.R.; Kim, Y.K.; Youn, Y.N.; Yu, S.H. Endophytic fungi from Lycium chinense Mill and characterization of two new Korean records of Colletotrichum. Int. J. Mol. Sci. 2014, 15, 15272–15286. [Google Scholar] [CrossRef] [PubMed]
- Aminaee, M.M.; Zare, R.; Zohdi, H. An investigation on Lecanicillium muscarium as a biocontrol agent of stem borer pests of Rosa damascena in Kerman Province of Iran. Arch. Phytopathol. Plant Prot. 2010, 43, 678–688. [Google Scholar] [CrossRef]
- Asgari, B.; Zare, R. Hyphomycetous fungal community of barley phylloplane in East Azerbaijan province with emphasis on new taxa for Iranian fungal flora. Rostaniha 2004, 5, 67–70. [Google Scholar]
- Li, B.H.; Wang, C.C.; Dong, X.L.; Zhang, Z.F.; Wang, C.X. Acremonium Brown Spot, a New Disease Caused by Acremonium sclerotigenum on Bagged Apple Fruit in China. Plant Dis. 2014, 98, 1012. [Google Scholar] [CrossRef]
- Mirtalebi, M.; Banihashemi, Z.; Sabahi, F.; Mafakheri, H. Dieback of rose caused by Acremonium sclerotigenum as a new causal agent of rose dieback in Iran. Span. J. Agric. Res. 2016, 14, 2171–9292. [Google Scholar] [CrossRef]
- Zhang, B.; Li, X.; Wang, F.; Li, M.; Zhang, J.; Gu, L.; Zhang, L.; Tu, W.; Zhang, Z. Assaying the potential autotoxins and microbial community associated with Rehmannia glutinosa replant problems based on its ‘autotoxic circle’. Plant Soil 2016, 407, 307–322. [Google Scholar] [CrossRef]
- Hijikawa, Y.; Matsuzaki, M.; Suzuki, S.; Inaoka, D.K.; Tatsumi, R.; Kido, Y.; Kita, K. Re-identification of the ascofuranone-producing fungus Ascochyta viciae as Acremonium sclerotigenum. J. Antibiot. (Tokyo) 2017, 70, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.A.; Elliot, C.; Burri, L.; Kido, Y.; Kita, K.; Moore, A.L.; Keeling, P.J. A broad distribution of the alternative oxidase in microsporidian parasites. PLoS Pathog. 2010, 6, e1000761. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Hosokawa, T.; Sawada, M.; Ando, K. Isolation and structure of ascofuranone and ascofuranol, antibiotics with hypolipidemic activity. J. Antibiot. (Tokyo) 1973, 26, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Sardinas, J.L.; Pisano, M.A. Steroid transformations by species of Cephalosporium and other fungi. Appl. Microbiol. 1967, 15, 277–284. [Google Scholar] [PubMed]
- Cicatiello, P.; Dardano, P.; Pirozzi, M.; Gravagnuolo, A.M.; De Stefano, L.; Giardina, P. Self-assembly of two hydrophobins from marine fungi affected by interaction with surfaces. Biotechnol. Bioeng. 2017, 114, 2173–2186. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.N.; Vitoreli, G.A.; de Menezes, G.C.A.; Mendes, C.R.B.; Secchi, E.R.; Rosa, C.A.; Rosa, L.H. Taxonomy, phylogeny and ecology of cultivable fungi present in seawater gradients across the Northern Antarctica Peninsula. Extremophiles 2017, 21, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Huang, X.; Xie, T.N.; Duan, N.; Xue, Y.R.; Zhao, T.X.; Lever, M.A.; Hinrichs, K.U.; Inagaki, F. Exploration of cultivable fungal communities in deep coal-bearing sediments from ∼1.3 to 2.5 km below the ocean floor. Environ. Microbiol. 2017, 19, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, E.; Arzanlou, M.; Babai-Ahari, A. Diversity of culturable fungi inhabiting petroleum-contaminated soils in Southern Iran. Antonie Van Leeuwenhoek 2017, 110, 903–923. [Google Scholar] [CrossRef] [PubMed]
- Schell, W.A.; Perfect, J.R. Fatal, disseminated Acremonium strictum infection in a neutropenic host. J. Clin. Microbiol. 1996, 34, 1333–1336. [Google Scholar] [PubMed]
- Jesenská, Z.; Piecková, E.; Bernát, D. Heat resistance of fungi from soil. Int. J. Food Microbiol. 1993, 19, 187–192. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Taylor, A.F.; Adams, R.I.; Baschien, C.; Bengtsson-Palme, J.; Cangren, P.; Coleine, C.; Daniel, H.-M.; Glassman, S.I.; Hirooka, Y.; et al. Taxonomic annotation of public fungal ITS sequences from the built environment—A report from an April 10–11, 2017 workshop (Aberdeen, UK). MycoKeys 2018, 8, 65–82. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.C. Introduction to Food-Borne fungi, 7th ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2004. [Google Scholar]
- Giraldo, A.; Gené, J.; Cano, J.; de Hoog, S.; Guarro, J. Two new species of Acremonium from Spanish soils. Mycologia 2012, 104, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, A.; Gené, J.; Cano, J.; de Hoog, S.; Decock, C.; Guarro, J. Acremonium with catenate elongate conidia: Phylogeny of Acremonium fusidioides and related species. Mycologia 2014, 106, 328–338. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).