A Comparative Study on the Faecal Bacterial Community and Potential Zoonotic Bacteria of Muskoxen (Ovibos moschatus) in Northeast Greenland, Northwest Greenland and Norway
Abstract
1. Introduction
2. Materials and Methods
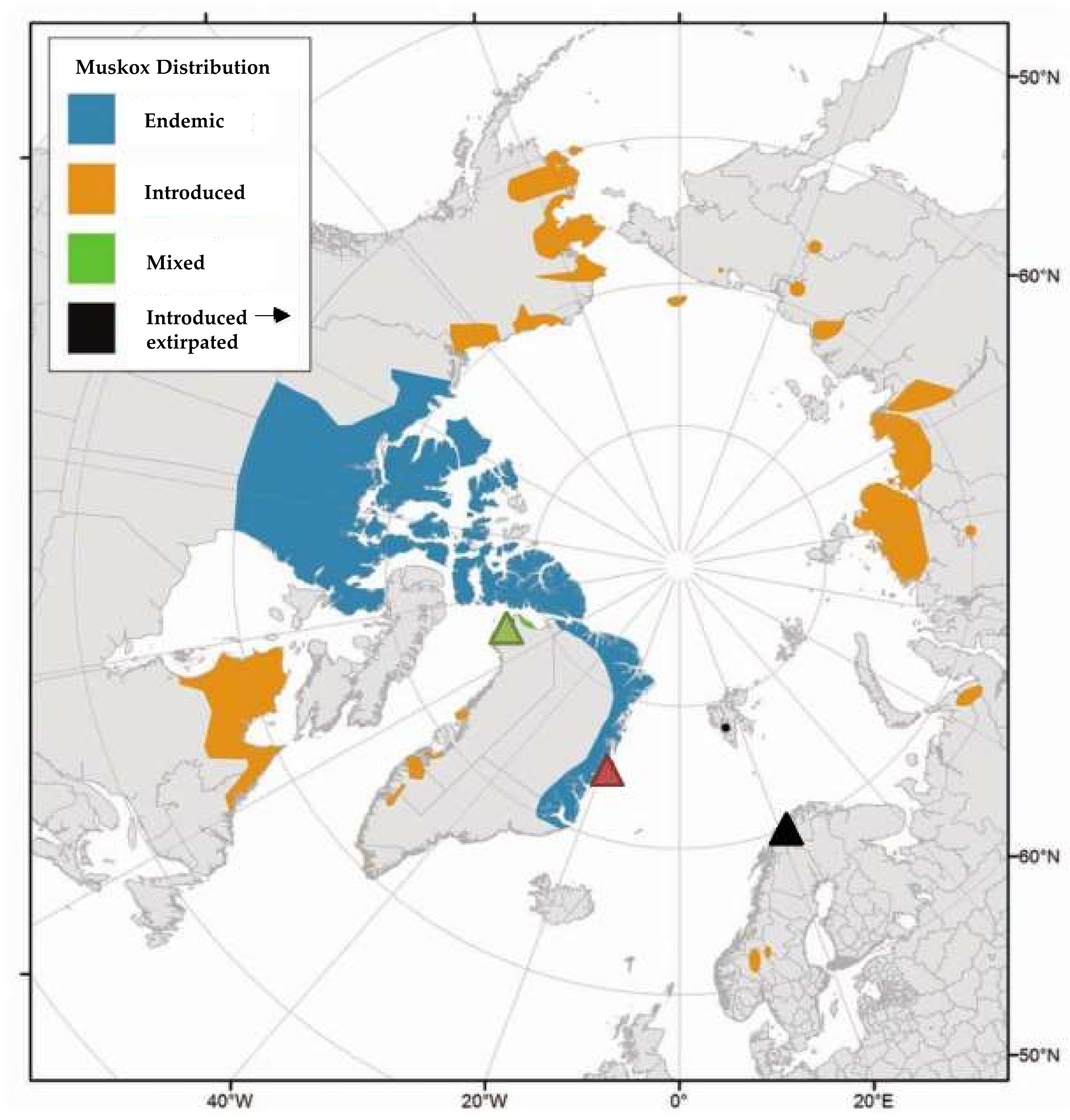
2.1. Sampling
2.2. Extraction and Library Construction
2.3. Bioinformatic Analysis
2.4. Statistical Analyses
3. Results
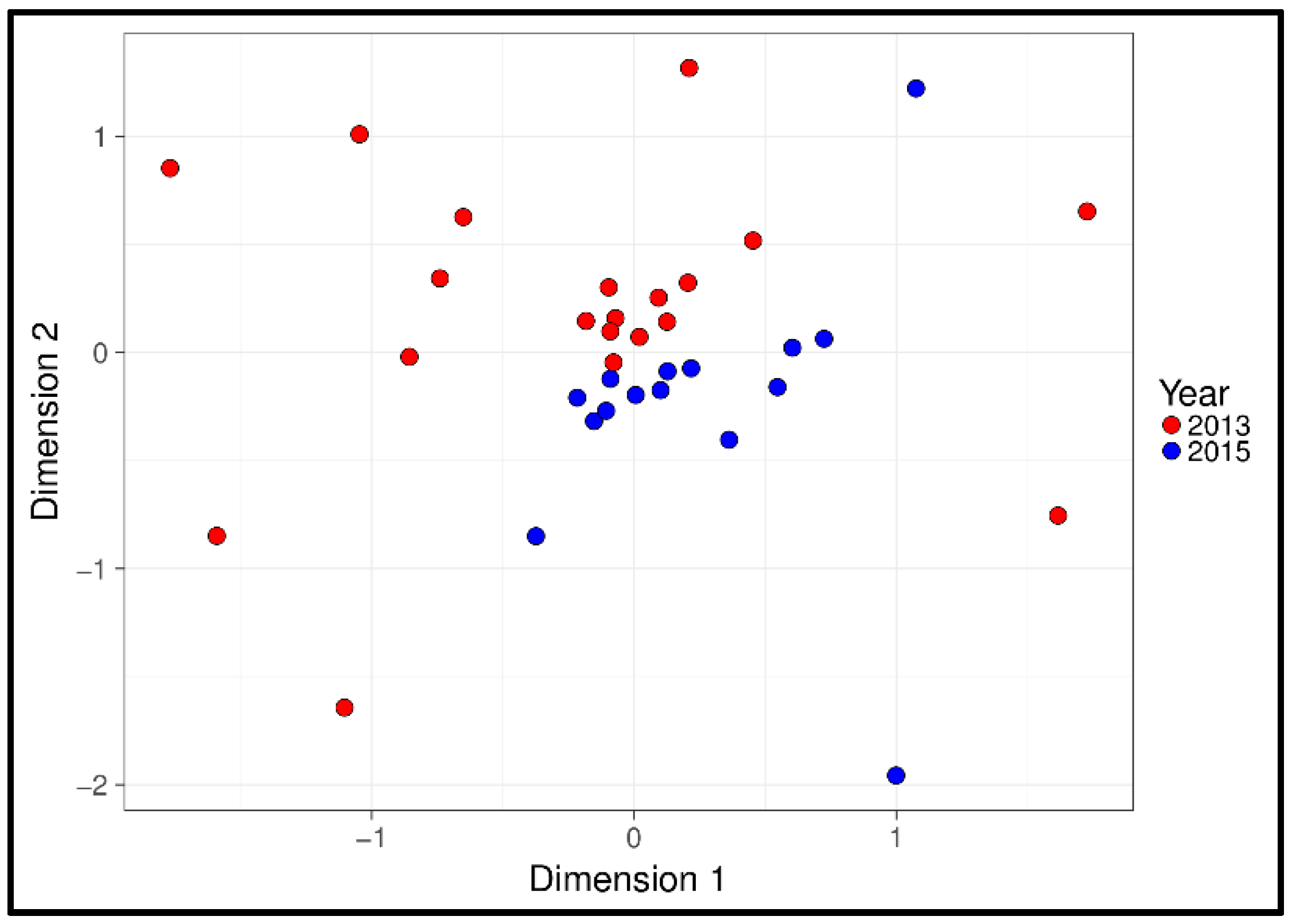
3.1. Physiological and Temporal Effects
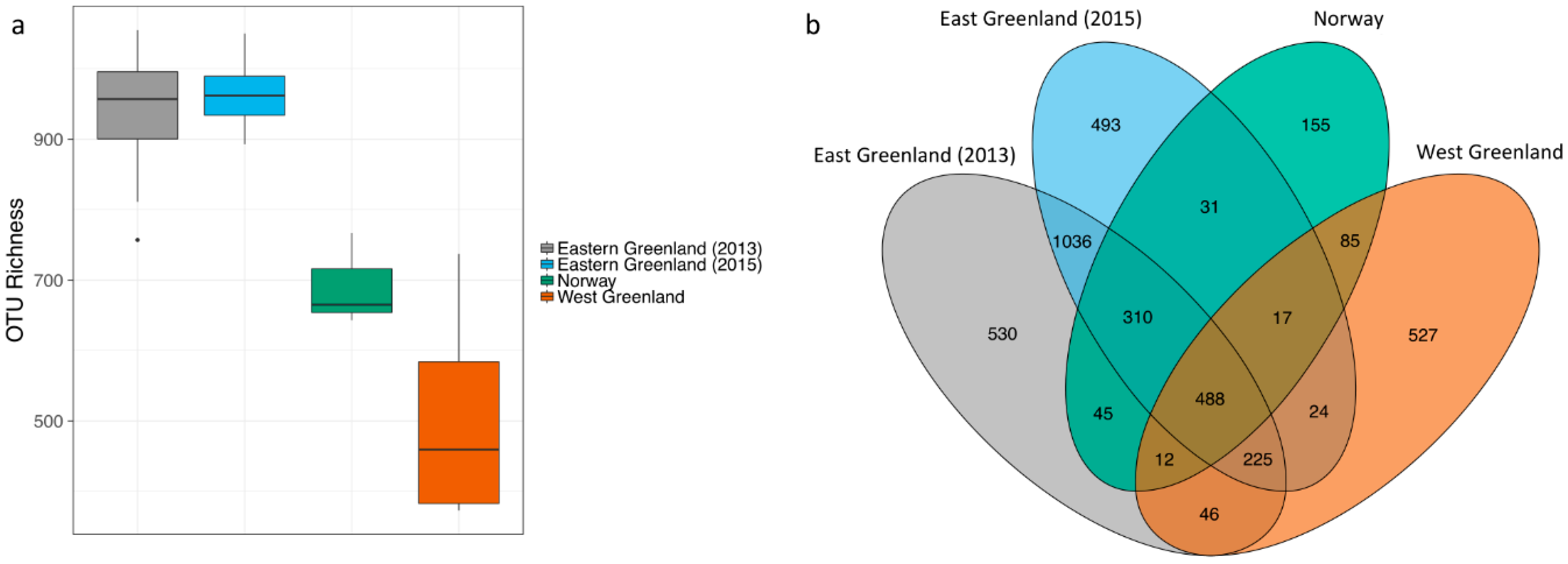
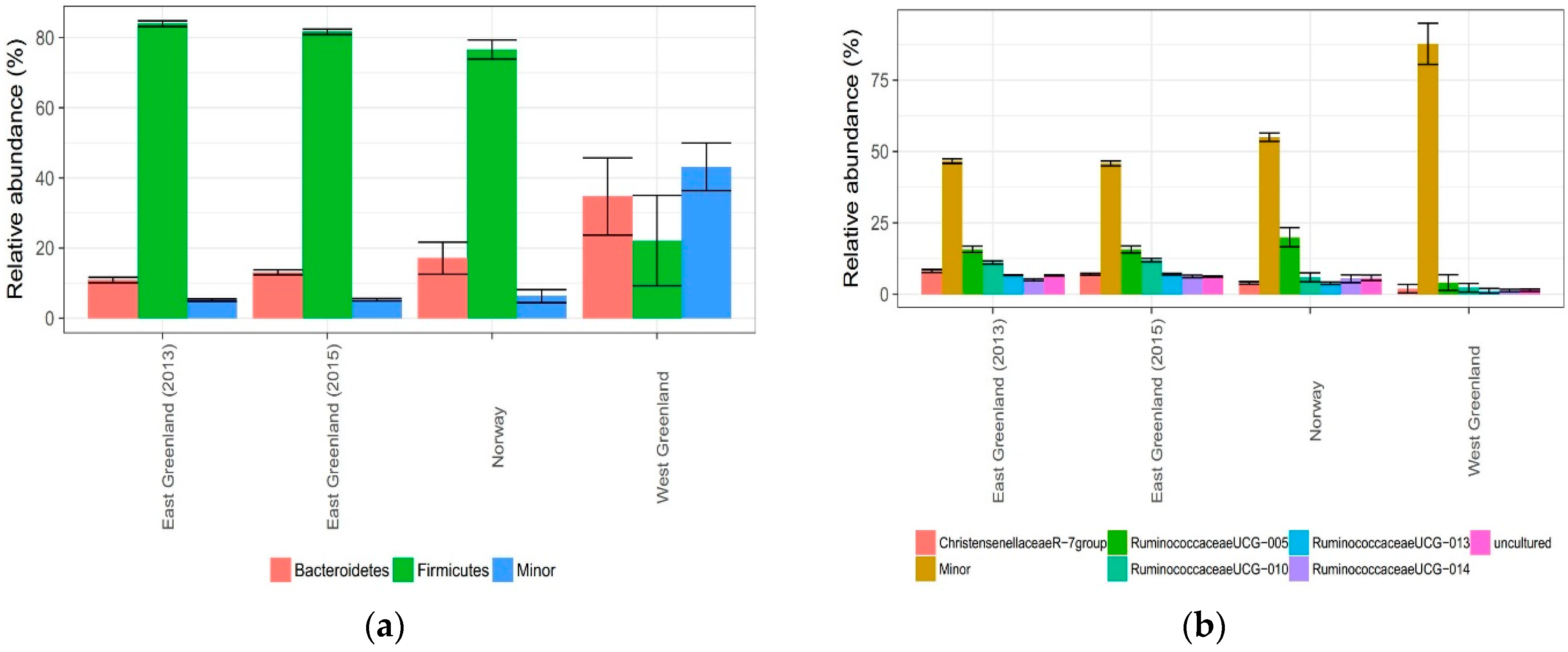
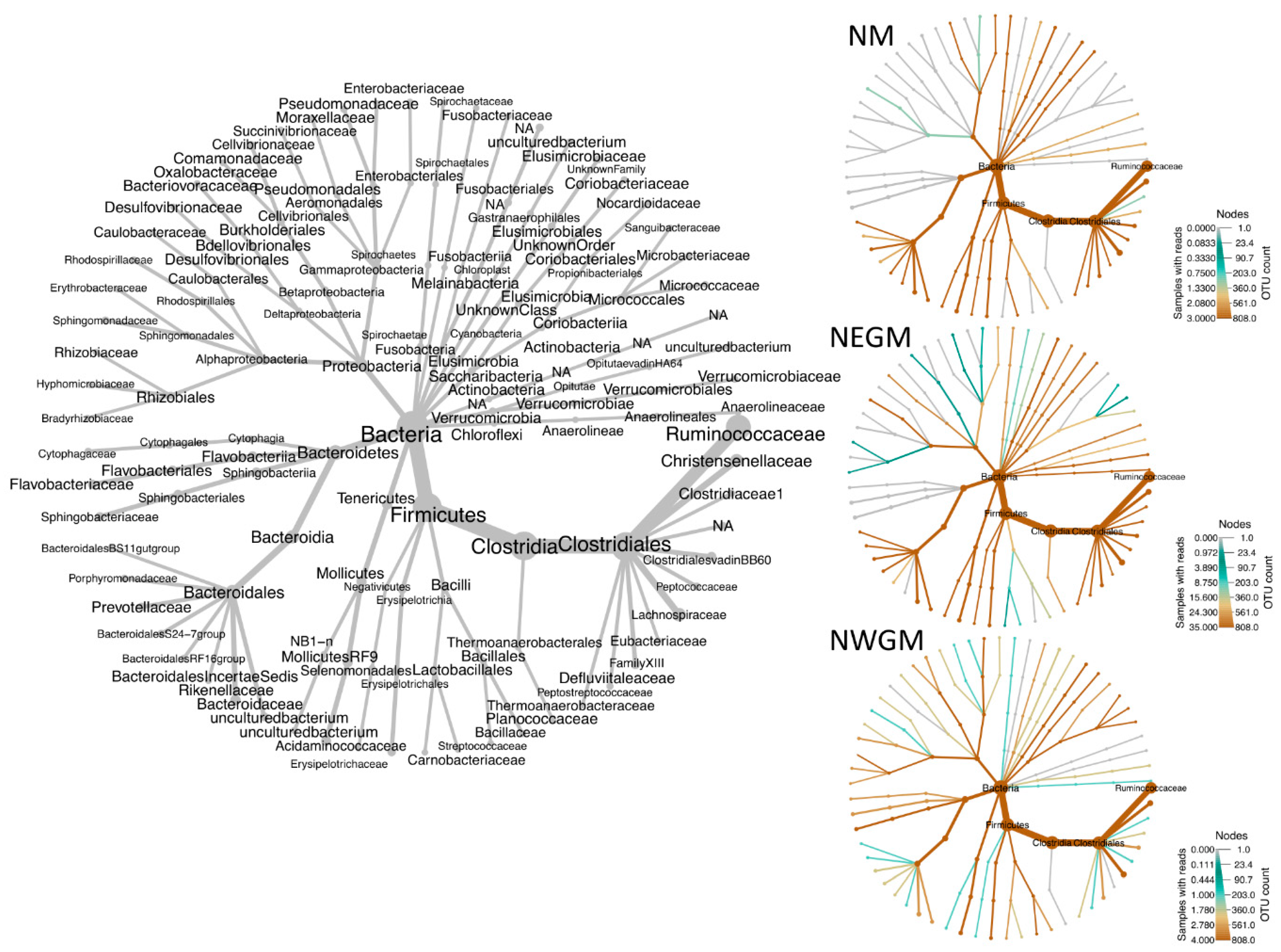
3.2. Effect of Location
3.3. Potential Zoonotic Bacteria
4. Discussion
4.1. Physiological and Temporal Effects
4.2. Effect of Location
4.3. Potential Zoonotic Bacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kutz, S.; Rowell, J.; Adamczewski, J.; Gunn, A.; Cuyler, C.; Aleuy, O.A.; Austin, M.; Berger, J.; Blake, J.; Bondo, K. Muskox Health Ecology Symposium 2016: Gathering to Share Knowledge on Umingmak in a Time of Rapid Change. Arctic 2017, 70, 225–236. [Google Scholar] [CrossRef]
- Mosbacher, J.B. Ecology of a High Arctic Key Species: Muskoxen in Northeast Greenland. Ph.D. Thesis, Aarhus University, Roskilde, Denmark, 2017. [Google Scholar]
- Klein, D.R.; Bay, C. Diet selection by vertebrate herbivores in the high arctic of Greenland. Ecography 1991, 14, 152–155. [Google Scholar] [CrossRef]
- Kristensen, D.K.; Kristensen, E.; Forchhammer, M.C.; Michelsen, A.; Schmidt, N.M. Arctic herbivore diet can be inferred from stable carbon and nitrogen isotopes in C3 plants, faeces, and wool. Can. J. Zool. 2011, 89, 892–899. [Google Scholar] [CrossRef]
- Thing, H.; Klein, D.R.; Jingfors, K.; Holt, S. Ecology of muskoxen in Jameson Land, northeast Greenland. Ecography 1987, 10, 95–103. [Google Scholar] [CrossRef]
- Mosbacher, J.B.; Kristensen, D.K.; Michelsen, A.; Stelvig, M.; Schmidt, N.M. Quantifying Muskox Plant Biomass Removal and Spatial Relocation of Nitrogen in a High Arctic Tundra Ecosystem. Arct. Antarct. Alp. Res. 2016, 48, 229–240. [Google Scholar] [CrossRef]
- Schmidt, N.M.; Mosbacher, J.B.; Vesterinen, E.J.; Roslin, T.; Michelsen, A. Limited dietary overlap amongst resident Arctic herbivores in winter: Complementary insights from complementary methods. Oecologia 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef] [PubMed]
- Carroll, E.J.; Hungate, R.E. The Magnitude of the Microbial Fermentation in the Bovine Rumen. Appl. Microbiol. 1954, 2, 205–214. [Google Scholar] [PubMed]
- Munn, A.J.; Barboza, P.S. Could a big gut be too costly for muskoxen (Ovibos moschatus) in their first winter? Zoology 2008, 111, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet Dominates Host Genotype in Shaping the Murine Gut Microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, J.; Kuehn, L.A.; Bono, J.L.; Berry, E.D.; Kalchayanand, N.; Freetly, H.C.; Benson, A.K.; Wells, J.E. Investigation of bacterial diversity in the feces of cattle fed different diets. J. Anim. Sci. 2014, 92, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Shanks, O.C.; Kelty, C.A.; Archibeque, S.; Jenkins, M.; Newton, R.J.; McLellan, S.L.; Huse, S.M.; Sogin, M.L. Community Structures of Fecal Bacteria in Cattle from Different Animal Feeding Operations. Appl. Environ. Microbiol. 2011, 77, 2992–3001. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Flores, A.; Hagen, L.H.; Ishaq, S.L.; Zamanzadeh, M.; Wright, A.-D.G.; Pope, P.B.; Sundset, M.A. Rumen and cecum microbiomes in reindeer (Rangifer tarandus tarandus) are changed in response to a lichen diet and may affect enteric methane emissions. PLoS ONE 2016, 11, e0155213. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, P.; O’Toole, N.; Barboza, P.S.; Ungerfeld, E.; Leigh, M.B.; Selinger, L.B.; Butler, G.; Tsang, A.; McAllister, T.A. Snapshot of the eukaryotic gene expression in muskoxen rumen—A metatranscriptomic approach. PLoS ONE 2011, 6, e20521. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Flores, A.; Bockwoldt, M.; Hagen, L.H.; Pope, P.B.; Sundset, M.A. First insight into the faecal microbiota of the high Arctic muskoxen (Ovibos moschatus). Microb. Genom. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Luo, H.; Meng, K.; Wang, Y.; Huang, H.; Shi, P.; Pan, X.; Yang, P.; Diao, Q.; Zhang, H.; et al. High Genetic Diversity and Different Distributions of Glycosyl Hydrolase Family 10 and 11 Xylanases in the Goat Rumen. PLoS ONE 2011, 6, e16731. [Google Scholar] [CrossRef] [PubMed]
- Davidson, R.; Simard, M.; Kutz, S.J.; Kapel, C.M.; Hamnes, I.S.; Robertson, L.J. Arctic parasitology: Why should we care? Trends Parasitol. 2011, 27, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J. Neglected Infections of Poverty among the Indigenous Peoples of the Arctic. PLoS Negl. Trop. Dis. 2010, 4, e606. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, E.J.; Castrodale, L.J.; de Rosemond, S.J.C.; Dixon, B.R.; Elmore, S.A.; Gesy, K.M.; Hoberg, E.P.; Polley, L.; Schurer, J.M.; Simard, M.; et al. Chapter Two—Tradition and Transition: Parasitic Zoonoses of People and Animals in Alaska, Northern Canada, and Greenland. In Advances in Parasitology; Rollinson, D., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 82, pp. 33–204. [Google Scholar]
- Koch, A.; Svendsen, C.B.; Christensen, J.J.; Bundgaard, H.; Vindfeld, L.; Christiansen, C.B.; Kemp, M.; Villumsen, S. Q Fever in Greenland. Emerg. Infect. Dis. 2010, 16, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Pufall, E.L.; Jones, A.Q.; McEwen, S.A.; Lyall, C.; Peregrine, A.S.; Edge, V.L. Perception of the importance of traditional country foods to the physical, mental, and spiritual health of Labrador Inuit. Arctic 2011, 64, 242–250. [Google Scholar] [CrossRef]
- Schmidt, N.M.; van Beest, F.M.; Mosbacher, J.B.; Stelvig, M.; Hansen, L.H.; Nabe-Nielsen, J.; Grøndahl, C. Ungulate movement in an extreme seasonal environment: Year-round movement patterns of high-arctic muskoxen. Wildl. Biol. 2016, 22, 253–267. [Google Scholar] [CrossRef]
- Olesen, C.R.; Thing, H. Guide to field classification by sex and age of the muskox. Can. J. Zool. 1989, 67, 1116–1119. [Google Scholar] [CrossRef]
- Thulin, C.-G.; Englund, L.; Ericsson, G.; Spong, G. The impact of founder events and introductions on genetic variation in the muskox Ovibos moschatus in Sweden. Acta Theriol. 2011, 56, 305–314. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Schnell, I.B.; Bohmann, K.; Gilbert, M.T.P. Tag jumps illuminated–reducing sequence-to-sample misidentifications in metabarcoding studies. Mol. Ecol. Resour. 2015, 15, 1289–1303. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 2010, 7, 335. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing. 2013. Available online: http://softlibre.unizar.es/manuales/aplicaciones/r/fullrefman.pdf (accessed on 1 November 2017).
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The vegan package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Wang, Y.I.; Naumann, U.; Wright, S.T.; Warton, D.I. mvabund—An R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 2012, 3, 471–474. [Google Scholar] [CrossRef]
- Wickham, H.; Wickham, M.H. The Ggplot Package. 2007. Available online: http://ftp.uni-bayreuth.de/math/statlib/R/CRAN/doc/packages/ggplot.pdf (accessed on 1 November 2017).
- Acha, P.N.; Szyfres, B. Zoonoses and Communicable Diseases Common to Man and Animals; Pan American Health Organization: Washington, DC, USA, 2001; Volume 3. [Google Scholar]
- Paul, J. A checklist of bacteria associated with infection in humans. In Oxford Textbook of Medicine: Infection, 5th ed.; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Indra, A.; Lassnig, H.; Baliko, N.; Much, P.; Fiedler, A.; Huhulescu, S.; Allerberger, F. Clostridium difficile: A new zoonotic agent? Wien. Klin. Wochenschr. 2009, 121, 91. [Google Scholar] [CrossRef] [PubMed]
- Elsden, S.R.; Hitchcock, M.W.S.; Marshall, R.A.; Phillipson, A.T. Volatile acids in the digesta of ruminants and other animals. J. Exp. Biol. 1946, 22, 191–202. [Google Scholar] [PubMed]
- Patni, N.K.; Jui, P.Y. Volatile fatty acids in stored dairy-cattle slurry. Agric. Wastes 1985, 13, 159–178. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Gillilland, M.G.; Young, V.B.; Huffnagle, G.B. Gastrointestinal microbial ecology with perspectives on health and disease. In Physiology of the Gastrointestinal Tract, 5th ed.; Elsevier: New York City, NY, USA, 2012; pp. 1119–1134. [Google Scholar]
- Jami, E.; White, B.A.; Mizrahi, I. Potential Role of the Bovine Rumen Microbiome in Modulating Milk Composition and Feed Efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, I. The Role of the Rumen Microbiota in Determining the Feed Efficiency of Dairy Cows. In Beneficial Microorganisms in Multicellular Life Forms; Springer: Berlin/Heidelberg, Germany, 2012; pp. 203–210. ISBN 978-3-642-21679-4. [Google Scholar]
- Saraswati, S.; Sitaraman, R. Aging and the human gut microbiota—From correlation to causality. Front. Microbiol. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Vrieze, A.; Holleman, F.; Zoetendal, E.G.; de Vos, W.M.; Hoekstra, J.B.L.; Nieuwdorp, M. The environment within: How gut microbiota may influence metabolism and body composition. Diabetologia 2010, 53, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Aagnes, T.H.; Sørmo, W.; Mathiesen, S.D. Ruminal microbial digestion in free-living, in captive lichen-fed, and in starved reindeer (Rangifer tarandus tarandus) in winter. Appl. Environ. Microbiol. 1995, 61, 583–591. [Google Scholar] [PubMed]
- Aivelo, T.; Laakkonen, J.; Jernvall, J. Population-and individual-level dynamics of the intestinal microbiota of a small primate. Appl. Environ. Microbiol. 2016, 82, 3537–3545. [Google Scholar] [CrossRef] [PubMed]
- Maurice, C.F.; Knowles, S.C.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015, 9, 2423. [Google Scholar] [CrossRef] [PubMed]
- Mathiesen, S.D.; Mackie, R.I.; Aschfalk, A.; Ringø, E.; Sundset, M.A.; Holzapfel, W.; Naughton, P. Microbial ecology of the gastrointestinal tract in reindeer—changes through season. Microb. Ecol. Grow. Anim. 2005, 73–100, 0444509267. [Google Scholar]
- Mosbacher, J.B.; Michelsen, A.; Stelvig, M.; Hendrichsen, D.K.; Schmidt, N.M. Show Me Your Rump Hair and I Will Tell You What You Ate—The Dietary History of Muskoxen (Ovibos moschatus) Revealed by Sequential Stable Isotope Analysis of Guard Hairs. PLoS ONE 2016, 11, e0152874. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.H.; Tamstorf, M.P.; Abermann, J.; Westergaard-Nielsen, A.; Lund, M.; Skov, K.; Sigsgaard, C.; Mylius, M.R.; Hansen, B.U.; Liston, G.E. Spatiotemporal characteristics of seasonal snow cover in Northeast Greenland from in situ observations. Arct. Antarct. Alp. Res. 2016, 48, 653–671. [Google Scholar] [CrossRef]
- Larsen, T.S.; Nilsson, N.Ö.; Blix, A.S. Seasonal changes in lipogenesis and lipolysis in isolated adipocytes from Svalbard and Norwegian reindeer. Acta Physiol. Scand. 1985, 123, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Crater, A.R.; Barboza, P.S. The rumen in winter: Cold shocks in naturally feeding muskoxen (Ovibos moschatus). J. Mammal. 2007, 88, 625–631. [Google Scholar] [CrossRef]
- Crater, A.R.; Barboza, P.S.; Forster, R.J. Regulation of rumen fermentation during seasonal fluctuations in food intake of muskoxen. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2007, 146, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Barboza, P.S.; Peltier, T.C.; Forster, R.J. Ruminal degradation increases with seasonal hyperphagia in muskoxen (Ovibos moschatus): A preliminary report. J. Anim. Feed Sci. 2004, 13, 711–714. [Google Scholar] [CrossRef]
- Tajima, K.; Nonaka, I.; Higuchi, K.; Takusari, N.; Kurihara, M.; Takenaka, A.; Mitsumori, M.; Kajikawa, H.; Aminov, R.I. Influence of high temperature and humidity on rumen bacterial diversity in Holstein heifers. Anaerobe 2007, 13, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.; Pope, P.B.; Denman, S.E.; McSweeney, C.S. Plant biomass degradation by gut microbiomes: More of the same or something new? Curr. Opin. Biotechnol. 2009, 20, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Steelman, S.M.; Chowdhary, B.P.; Dowd, S.; Suchodolski, J.; Janečka, J.E. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 2012, 8, 231. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Hehemann, J.-H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef] [PubMed]
- Naas, A.E.; Mackenzie, A.K.; Mravec, J.; Schückel, J.; Willats, W.G.T.; Eijsink, V.G.H.; Pope, P.B. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? MBio 2014, 5, e01401–e01414. [Google Scholar] [CrossRef] [PubMed]
- Salyers, A.A.; Vercellotti, J.R.; West, S.E.; Wilkins, T.D. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 1977, 33, 319–322. [Google Scholar] [PubMed]
- Rincón, M.T.; McCrae, S.I.; Kirby, J.; Scott, K.P.; Flint, H.J. EndB, a multidomain family 44 cellulase from Ruminococcus flavefaciens 17, binds to cellulose via a novel cellulose-binding module and to another R. flavefaciens protein via a dockerin domain. Appl. Environ. Microbiol. 2001, 67, 4426–4431. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, S.L.; Wright, A.-D. High-Throughput DNA Sequencing of the Ruminal Bacteria from Moose (Alces alces) in Vermont, Alaska, and Norway. Microb. Ecol. 2014, 68, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Vogtmann, E.; Chen, J.; Amir, A.; Shi, J.; Abnet, C.C.; Nelson, H.; Knight, R.; Chia, N.; Sinha, R. Comparison of collection methods for fecal samples in microbiome studies. Am. J. Epidemiol. 2017, 185, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Bernardet, J.; Bowman, J.P. Flavobacterium. Bergey's Man. Syst. Archaea Bact. 2015. [Google Scholar] [CrossRef]
- Kämpfer, P.; Order, I. Sphingobacteriales ord. nov. Bergey's Man. Syst. Archaea Bact. 2011, 4, 330. [Google Scholar]
- Alderisio, K.A.; DeLuca, N. Seasonal Enumeration of Fecal Coliform Bacteria from the Feces of Ring-Billed Gulls (Larus delawarensis) and Canada Geese (Branta canadensis). Appl. Environ. Microbiol. 1999, 65, 5628–5630. [Google Scholar] [PubMed]
- French, N.P.; Midwinter, A.; Holland, B.; Collins-Emerson, J.; Pattison, R.; Colles, F.; Carter, P. Molecular Epidemiology of Campylobacter jejuni Isolates from Wild-Bird Fecal Material in Children’s Playgrounds. Appl. Environ. Microbiol. 2009, 75, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Tian, P.; Luo, Y.; Tian, J.; Hua, C.; Geng, Y.; Cong, R.; Ni, Y.; Zhao, R. Microbiome-Metabolome Responses to a High-Grain Diet Associated with the Hind-Gut Health of Goats. Front. Microbiol. 2017, 8, 1764. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.P.; Gratz, S.W.; Sheridan, P.O.; Flint, H.J.; Duncan, S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013, 69, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Zhang, R.; Wang, D.; Zhu, W. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet. Res. 2012, 8, 237. [Google Scholar] [CrossRef] [PubMed]
- Heiman, M.L.; Greenway, F.L. A healthy gastrointestinal microbiome is dependent on dietary diversity. Mol. Metab. 2016, 5, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Blix, A.S.; Ness, J.; Lian, H. Experiences from 40 years of muskox (Ovibos moschatus) farming in Norway. Rangifer 2011, 31, 1–6. [Google Scholar] [CrossRef]
- Norwegian Meteorological Institute (MET). Tromsø weather data: eKlima. Available online: http://sharki.oslo.dnmi.no/portal/page?_pageid=73,39035,73_39049&_dad=portal&_schema=PORTAL (accessed on 1 December 2017).
- Qaanaaq weather data: yr.no. Available online: https://www.yr.no/sted/Gr%C3%B8nland/Annet/Qaanaaq/statistikk.html (accessed on 1 December 2017).
- Groves, P. Intraspecific variation in mitochondrial DNA of muskoxen, based on control-region sequences. Can. J. Zool. 1997, 75, 568–575. [Google Scholar] [CrossRef]
- Holm, L.-E.; Forchhammer, M.C.; Boomsma, J.J. Low genetic variation in muskoxen (Ovibos moschatus) from western Greenland using microsatellites. Mol. Ecol. 1999, 8, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Deehan, E.C.; Walter, J. The fiber gap and the disappearing gut microbiome: Implications for human nutrition. Trends Endocrinol. Metab. 2016, 27, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Furet, J.-P.; Bensaada, M.; Philippe, C.; Roth, H.; Rabot, S.; Lakhdari, O.; Lombard, V.; Henrissat, B.; Corthier, G. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ. Microbiol. 2015, 17, 4954–4964. [Google Scholar] [CrossRef] [PubMed]
- Martínez, I.; Lattimer, J.M.; Hubach, K.L.; Case, J.A.; Yang, J.; Weber, C.G.; Louk, J.A.; Rose, D.J.; Kyureghian, G.; Peterson, D.A. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013, 7, 269. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Li, S.; Danscher, A.M.; Derakshani, H.; Andersen, P.H.; Khafipour, E. Changes in Microbiota in Rumen Digesta and Feces Due to a Grain-Based Subacute Ruminal Acidosis (SARA) Challenge. Microb. Ecol. 2017, 74, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Abnous, K.; Brooks, S.P.; Kwan, J.; Matias, F.; Green-Johnson, J.; Selinger, L.B.; Thomas, M.; Kalmokoff, M. Diets enriched in oat bran or wheat bran temporally and differentially alter the composition of the fecal community of rats. J. Nutr. 2009, 139, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Middelbos, I.S.; Boler, B.M.V.; Qu, A.; White, B.A.; Swanson, K.S.; Fahey, G.C., Jr. Phylogenetic Characterization of Fecal Microbial Communities of Dogs Fed Diets with or without Supplemental Dietary Fiber Using 454 Pyrosequencing. PLoS ONE 2010, 5, e9768. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wu, Q.; Dai, J.; Zhang, S.; Wei, F. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 17714–17719. [Google Scholar] [CrossRef] [PubMed]
- Kocherginskaya, S.A.; Aminov, R.I.; White, B.A. Analysis of the Rumen Bacterial Diversity under two Different Diet Conditions using Denaturing Gradient Gel Electrophoresis, Random Sequencing, and Statistical Ecology Approaches. Anaerobe 2001, 7, 119–134. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Vitorino, F.; Goldfarb, K.C.; Karaoz, U.; Leal, S.; Garcia-Amado, M.A.; Hugenholtz, P.; Tringe, S.G.; Brodie, E.L.; Dominguez-Bello, M.G. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J. 2012, 6, 531. [Google Scholar] [CrossRef] [PubMed]
- Callaway, T.R.; Dowd, S.E.; Edrington, T.S.; Anderson, R.C.; Krueger, N.; Bauer, N.; Kononoff, P.J.; Nisbet, D.J. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 2010, 88, 3977–3983. [Google Scholar] [CrossRef] [PubMed]
- Dougal, K.; Harris, P.A.; Edwards, A.; Pachebat, J.A.; Blackmore, T.M.; Worgan, H.J.; Newbold, C.J. A comparison of the microbiome and the metabolome of different regions of the equine hindgut. FEMS Microbiol. Ecol. 2012, 82, 642–652. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.N.V.; Jewell, K.A.; Freitas, F.S.; Benjamin, L.A.; Tótola, M.R.; Borges, A.C.; Moraes, C.A.; Suen, G. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet. Microbial. 2013, 164, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Davidson, R.K.; Amundsen, H.; Lie, N.O.; Luyckx, K.; Robertson, L.J.; Verocai, G.G.; Kutz, S.J.; Ytrehus, B. Sentinels in a climatic outpost: Endoparasites in the introduced muskox (Ovibos moschatus wardi) population of Dovrefjell, Norway. Int. J. Parasitol. Parasit. Wildl. 2014, 3, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Kutz, S.J.; Thompson, R.A.; Polley, L.; Kandola, K.; Nagy, J.; Wielinga, C.M.; Elkin, B.T. Giardia assemblage A: Human genotype in muskoxen in the Canadian Arctic. Parasit. Vectors 2008, 1, 32. [Google Scholar] [CrossRef] [PubMed]
- Davidson, R.K.; Lavikainen, A.; Konyaev, S.; Schurer, J.; Miller, A.L.; Oksanen, A.; Skírnisson, K.; Jenkins, E. Echinococcus across the north: Current knowledge, future challenges. Food Waterborne Parasitol. 2016, 4, 39–53. [Google Scholar] [CrossRef]
- Schurer, J.; Shury, T.; Leighton, F.; Jenkins, E. Surveillance for Echinococcus canadensis genotypes in Canadian ungulates. Int. J. Parasitol. Parasit. Wildl. 2013, 2, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Woldehiwet, Z. Q fever (coxiellosis): Epidemiology and pathogenesis. Res. Vet. Sci. 2004, 77, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Billinis, C. Wildlife diseases that pose a risk to small ruminants and their farmers. Small Rumin. Res. 2013, 110, 67–70. [Google Scholar] [CrossRef]
- Gates, C.C.; Wobeser, G.; Forbes, L.B. Rangiferine brucellosis in a muskox, Ovibos moschatus moschatus (Zimmermann). J. Wildl. Dis. 1984, 20, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Kutz, S.; Bollinger, T.; Branigan, M.; Checkley, S.; Davison, T.; Dumond, M.; Elkin, B.; Forde, T.; Hutchins, W.; Niptanatiak, A.; et al. Erysipelothrix rhusiopathiae associated with recent widespread muskox mortalities in the Canadian Arctic. Can. Vet. J. 2015, 56, 560–563. [Google Scholar] [PubMed]
- Nymo, I.H.; Beckmen, K.; Godfroid, J. Anti-Brucella Antibodies in Moose (Alces alces gigas), Muskoxen (Ovibos moschatus), and Plains Bison (Bison bison bison) in Alaska, USA. J. Wildl. Dis. 2015, 52, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Hancock, D.; Besser, T.; Lejeune, J.; Davis, M.; Rice, D. The control of VTEC in the animal reservoir. Int. J. Food Microbiol. 2001, 66, 71–78. [Google Scholar] [CrossRef]
- Wasteson, Y.; Arnemo, J.M.; Johansen, B.K.; Vold, L.; Mathiesen, S.D.; Olsen, M.A.; Wiig, Ø.; Derocher, A.E. Analysis of faecal samples from wild animals for verocytotoxin producing Escherichia coli and E. coli 0157. Vet. Rec. 1999, 144, 646–647. [Google Scholar] [CrossRef] [PubMed]
- Wahlstrom, H.; Tysen, E.; Olsson Engvall, E.; Brandstrom, B.; Eriksson, E.; Morner, T.; Vagsholm, I. Survey of Campylobacter species, VTEC 01 57 and Salmonella species in Swedish wildlife. Vet. Rec. 2003, 153, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Lukjancenko, O.; Wassenaar, T.M.; Ussery, D.W. Comparison of 61 Sequenced Escherichia coli Genomes. Microb. Ecol. 2010, 60, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Wasteson, Y. Zoonotic Escherichia coli. Acta Vet. Scand. 2002, 43, S79. [Google Scholar] [CrossRef]
- Forde, T.L.; Orsel, K.; Zadoks, R.N.; Biek, R.; Adams, L.G.; Checkley, S.L.; Davison, T.; De Buck, J.; Dumond, M.; Elkin, B.T.; et al. Bacterial Genomics Reveal the Complex Epidemiology of an Emerging Pathogen in Arctic and Boreal Ungulates. Front. Microbiol. 2016, 7, 1759. [Google Scholar] [CrossRef] [PubMed]
- Mair, N.S. Yersiniosis in wildlife and its public health implications. J. Wildl. Dis. 1973, 9, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Nishi, J.S.; Shury, T.; Elkin, B.T. Wildlife reservoirs for bovine tuberculosis (Mycobacterium bovis) in Canada: Strategies for management and research. Vet. Microbiol. 2006, 112, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Handeland, K.; Tengs, T.; Kokotovic, B.; Vikøren, T.; Ayling, R.D.; Bergsjø, B.; Sigurðardóttir, Ó.G.; Bretten, T. Mycoplasma ovipneumoniae—A Primary Cause of Severe Pneumonia Epizootics in the Norwegian Muskox (Ovibos moschatus) Population. PLoS ONE 2014, 9, e106116. [Google Scholar] [CrossRef] [PubMed]
- Ytrehus, B.; Bretten, T.; Bergsjø, B.; Isaksen, K. Fatal pneumonia epizootic in musk ox (Ovibos moschatus) in a period of extraordinary weather conditions. EcoHealth 2008, 5, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Båverud, V.; Gustafsson, A.; Franklin, A.; Aspán, A.; Gunnarsson, A. Clostridium difficile: Prevalence in horses and environment, and antimicrobial susceptibility. Equine Vet. J. 2003, 35, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Bredy, J.P.; Botzler, R.G. The effects of six environmental variables on Pasteurella multocida populations in water. J. Wildl. Dis. 1989, 25, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Brooke, C.J.; Riley, T.V. Erysipelothrix rhusiopathiae: Bacteriology, epidemiology and clinical manifestations of an occupational pathogen. J. Med. Microbiol. 1999, 48, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Jensen, G.B.; Hansen, B.M.; Eilenberg, J.; Mahillon, J. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 2003, 5, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Whittington, R.J.; Marshall, D.J.; Nicholls, P.J.; Marsh, I.B.; Reddacliff, L.A. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 2004, 70, 2989–3004. [Google Scholar] [CrossRef] [PubMed]
- Seidel, K.B.; Rowell, J.E. Canadian Muskoxen in Central Europe-A Zoo Veterinary Review. Rangifer 1996, 16, 79–85. [Google Scholar] [CrossRef]
- Mikko, S.; Røed, K.; Schmutz, S.; Andersson, L. Monomorphism and polymorphism at Mhc DRB loci in domestic and wild ruminants. Immunol. Rev. 1999, 167, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.D.; Hughes, K.L. Is Rhodococcus equi a soil organism? J. Reprod. Fertil. Suppl. 1982, 32, 481–489. [Google Scholar] [PubMed]
| NEGM Only (n = 35), Biotic and Temporal Effects | |||||
|---|---|---|---|---|---|
| MGLM-ANOVA (Composition) | |||||
| Variable | Df | Deviance | p-value | ||
| Year | 33 | 5584 | 0.002 | ||
| Sex | 32 | 3421 | 0.080 | ||
| Body mass (kg) | 31 | 3951 | 0.048 | ||
| GLM (OTU Richness) | |||||
| Variable | Df | Deviance | AIC | p-value | |
| Year | 1 | 120,604 | 392.4 | 0.362 | |
| Sex | 1 | 119,037 | 391.94 | 0.541 | |
| Body mass (kg) | 1 | 117,833 | 391.58 | 0.895 | |
| Location Effects, All Groups (±NWGM) | |||||
| MGLM-ANOVA (Composition) | |||||
| Variable | Df | Deviance | p-value | ||
| Including NWGM | Location | 35 | 3.161 | 0.002 | |
| Excluding NWGM | Location | 35 | 3.161 | 0.002 | |
| GLM (OTU Richness) | |||||
| Variable | Df | Deviance | AIC | p-value | |
| Including NWGM | Location | 2 | 2.25 | 64.134 | >0.001 |
| Excluding NWGM | Location | 2 | 0.464 | −55.579 | >0.001 |
| Genera | Location/No. Animals | Zoonotic Species Examples | Disease in Man | Risk and Epizootiology | ||
|---|---|---|---|---|---|---|
| NEGM | NM | NWGM | ||||
| Actinomyces | 4/35 | 0/3 | 0/4 | A. bovis; A. pyogenes | Abscesses; chronic bronchopneumonia; sepsis; endocarditis. | Rare infection in people. Worldwide distribution. Cows are the main carrier. |
| Bacillus | 16/35 | 1/3 | 4/4 | B. anthracis | “Anthrax” | For man, the source of infection is always infected animals, contaminated animal products or spores from originating from infected animals. (Cutaneous) Infection is for example, known to occur when skinning or butchering an animal or by contact with infected leather, pelts, wool, or fur. Broken dermis favours transmission. (Gastrointestinal) infection can be acquired from consumption of domestic and wild animals. Animals mainly become infected by ingestion of pasture or water that have been contaminated with spores—typically in the vicinity of anthrax-infected carcasses as dead and dying animals are the vessel of high rate of replication of B. anthracis. The bacilli sporulate if the carcass is opened and they contaminate the surrounding environment, leading to new infections in particularly grazing animals. Infected animals and especially birds can effectively transport the infection to other areas. Transmission between humans is a rare form of infection for humans. |
| Bacteroides | 35/35 | 3/3 | 3/4 | Bacteroides sp.; B. fragilis; B. tectus; B. ovatus | Wound infections | Unknown species of Bacteroides associated with purulent wound infection from a horse bite. Wound infections with other species of Bacteroides associated with cats and dogs. |
| Clostridium | 33/35 | 1/3 | 1/4 | C. difficile; C. perfringens | Gastroenteritis | Both types are regarded as potential zoonoses. Infection is acquired from the environment, from contact or ingestion of contaminated meat/animals/animal products [39]. |
| Erysipelothrix | 15/35 | 0/3 | 0/4 | Dermatitis; septic arthritis; endocarditis; sepsis | The course of disease usually only lasts for up to four weeks but sepsis with potential subsequent endocarditis can occur. Distributed Worldwide in a wide range of species incl. domestic animals, for example, swine and ruminants and frequently associated with fish. Many animals carry the disease non-symptomatically. People can acquire the infection through handling of infected animal products. | |
| Escherichia/Shigella | 23/35 | 3/3 | 2/4 | E. coli (VTEC and EHEC) 1 | Enteritis | Infection through faecal-oral route, commonly from contaminated animal products. Airborne transmission via dust is also possible. EHEC infections in humans often associated with cattle which are considered a reservoir species. VTEC infections associated with both cattle, sheep among other. |
| Fusobacterium | 10/35 | 0/3 | 0/4 | F. nucleatum; F. necrophorum (not confirmed) | Wound infections | Infection risk associated biting events or contamination of open skin lesions or through mucus membranes. Previously predominantly associated with dog bite. |
| Mycobacterium | 2/35 | 0/3 | 2/4 | Principally M. bovis in terms of zoonoses; M. avium subsp. paratuberculosis; M. simiae; M. kansasii; M. ulcerans | Pulmonary and extrapulmonary forms. The latter affecting glands, bones and joints, meninges, urinary tracts and more. | Mycobacterium spp. vary with host-species but zoonotic Mycobacterium spp. can generally infect a wide range of animal species and are considered to be distributed virtually Worldwide. These bacteria can be difficult to kill in the environment as they are resistant to many common disinfectants as well as desiccation. Pasteurization of milk has however reduced the incidences of zoonotically acquired infections. Infection can however also be acquired through inhalation. Main reservoir of M. bovis is cattle. The infection with M. bovis comes from animal sources since human to human transmission is considered rare. |
| Pasteurella | 0/35 | 1/3 | 0/4 | P. multocida | Disease often associated with bite wounds; abscesses; cellulitis; meningitis; septic arthritis; osteomyelitis; respiratory tract disease; sepsis and endocarditis are rare | Infection is typically acquired through bite wounds but can also occur through inhalation and the digestive tract. Cats and dogs are frequent carriers but cattle and sheep and other also present important asymptomatic reservoirs. Many wild animals are also asymptomatic carriers and outbreaks in wildlife occur occasionally. Pasteurella is believed only to survive shortly in the environment and animals therefore play a key role in the epidemiology and infection of Pasteurella in people. Infection between humans are also thought to occur. |
| Rhodococcus (syn. Coryne-bacterium) | 0/35 | 0/3 | 2/4 | R. equi | A rare disease in humans and often associated with immuno-suppressive states; but neglected reporting is suspected; granulomatous and suppurative lung disease; Pulmonary (most prevalent form) and extra-pulmonary form including osteomyelitis; cachexia; bloody diarrhoea; abscessation and other. | A saprophyte reported Worldwide which replicates effectively in faeces from herbivores such as goats, sheep, cows, deer, horses, cats and dogs. Often isolated from horses and a significant disease of foals. Infection in humans occurs through inhalation of the pathogen, often mediated by dust-particles or through infection of for example, infected sputum. Acetic acid in faeces is believed to favour effective replication of Rhodococcus in faeces. The main reservoir is as such soil exposed to faeces from herbivores which shed high amount of acetic acid in their faeces [40,41]. |
| Streptococcus | 11/35 | 3/3 | 2/4 | S. bovis; S. suis; S. zooepidemicus (Lancefield group D and C respectively) | S. suis: meningitis; arthritis and endophthalmitis; S. zooepidemicus: pneumonia; endocarditis; meningitis; pericarditis; exudative pharyngitis; tonsillitis. | Ingestion of for example, raw milk or pork meat and handling of infected animals are related risks. S. zooepidemicus is a commensal of the skin on, upper respiratory tract and in the tonsils of various animal species. It causes multiples diseases in horses and is implicated in mastitis of cattle. S. suis is a highly occupational disease in relation to slaughterhouses and butchering and believed most often to be acquired through the skin. |
| Yersinia | 0/35 | 0/3 | 2/4 | Y. enterocolitica | Enteritis and acute diarrhoea; reactive arthritis; nodular erythema | Worldwide distribution and isolated broadly in the environment: from animals, food and water. Can occur sporadically or as epidemics. Faecal-oral route of transmission from other people or from contaminated animal products. Occupation involving swine, consumption of pork and milk products are associated with zoonotic transmission to people. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersen-Ranberg, E.U.; Barnes, C.J.; Rasmussen, L.; Salgado-Flores, A.; Grøndahl, C.; Mosbacher, J.B.; Hansen, A.J.; Sundset, M.A.; Schmidt, N.M.; Sonne, C. A Comparative Study on the Faecal Bacterial Community and Potential Zoonotic Bacteria of Muskoxen (Ovibos moschatus) in Northeast Greenland, Northwest Greenland and Norway. Microorganisms 2018, 6, 76. https://doi.org/10.3390/microorganisms6030076
Andersen-Ranberg EU, Barnes CJ, Rasmussen L, Salgado-Flores A, Grøndahl C, Mosbacher JB, Hansen AJ, Sundset MA, Schmidt NM, Sonne C. A Comparative Study on the Faecal Bacterial Community and Potential Zoonotic Bacteria of Muskoxen (Ovibos moschatus) in Northeast Greenland, Northwest Greenland and Norway. Microorganisms. 2018; 6(3):76. https://doi.org/10.3390/microorganisms6030076
Chicago/Turabian StyleAndersen-Ranberg, Emilie U., Christopher J. Barnes, Linett Rasmussen, Alejandro Salgado-Flores, Carsten Grøndahl, Jesper B. Mosbacher, Anders J. Hansen, Monica Alterskjær Sundset, Niels Martin Schmidt, and Christian Sonne. 2018. "A Comparative Study on the Faecal Bacterial Community and Potential Zoonotic Bacteria of Muskoxen (Ovibos moschatus) in Northeast Greenland, Northwest Greenland and Norway" Microorganisms 6, no. 3: 76. https://doi.org/10.3390/microorganisms6030076
APA StyleAndersen-Ranberg, E. U., Barnes, C. J., Rasmussen, L., Salgado-Flores, A., Grøndahl, C., Mosbacher, J. B., Hansen, A. J., Sundset, M. A., Schmidt, N. M., & Sonne, C. (2018). A Comparative Study on the Faecal Bacterial Community and Potential Zoonotic Bacteria of Muskoxen (Ovibos moschatus) in Northeast Greenland, Northwest Greenland and Norway. Microorganisms, 6(3), 76. https://doi.org/10.3390/microorganisms6030076






