Novel and Unexpected Microbial Diversity in Acid Mine Drainage in Svalbard (78° N), Revealed by Culture-Independent Approaches
Abstract
:1. Introduction
2. Experimental Section
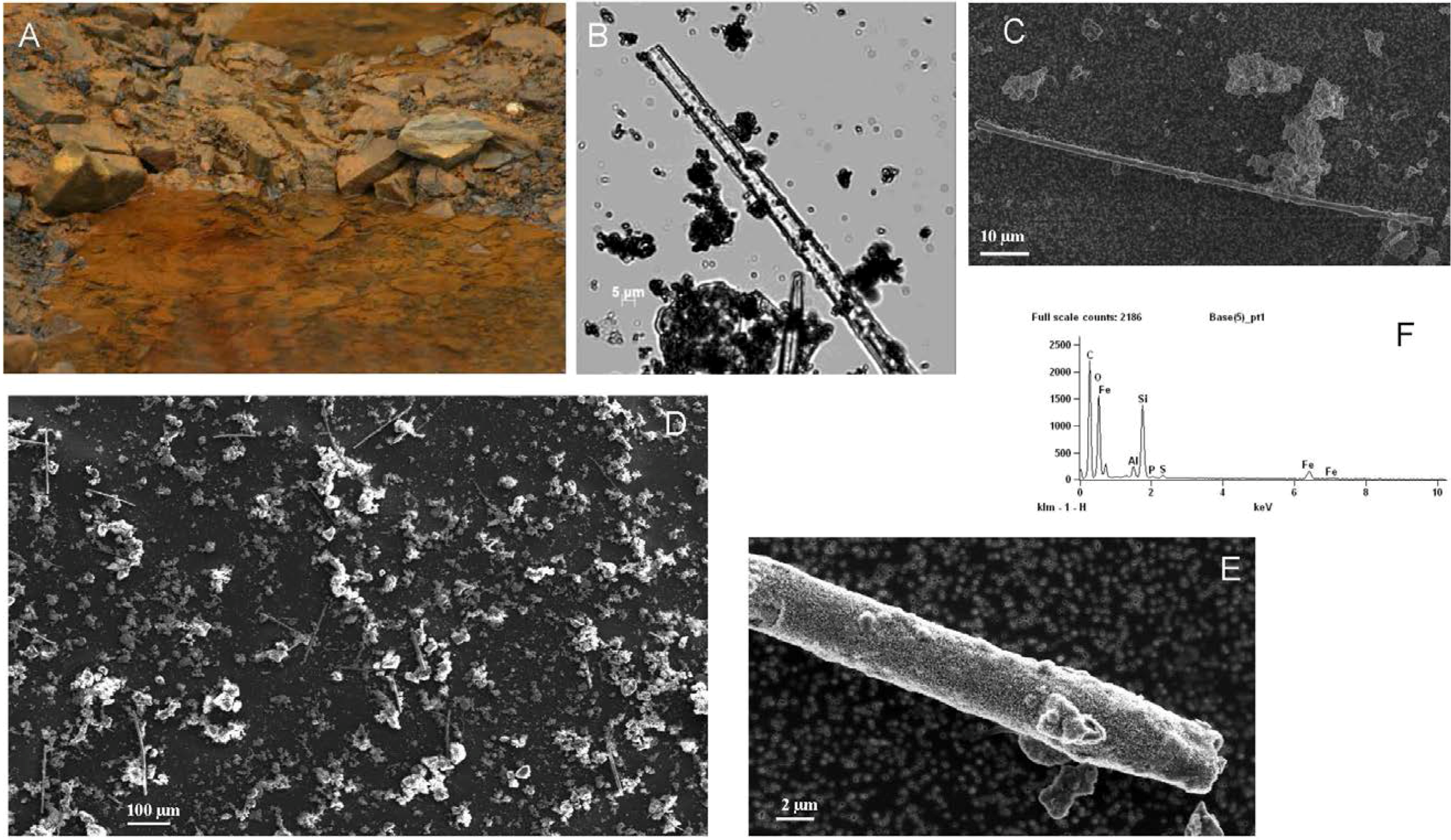
2.1. Area Description and Sample Collection
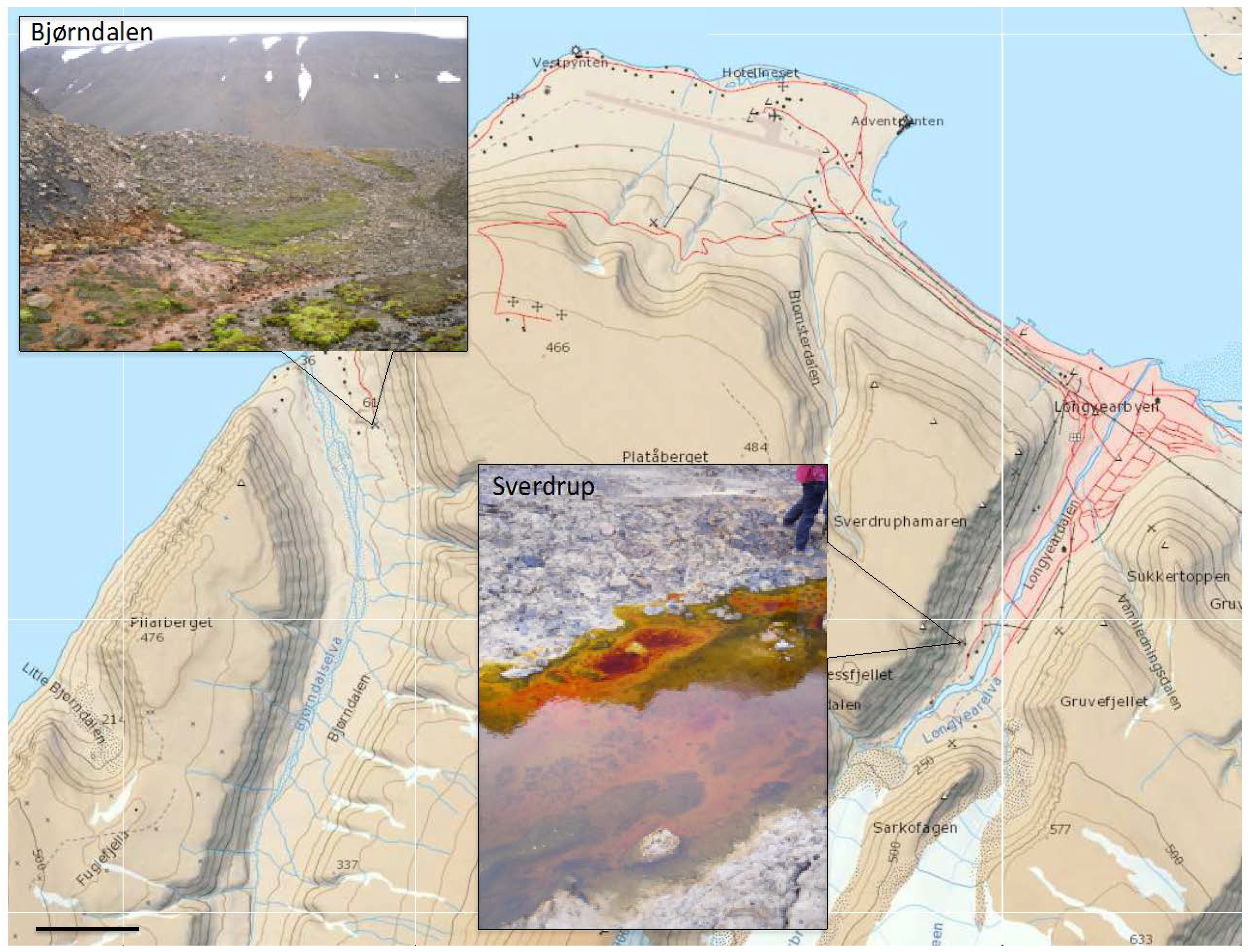
2.2. Geochemical Analyses
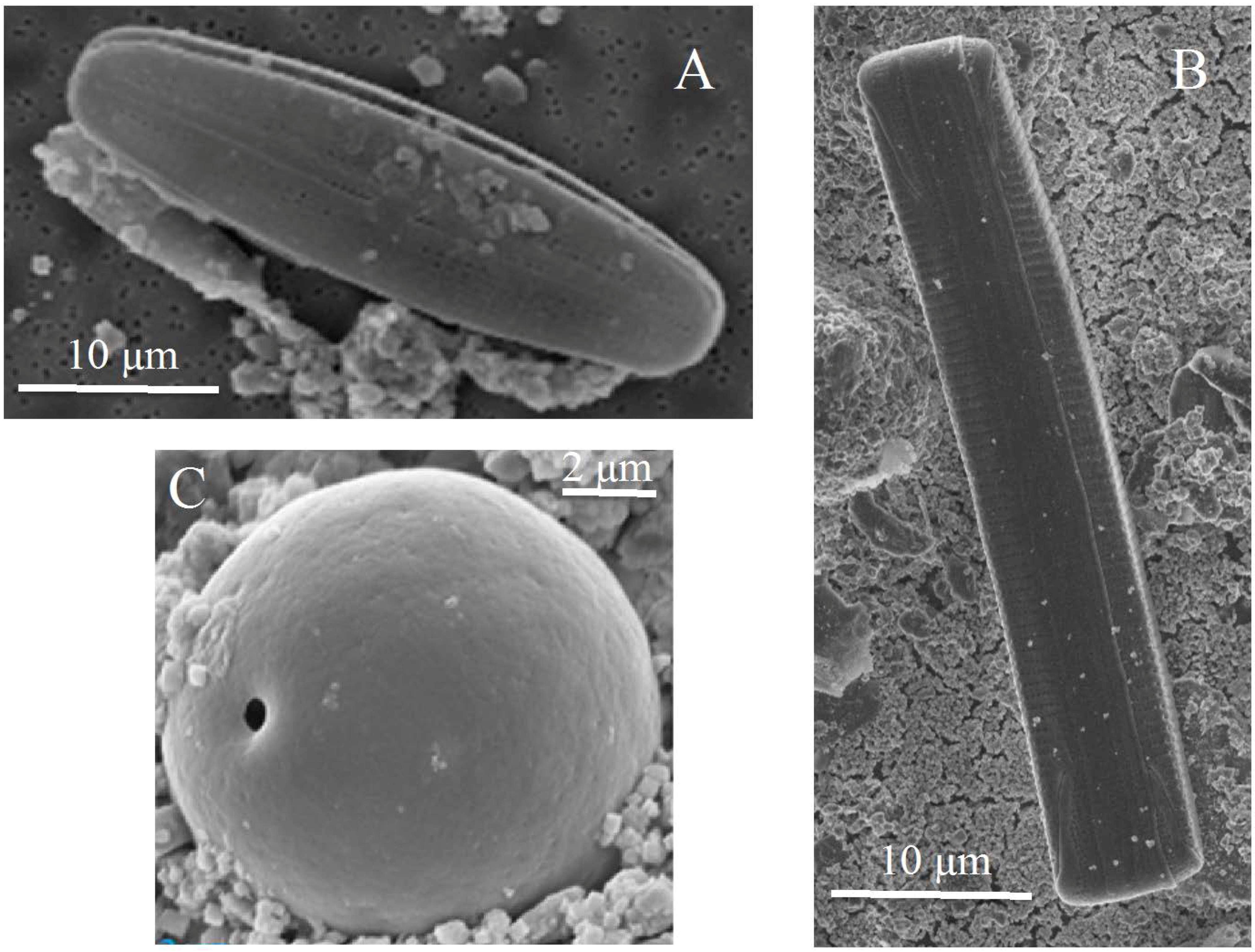
2.3. Scanning Electron Microscopy (SEM)
2.4. DNA Extraction
2.5. RNA Gene Amplification and Cloning
2.6. Phylogenetic Analyses
2.7. qPCR
2.8. Amplicon Library Construction and 454 Sequencing
2.9. Noise Removal and Taxonomic Clustering
3. Results
3.1. Observations and Chemical Analysis
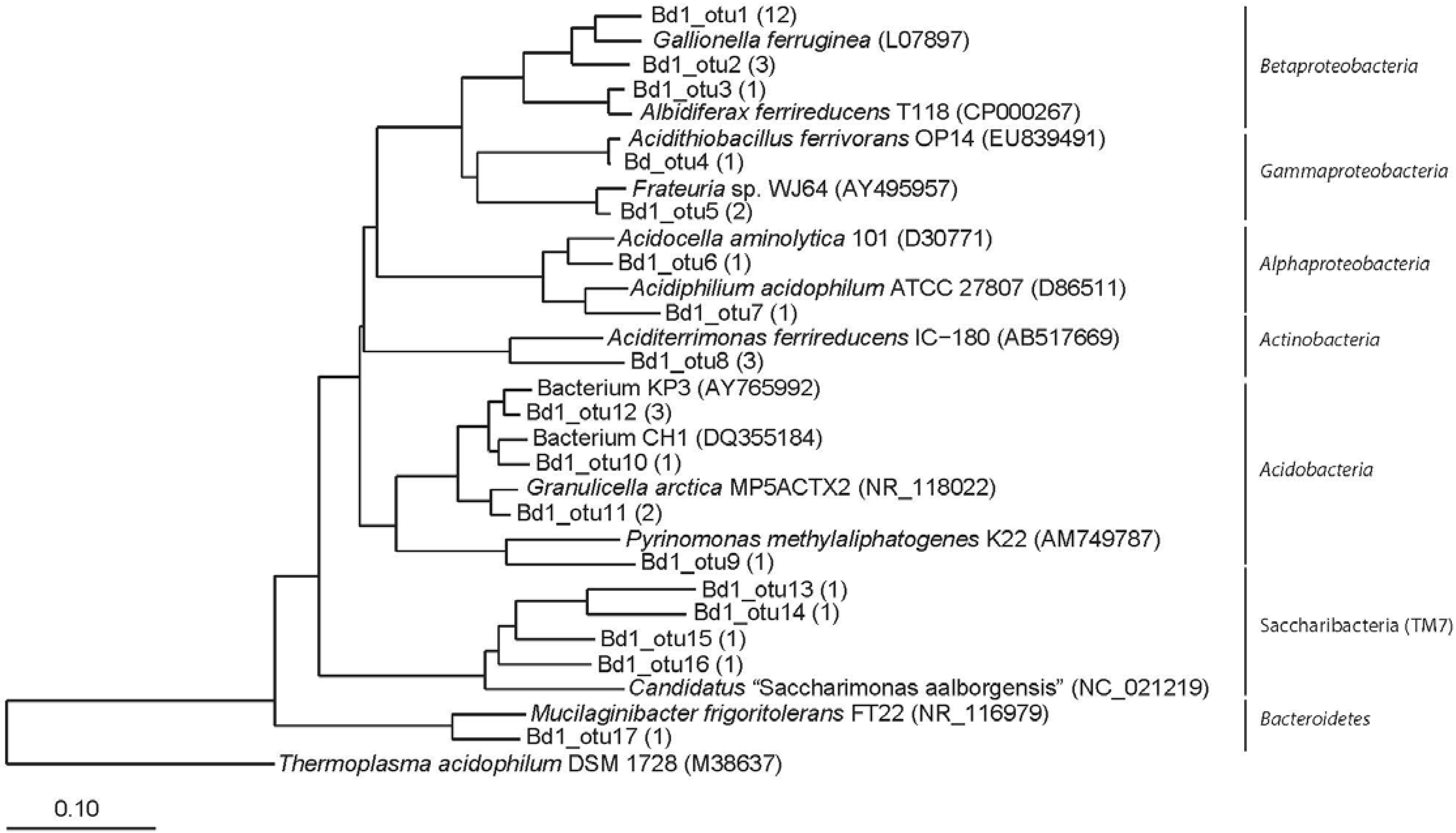
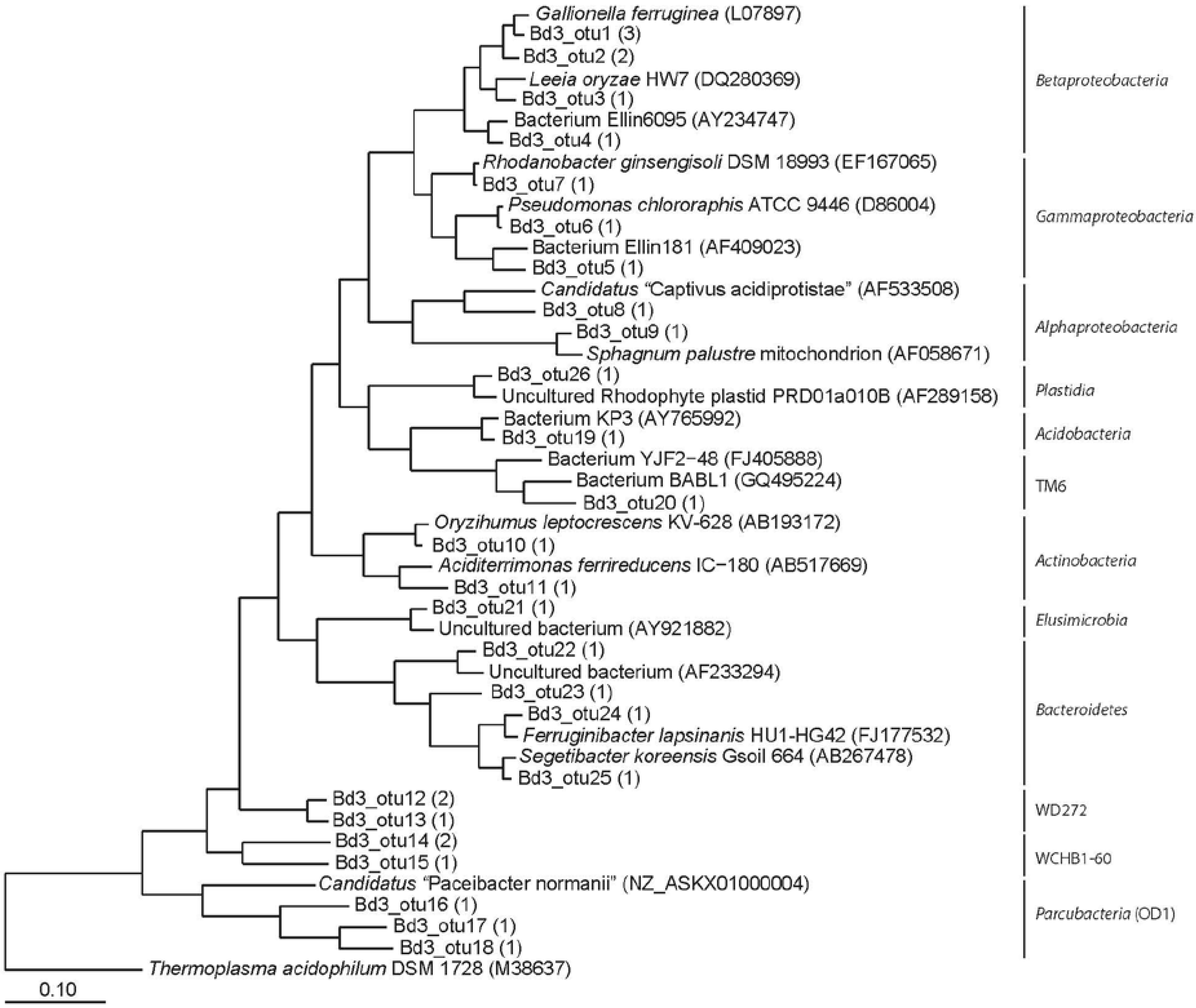
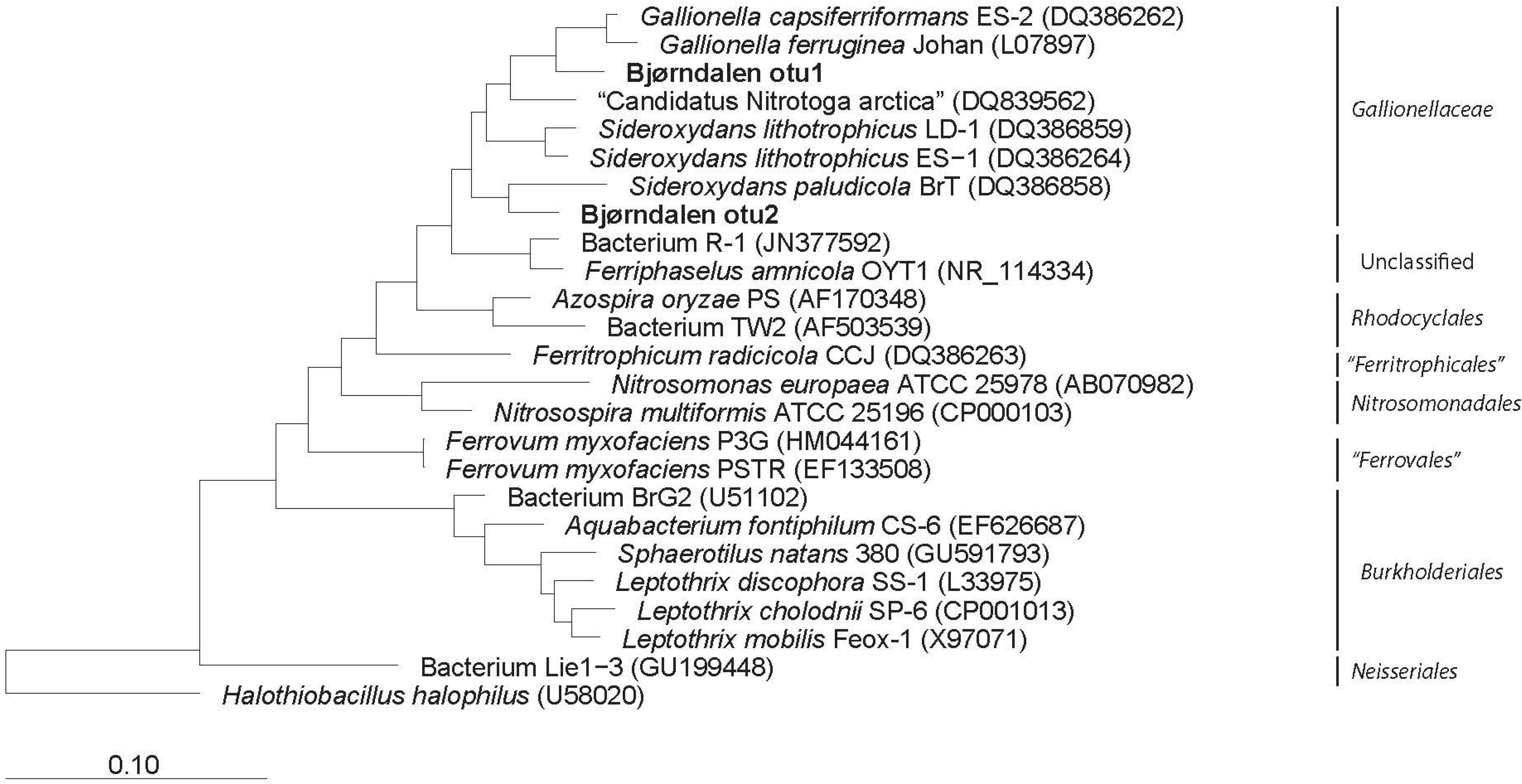
3.2. Clone Libraries and Phylogeny
| Area | Bjørndalen | Sverdrup | |||
|---|---|---|---|---|---|
| Samples | Bd1 | Bd2 | Bd3 | Bd4 | Sv1, Sv2 |
| Type | Spring water | Thaw water | Mix upstream | Mix downstream | Mat and sediment |
| pH | 2.8 | 6.2 | 3.5 | 3.8 | 2.3 |
| °C (water) | 9 | 7 | 7 | 7 | 4 |
| °C (air) | 6 | 6 | 6 | 6 | 4 |
| Na | 3.1 | 2.5 | 3 | 2.8 | 42 |
| K | 1.1 | 0.5 | 1.6 | 1.3 | 0.2 |
| Zn | 0.3 | Na | 0.1 | 0.07 | 5.1 |
| Ni | 0.1 | Na | 0.03 | 0.02 | 1.3 |
| Mg | 5.4 | 1.3 | 2.6 | 2.3 | 145 |
| Ca | Na | Na | Na | Na | 10 |
| Mn | 0.4 | Na | 0.2 | 0.1 | 9.7 |
| Cu | 0.03 | Na | Na | Na | 0.8 |
| Al | 3.4 | Na | 1.8 | 0.9 | 76 |
| Fe total | 15 | Na | 2 | 2 | 883 |
| Fe2+ | 9 | Na | 1 | 1 | 183 |
| Fe3+ | 6 | Na | 1 | 1 | 700 |
| SO42− | 110 | 10 | 54 | 32 | 1543 |
3.3. Community Composition and Diversity
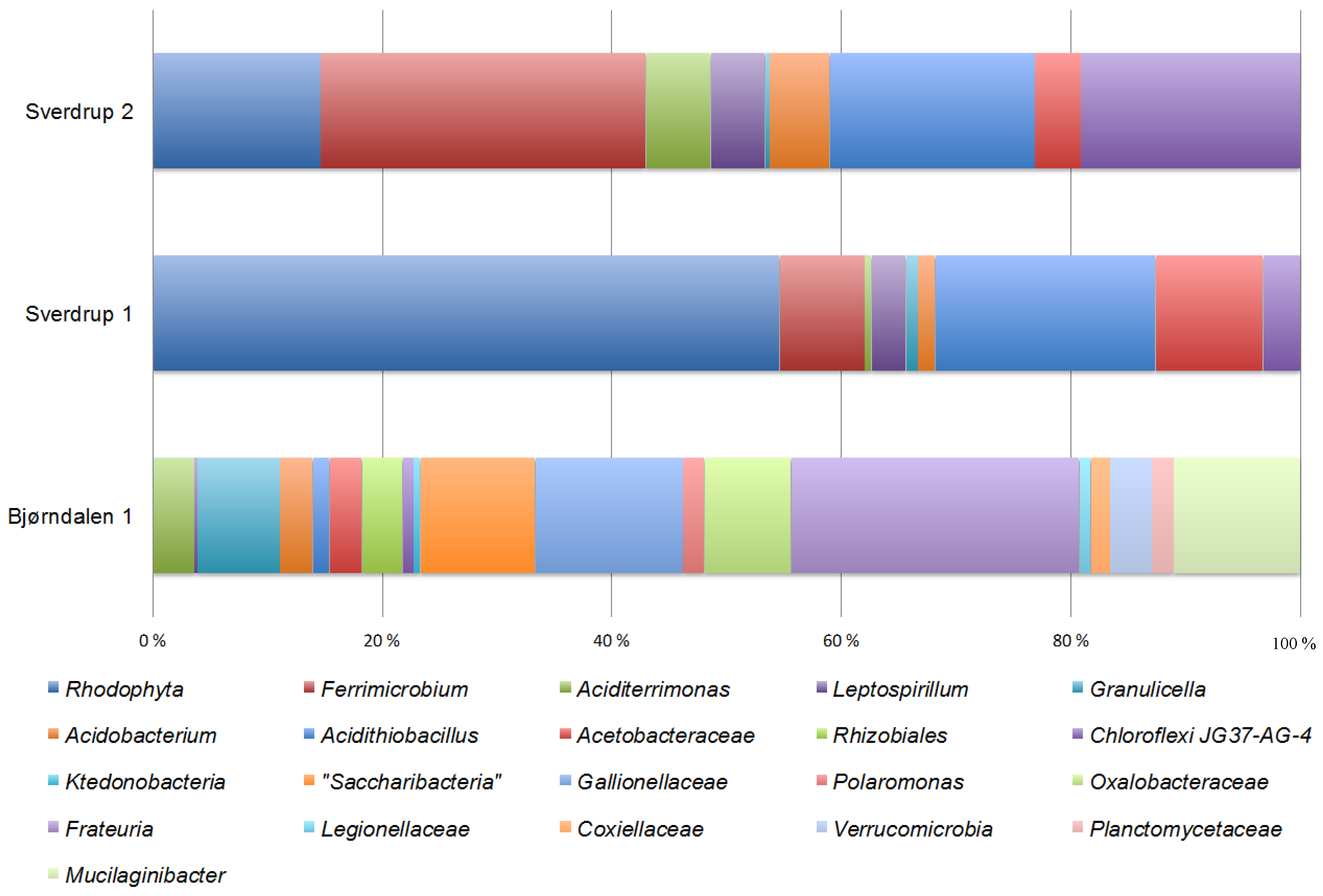
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix

| Site name | PCR Cycles * | Raw reads | Filtered reads | % Chimeric reads | Unique reads | 3% OTUs | Chao1 (3%) | (1−D) ** | Hʹ ** | Rarefied Richness |
|---|---|---|---|---|---|---|---|---|---|---|
| Bd1 | 25 | 21,698 | 18,324 | 1.33% | 3693 | 2209 | 3569 | 0.98 | 6.06 | 2 034 |
| Sv1 | 25 | 17,409 | 15,368 | 0.02% | 92 | 72 | 116 | 0.83 | 2.11 | 65.9 |
| Sv2 | 25 | 17,716 | 15,670 | 0.05% | 100 | 80 | 152 | 0.67 | 1.69 | 73.2 |
| Sample | Archaeal 16S rDNA Copies | Average Bacterial 16S rDNA Copies | Gallionella/Sideroxydans 16S rDNA Copies | Ferrovum 16S rDNA Copies | Estimated Bacterial Cells per Gram * | Gallionella/Sideroxydans % Copies of Total |
|---|---|---|---|---|---|---|
| Bd1 | not detected | 3.27 × 108 | 4.49 × 107 ± 9.7% | not detected | 3.8 × 107 ± 0.4% | 13.7% ± 1.32 |
| Bd3 | not detected | 1.62 × 108 | 8.82 × 106 ± 37% | not detected | 1.9 × 107 ± 31% | 5.5% ± 2.03 |
| Sv1 | not detected | 5.27 × 107 | 2.31 × 104 ± 3.3% | not detected | 6.1 × 106 ± 28% | <1% ± 0.03 |
| Sv2 | not detected | 2.64 × 108 | 2.30 × 106 ± 9.9% | not detected | 3.06 × 107 ± 5% | <1% ± 0.1 |
References
- Elberling, B.; Søndergaard, J.; Jensen, L.A.; Schmidt, L.B.; Hansen, B.U.; Asmud, G.; Baliczunic, T.; Hollesen, J.; Hanson, S.; Jansson, P.-E.; et al. Arctic vegetation damage by winter-generated coal mining pollution released upon thawing. Environ. Sci. Technol. 2007, 41, 2407–2413. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Hallberg, K. The microbiology of acidic mine waters. Res. Microbiol. 2003, 154, 466–473. [Google Scholar] [CrossRef]
- Hallberg, K.B. New perspectives in acid mine drainage microbiology. Hydrometallurgy 2010, 104, 448–453. [Google Scholar] [CrossRef]
- Rawlings, D.E.; Johnson, D.B. The microbiology of biomining: Development and optimization of mineral-oxidizing microbial consortia. Microbiology 2007, 153, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Kupka, D.; Rzhepishevska, O.I.; Dopson, M.; Lindström, E.B.; Karnachuk, O.V.; Tuovinen, O.H. Bacterial oxidation of ferrous iron at low temperatures. Biotechnol. Bioeng. 2007, 97, 1470–1478. [Google Scholar] [CrossRef] [PubMed]
- Elberling, B.; Schippers, A.; Sand, W. Bacterial and chemical oxidation of pyritic mine tailings at low temperatures. J. Contam. Hydrol. 2000, 41, 225–238. [Google Scholar] [CrossRef]
- Søndegaard, J.; Elberling, B.; Asmud, G.; Gudum, C.; Iversen, K.M. Temporal trends of dissolved weathering products released from a high Arctic coal mine waste rock pile in Svalbard (78° N). Appl. Geochem. 2007, 22, 1025–1038. [Google Scholar] [CrossRef]
- Hollesen, J.; Elberling, B.; Jansson, P.E. Modelling temperature-dependent heat production over decades in High Arctic coal waste rock piles. Cold Reg. Sci. Technol. 2011, 65, 258–268. [Google Scholar] [CrossRef]
- Askaer, L.; Schmidt, L.B.; Elberling, B.; Asmund, G.; Jónsdóttir, I.S. Environmental impact on an Arctic soil-plant system resulting from metals released from coal mine waste in Svalbard (78° N). Water Air Soil Pollut. 2008, 195, 99–114. [Google Scholar] [CrossRef]
- Humlum, O.; Instanes, A.; Sollid, J.L. Permafrost in Svalbard: A review of research history, climatic background and engineering challenges. Polar Res. 2003, 22, 191–215. [Google Scholar] [CrossRef]
- Hollesen, J.; Elberling, B.; Hansen, B.U. Modelling subsurface temperatures in a heat producing coal waste rock pile, Svalbard (78° N). Cold Reg. Sci. Technol. 2009, 58, 68–76. [Google Scholar] [CrossRef]
- Easton, A.J. The colorimetric determination of iron. In Chemical Analysis of Silicate Rocks; Easton, A.J., Ed.; Elsevier: Amstedam, The Nethelands, 1972. [Google Scholar]
- Kolmert, Å.; Wikström, P.; Hallberg, K.B. A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J. Microbiol. Method 2000, 41, 179–184. [Google Scholar]
- Lane, D.J. 16S/23S rDNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Rincón, B.; Raposo, F.; Borja, R.; González, J.M.; Portillo, M.C.; Saiz-Jiménez, C. Performance and microbial communities of a continuous stirred tank anaerobic reactor treating two-phase olive mill solid wasters at low organic loading rates. J. Biotechnol. 2006, 121, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.; Feldgarden, M.; Ward, D.V.; Giannokous, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar, A.B.; Lai, T.; Steppi, S.; Jobb, G.; Förster, W.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.; Ludwig, W.; Peplies, J.; Glökner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Øvreås, L.; Torsvik, V.V. Microbial diversity and community structure in two different agricultural soil communities. Microbiol. Ecol. 1998, 36, 303–315. [Google Scholar]
- Takai, K.; Horikoshi, K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 2000, 66, 5066–5072. [Google Scholar] [CrossRef] [PubMed]
- Juanjuan, W.; Vollrath, S.; Behrends, T.; Bodelier, P.L.E.; Muyzer, G.; Meima-Franke, M.; den Oudsten, F.; van Capellen, P.; Laanbroek, H.J. Distribution and diversity of Gallionella-like neutrophilic iron oxidizers in a tidal freshwater marsh. Appl. Environ. Microbiol. 2011, 77, 2337–2344. [Google Scholar]
- Heinzel, E.; Janneck, E.; Glombitza, F.; Schlomann, M.; Seifert, J. Population dynamics of iron-oxidizing communities in pilot plants for the treatment of acid mine waters. Environ. Sci. Technol. 2009, 43, 6138–6144. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.K.; Schmidt, T.M. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Fabisch, M.; Beulig, F.; Akob, D.M.; Küsel, K. Surprising abundance of Gallionella-related iron oxidizers in creek sediments at pH 4.4 or at high heavy metal concentrations. Front. Microbiol. 2013, 4, 390. [Google Scholar] [CrossRef] [PubMed]
- Roesch, L.F.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.M.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, M.; Sjøtun, K.; Lanzén, A.; Øvreås, L. Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J. 2012, 6, 2188–2198. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Lanzén, A.; Davenport, R.; Turnbaugh, J. Removing noise from pyrosequenced amplicons. BMC Bioinform. 2011, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Lanzén, A.; Jørgensen, S.L.; Huson, D.H.; Gorfer, M.; Grindhaug, S.H.; Jonassen, I.; Øvreås, L.; Urich, T. CREST—Classification Resources for Environmental Sequence Tags. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Souza-Egipsy, V.; González-Toril, E.; Zettler, E.; Amaral-Zettler, L.; Aguilera, A.; Amils, R. Prokaryotic community structure in algal photosynthetic biofilms from extreme acidic streams in Río Tinto (Huelva, Spain). Int. Microbiol. 2008, 4, 251–260. [Google Scholar]
- González-Toril, E.; Aguilera, A.; Souza-Egipsy, V.; López-Pamo, E.; Sánchez-Espana, J.; Amils, R. Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain). Appl. Environ. Microbiol. 2011, 77, 2685–2694. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Zettler, E.; Gómez, F.; Amaral-Zettler, L.; Rodríguez, N.; Amils, R. Distribution and seasonal variability in benthic eukaryotic community of Río Tinto (SW, Spain), an acidic, high metal extreme environment. Syst. Appl. Microbiol. 2007, 30, 531–546. [Google Scholar] [CrossRef] [PubMed]
- García-Moyano, A.; González-Toril, E.; Aguilera, A.; Amils, R. Comparative microbial ecology study of the sediments and the water column of the Río Tinto, an extreme acidic environment. FEMS Microbiol. Ecol. 2012, 81, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Senko, J.M.; Wanjugi, P.; Lucas, M.; Bruns, M.A.; Burgos, W.D. Characterization of Fe(II) oxidizing bacterial activities and communities at two acidic Appalachian coalmine drainage-impacted sites. ISME J. 2008, 2, 1134–1145. [Google Scholar] [CrossRef] [PubMed]
- Santofimia, E.; González-Toril, E.; López-Pamo, E.; Gomariz, M.; Amils, R.; Aguilera, A. Microbial diversity and its relationship to physicochemical characteristics of the water in two extreme acidic pit lakes from the Iberian Pyrite Belt (SW Spain). PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Frischkorn, K.R.; Williams, K.H.; Tringe, S.G.; Banfield, J.F. Community genomic analysis constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 2013, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.X.; Li, J.T.; Chen, Y.T.; Huang, L.N.; Hua, Z.N.; Hu, M.; Shu, W.S. Shifts in microbial community composition and function in the acidification of a lead/zinc mine tailings. Environ. Microbiol. 2013, 15, 2431–2444. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hua, Z.S.; Chen, Z.N.; Kuang, J.L.; Li, S.J.; Shu, W.S. Correlating microbial diversity patterns with geochemistry in an extreme and heterogeneous mine tailings environment. Appl. Environ. Microbiol. 2014, 80, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Korehi, H.; Blöthe, M.; Schippers, A. Microbial diversity at the moderate acidic stage in three different sulfidic mine tailings dumps generating acid mine drainage. Res. Microbiol. 2014, 165, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Peura, S.; Eiler, A.; Bertilsson, S.; Nykänen, H.; Tiirola, M.; Jones, R.I. Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J. 2012, 6, 1640–1652. [Google Scholar] [CrossRef] [PubMed]
- Bertin, P.N.; Heinrich-Salmeron, A.; Plletier, E.; Goulhen-Chollet, F.; Arsene-Ploetze, F.; Gallien, S.; Lauga, B.; Casiot, C.; Calteau, A.; Vallene, D.; et al. Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta- and proteogenomics. ISME J. 2011, 5, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Dojka, M.A.; Hugengoltz, P.; Haack, S.; Pace, N. Microbial Diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 1998, 64, 3869–3877. [Google Scholar] [PubMed]
- McLean, J.S.; Lombardo, M.J.; Badger, J.H.; Edlund, A.; Novotny, M.; Yee-Greenbaum, J.; Vyahhi, N.; Hall, A.P.; Yang, Y.; Dupont, C.L.; et al. Candidate phylum TM6 genome recovered from a hospital sink provides genomic insights into this uncultivated phylum. Proc. Nat. Acad. Sci. USA 2012, 110, 2390–2399. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Qing, Y.; Guo, X.; Warren, A. Candidatus “Sonnerbornia yantaiensis”, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea). Syst. Appl. Microbiol. 2014, 37, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Hugenholtz, P.; Dawson, S.C.; Banfield, J.F. Extremely acidophilic protists from acid mine drainage host Rickettsiales-lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl. Environ. Microbiol. 2003, 69, 5512–5518. [Google Scholar] [CrossRef] [PubMed]
- Herlemann, D.P.R.; Geissinger, O.; Ikeda-Ohtsubo, W.; Kunin, V.; Sun, H.; Lapidus, A.; Hugenholtz, P.; Brune, A. Genomic analysis of “Elusimicrobium minutum”, the first cultivated representative of the phylum “Elusimicrobia” (formerly Termite Group 1). Appl. Environ. Microbiol. 2009, 75, 2841–2849. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, B.; Winsley, T.; Ji, M.; Neilan, B. Insights into the distribution and abundance of the ubiquitous Candidatus Saccharibacteria phylum following tag pyrosequencing. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Larose, C.; Bergen, S.; Ferrari, C.; Navarro, E.; Dommergue, A.; Schneider, D.; Vogel, T.M. Microbial sequences retrieved from environmental samples from seasonal Arctic snow and meltwater from Svarlbard, Norway. Extremophiles 2010, 14, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Mänistö, M.K.; Tiirola, M.; McConnell, J.; Häggblom, M.M. Mucilaginibacter frigotolerans sp. nov., Mucilaginibacter lappiensis sp. nov. and Mucilaginibacter mallensis sp. nov., isolated from soild and lichen samples. Int. J. Syst. Evolut. Microbiol. 2010, 60, 2849–2856. [Google Scholar] [CrossRef] [PubMed]
- Mänistö, M.K.; Rawat, S.; Starovoytov, V.; Häggblom, M.M. Granulicella arctica sp. nov, Granulicella mallensis sp. nov, Granulicella tundricola sp. nov and Granulicella sapmiensis sp. nov, novel acidobacteria from tundra soil. Int. J. Syst. Evolut. Microbiol. 2012, 62, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.X.; Yan, M.; Yu, Y.; Li, H.R.; He, J.F.; Sun, K.; Zhang, F. Diversity of bacteria in surface ice of Austre Lovénbreen glacier, Svalbard. Arch. Microbiol. 2013, 195, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Dold, B.; González-Toril, E.; Aguilera, A.; Lopez-Palmo, E.; Cisternas, M.E.; Bucchi, F.; Amils, R. Acido rock drainage and rock weathering in Antarctica: Important sources for iron cycling in the Souther Ocean. Environ. Sci. Technol. 2013, 47, 6129–6136. [Google Scholar] [PubMed]
- Kaden, R.; Spröer, C.; Beyer, D.; Krolla-Sidenstein, P. Rhodoferax saidenbachensis sp. nov. a psychrotolerant, very slowly growing bacterium within the family Comamonadaceae, proposal of appropriate taxonomic position of Albidiferax ferrireducens strain TI18T in the genus Rhodoferax and emended description of the genus Rhodoferax. Int. J. Syst. Evolut. Microbiol. 2014, 64, 1186–1193. [Google Scholar]
- Hallberg, K.B.; González-Toril, E.; Johnson, D.B. Acidithiobacillus ferrivorans, sp. nov.: Facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 2010, 14, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Coupland, K.; Johnson, D.B. Evidence that the potential for dissimilatory ferric iron reduction is widespread among acidophilic heterotrophic bacteria. FEMS Microbiol. Lett. 2008, 279, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Yamanoi, K.; Kudo, T.; Ohkuma, M.; Takashina, T. Aciditerrimonas ferrireducens gen. nov., sp. nov., an iron-reducing thermoacidophilic actinobacterium isolated from a solfataric field. Int. J. Syst. Evolut. Microbiol. 2011, 61, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Kock, D.; Schippers, A. Quantitative microbial community analysis of three different sulfidic mine tailings dumps generating acid mine drainage. Appl. Environ. Microbiol. 2008, 74, 5211–5219. [Google Scholar] [CrossRef] [PubMed]
- Winsley, T.J.; Snape, I.; McKinlay, J.; Stark, J.; van Dorst, J.M.; Ji, M.; Ferrari, B.C.; Siciliano, S. The ecological controls on the prevalence of candidate division TM7 in Polar regions. Front. Microbiol. 2014, 5, 345. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, S.; Schlömann, M.; Johnson, D.B. The iron-oxidizing proteobacteria. Microbiology 2011, 157, 1551–1564. [Google Scholar] [CrossRef] [PubMed]
- Hallbeck, L.; Pedersen, K. Culture parameters regulating stalk formation and growth rate of Gallionella ferruginea. J. General Microbiol. 1990, 136, 1675–1680. [Google Scholar] [CrossRef]
- Fleming, E.J.; Langdon, A.E.; Martinez-Garcia, M.; Stepanasukas, R.; Poulton, N.J.; Masland, E.D.P.; Emerson, D. What’s new is old: Resolving the identity of Leptothrix ochracea using single cell genomics, pyrosequencing and FISH. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Krepski, S.T.; Hanson, T.E.; Chan, C.S. Isolation and characterization of a novel biomineral stalk-forming iron-oxidizing bacterium from a circumneutral groundwater seep. Environ. Microbiol. 2012, 14, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Krepski, S.; Chan, C.; Itoh, T.; Ohkuma, M. Ferriphaselus amnicola gen. nov. sp. nov., a neutrophilic, stalk-forming, iron-oxidizing bacterium isolated from an iron-rich groundwater seep. Int. J. Sys. Evol. Microbiol. 2014, 64, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Kim, S.J.; Lee, S.S. Gallionella ferruginea in ochreous precipitates from acid mine drainage in Donghae coal mine area, Korea. Geosci. J. 2003, 7, 289–292. [Google Scholar] [CrossRef]
- Hallberg, K.B.; Coupland, K.; Kimura, S.; Johnson, D.B. Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 2006, 72, 2022–2030. [Google Scholar] [CrossRef] [PubMed]
- Lear, G.; Niyogi, D.; Harding, J.; Dong, Y.; Lewis, G. Biofilms bacterial community structure in streams affected by acid mine drainage. Appl. Environ. Microbiol. 2009, 75, 3455–3460. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Bryan, C.G.; Hallberg, K.B.; Johnson, D.B. Biodiversity and geochemistry of an extremely acidic, low-temperature subterranean environment sustained by chemolithotrophy. Environ. Microbiol. 2011, 13, 2092–2104. [Google Scholar] [CrossRef] [PubMed]
- Borin, S.; Ventura, S.; Tambone, F.; Mapelli, F.; Schubotz, F.; Brusetti, L.; Scaglia, B.; D’Acqui, L.P.; Solheim, B.; Turicchia, S.; et al. Rock weathering creates oases of life in a High Arctic desert. Environ. Microbiol. 2010, 12, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Mapelli, F.; Marasco, R.; Rizzi, A.; Baldi, F.; Ventura, S.; Daffonchio, D.; Borin, S. Bacterial communities involved in soil formation and plant establishment triggered by pyrite bioweathering on Arctic moraines. Microb. Ecol. 2011, 61, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.S.; Hamilton, T.L.; Havig, J.R.; Skidmore, M.L.; Shock, E.L. Chemolithotrophic primary production in a subglacial ecosystem. Appl. Environ. Microbiol. 2014, 80, 6146–6153. [Google Scholar] [CrossRef] [PubMed]
- Bruneel, O.; Duran, R.; Casiot, C.; Elbaz-Poulichet, F.; Personné, J.C. Diversity of microorganisms in Fe-As-rich acid mine drainage water of Carnoulès, France. Appl. Environ. Microbiol. 2006, 72, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.J.; Kohl, C.; Grettenberge, C.; Larson, L.N.; Burgos, W.D.; Macalady, J.L. Geochemical niches for Fe-oxidizing acidophiles in an acidic coal mine drainage. Appl. Environ. Microbiol. 2015, 81, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Hallberg, K.; Hedrich, S. Uncovering a microbial enigma: Isolation and characterization of the streamer-generating, iron-oxidizing acidophilic bacterium “Ferrovum myxofaciens”. Appl. Environ. Microbiol. 2014, 80, 672–680. [Google Scholar] [CrossRef] [PubMed]
- García-Moyano, A.; González-Toril, E.; Aguilera, A.; Amils, R. Prokaryotic community composition and ecology of floating macroscopic filaments from an extreme acidic environment, Río Tinto (SW, Spain). Syst. Appl. Microbiol. 2007, 30, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Dold, B. Dissolution kinetics of schewertmannite and ferryhydrite in oxidized mine samples and their detection by differential X-ray diffraction (DXRD). Appl. Geochem. 2003, 18, 1531–1540. [Google Scholar] [CrossRef]
- Wang, H.; Bigham, J.M.; Jones, F.S.; Tuovinen, O.H. Synthesis and properties of ammoniojarosites prepared with iron-oxidizing acidophilic microorganisms at 22–65 °C. Geochim. Cosmochim. Acta 2007, 71, 155–164. [Google Scholar] [CrossRef]
- Elberling, B. Temperature and oxygen control on pyrite oxidation in frozen mine tailings. Cold Reg. Sci. Technol. 2005, 4, 121–133. [Google Scholar] [CrossRef]
- Auvinen, H.; Nevatalo, L.M.; Kaksonen, A.H.; Puhakka, J.A. Low-temperature (9 °C) AMD treatment in a sulfidogenic bioreactor dominated by a mesophilic Desulfomicrobium species. Biotechnol. Bioeng. 2009, 104, 740–751. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Moyano, A.; Austnes, A.E.; Lanzén, A.; González-Toril, E.; Aguilera, Á.; Øvreås, L. Novel and Unexpected Microbial Diversity in Acid Mine Drainage in Svalbard (78° N), Revealed by Culture-Independent Approaches. Microorganisms 2015, 3, 667-694. https://doi.org/10.3390/microorganisms3040667
García-Moyano A, Austnes AE, Lanzén A, González-Toril E, Aguilera Á, Øvreås L. Novel and Unexpected Microbial Diversity in Acid Mine Drainage in Svalbard (78° N), Revealed by Culture-Independent Approaches. Microorganisms. 2015; 3(4):667-694. https://doi.org/10.3390/microorganisms3040667
Chicago/Turabian StyleGarcía-Moyano, Antonio, Andreas Erling Austnes, Anders Lanzén, Elena González-Toril, Ángeles Aguilera, and Lise Øvreås. 2015. "Novel and Unexpected Microbial Diversity in Acid Mine Drainage in Svalbard (78° N), Revealed by Culture-Independent Approaches" Microorganisms 3, no. 4: 667-694. https://doi.org/10.3390/microorganisms3040667
APA StyleGarcía-Moyano, A., Austnes, A. E., Lanzén, A., González-Toril, E., Aguilera, Á., & Øvreås, L. (2015). Novel and Unexpected Microbial Diversity in Acid Mine Drainage in Svalbard (78° N), Revealed by Culture-Independent Approaches. Microorganisms, 3(4), 667-694. https://doi.org/10.3390/microorganisms3040667







