Molecular Insights into Ammonium Sulfate-Induced Secretome Reprogramming of Bacillus subtilis Czk1 for Enhanced Biocontrol Against Rubber Tree Root Rot
Abstract
1. Introduction
2. Materials and Methods
2.1. Extraction and Activity Detection of Antifungal Protein Crude Extract
2.2. Protein Extraction
2.3. Protein Digestion and Peptide Desalting
2.4. LC-MS/MS Analysis
2.5. Protein Discovery Results
2.6. Bioinformatics Analysis
2.6.1. Functional Annotation and Enrichment Analysis
2.6.2. Metabolic Pathway Visualization
3. Results
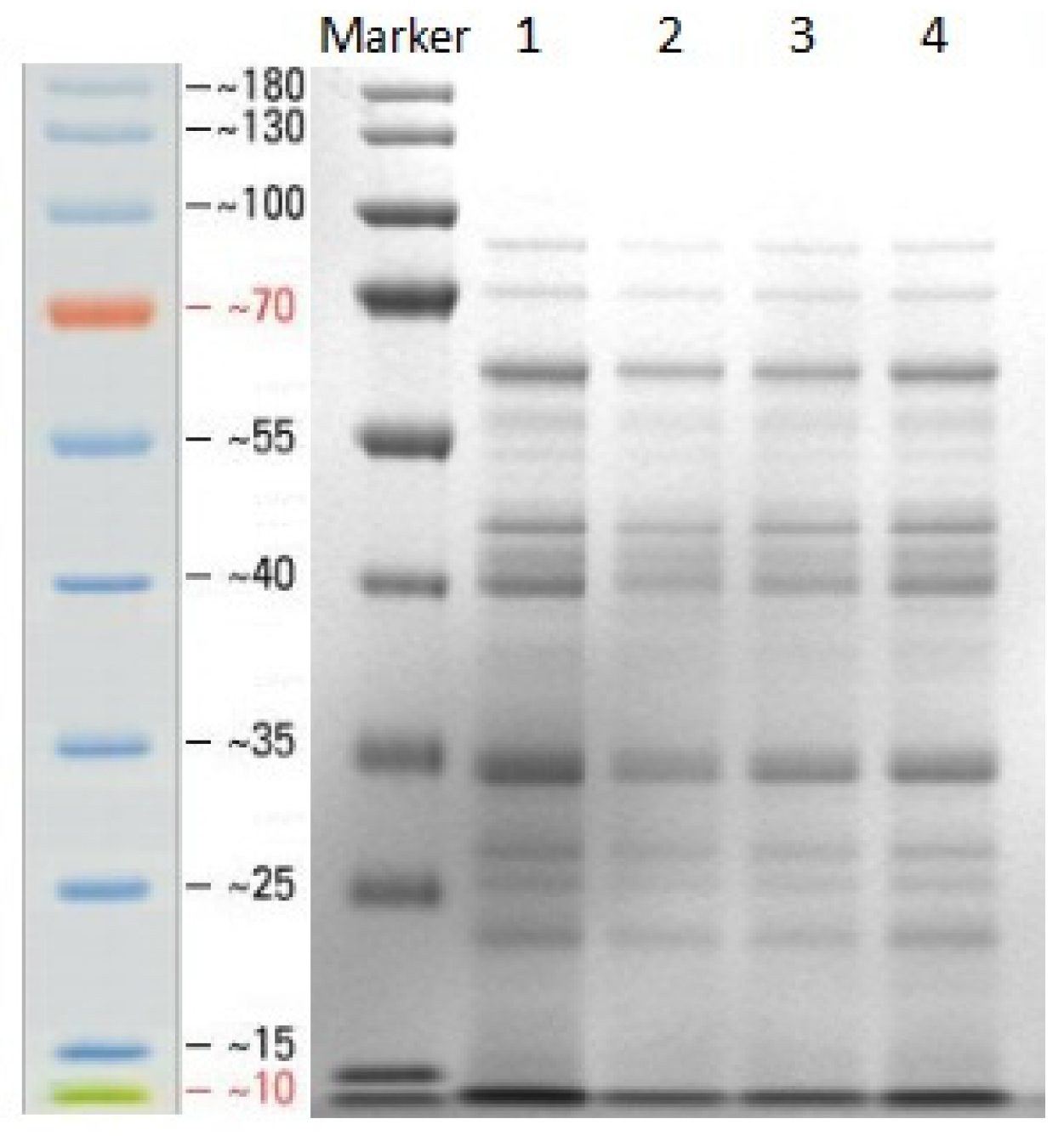
3.1. Extraction and Identification of Secreted Protein
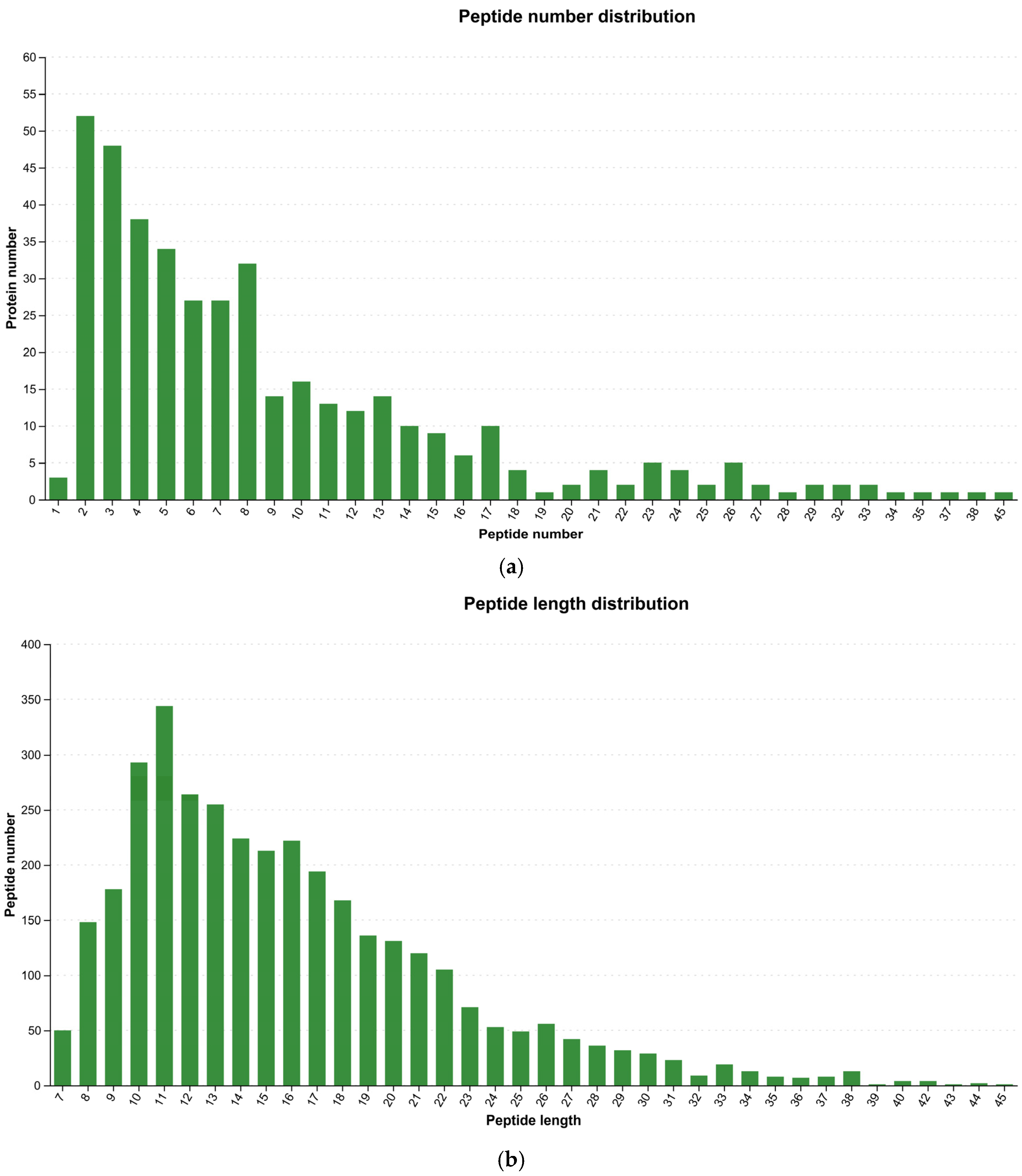
3.2. Peptide Segment Information
3.3. Distribution of Protein Identified by B. subtilis Czk1
3.4. Sample Correlation Heat Map
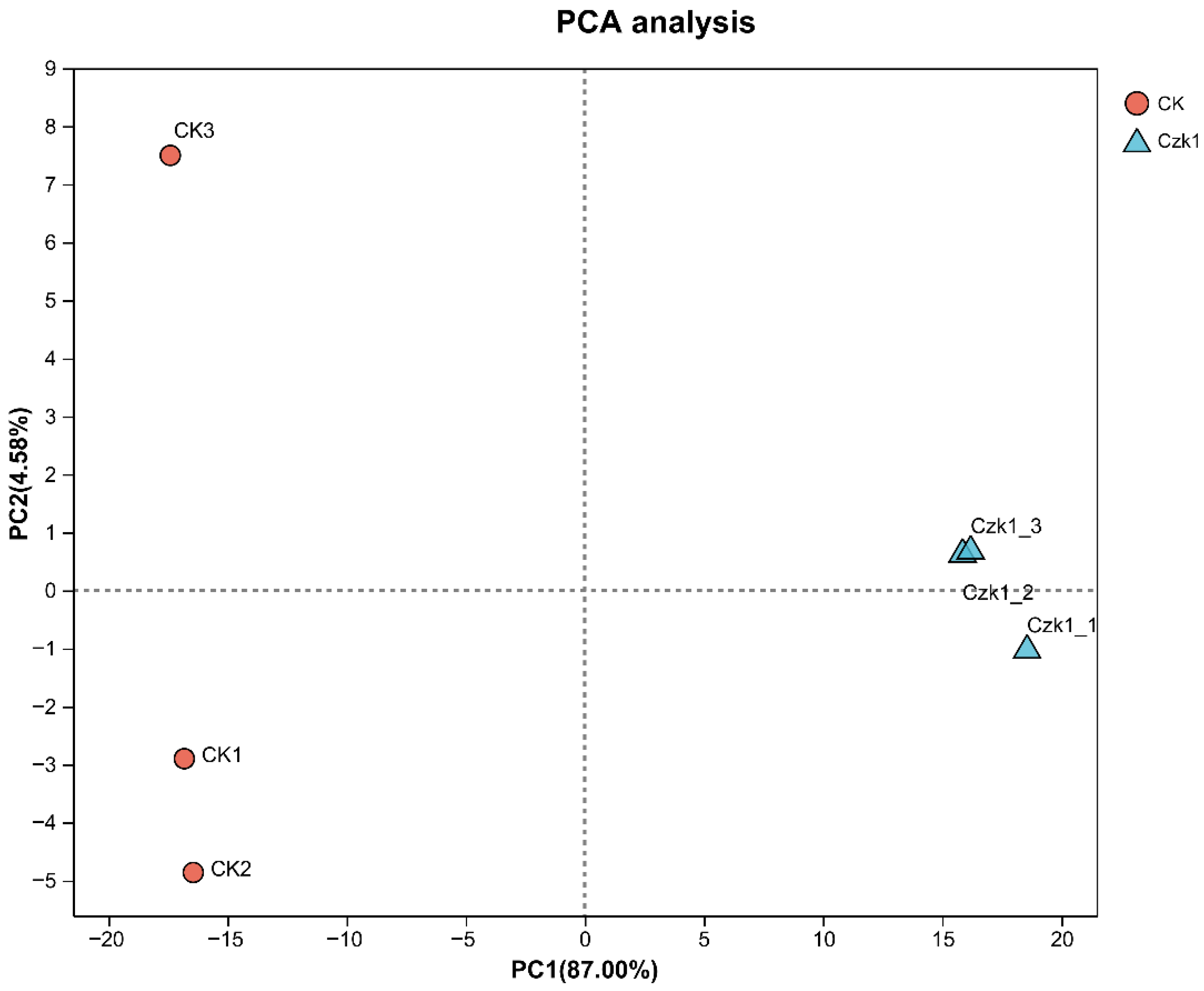
3.5. Principal Component Analysis
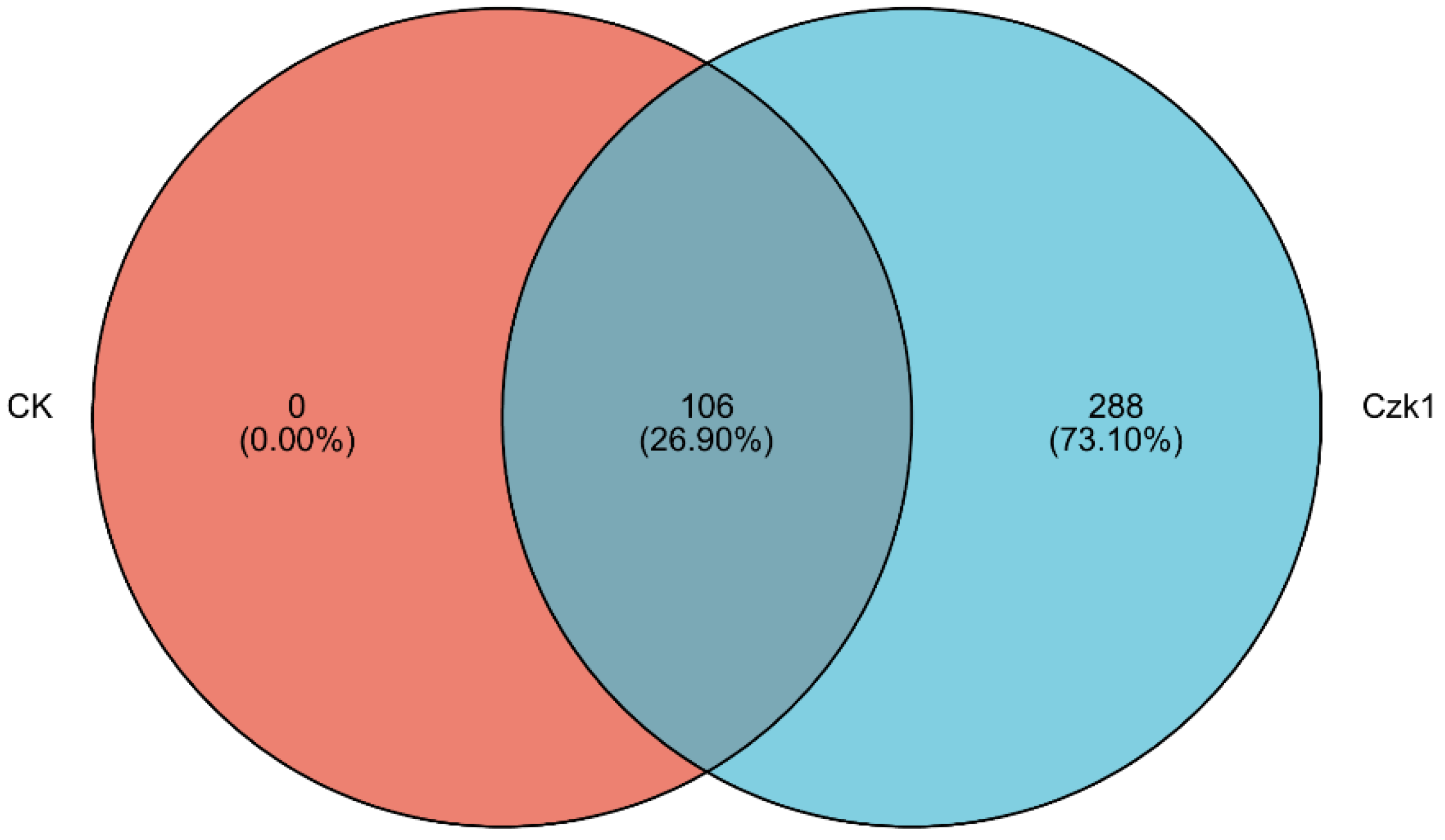
3.6. Analysis of Differentially Expressed Proteins
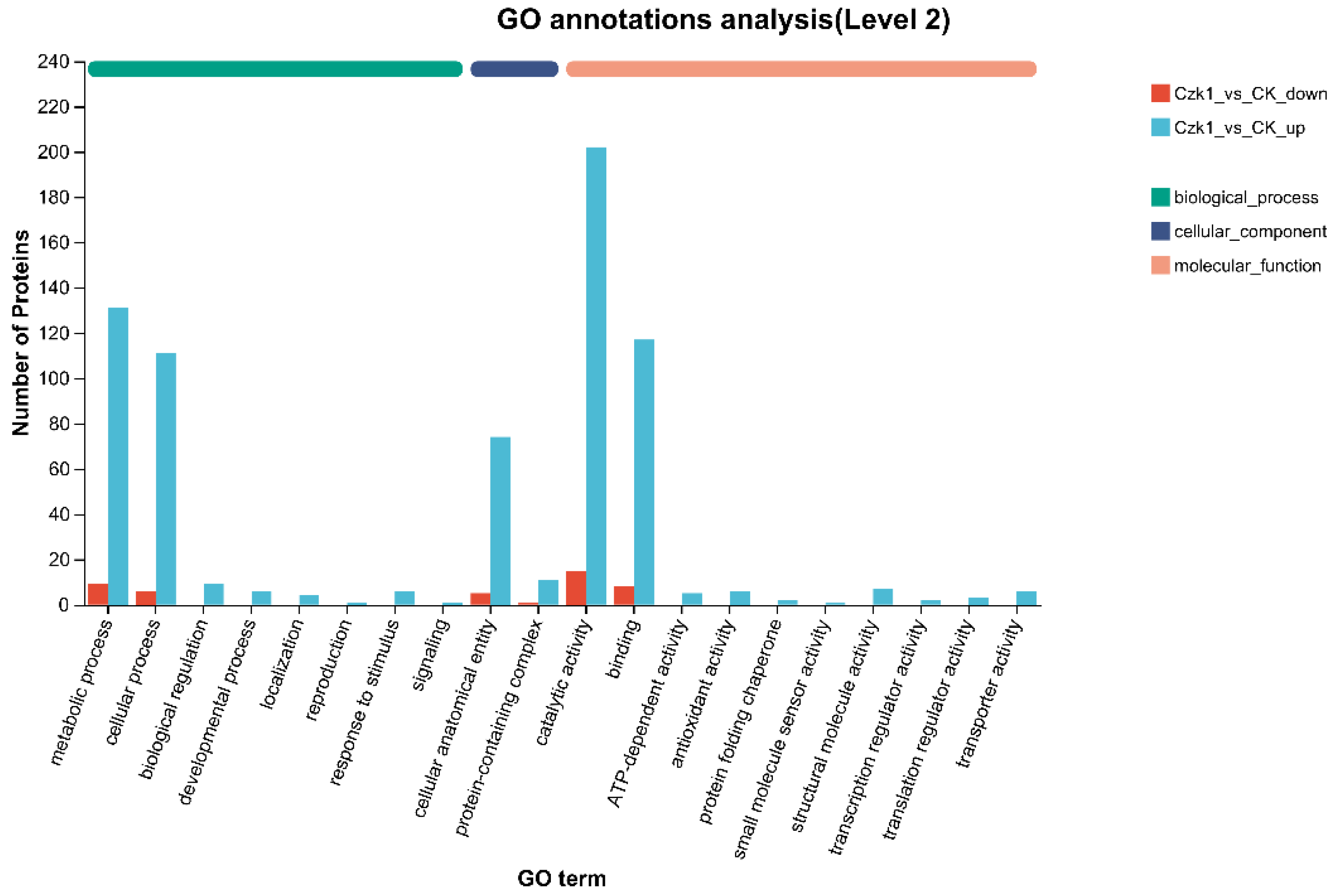
3.7. GO Function Annotation of Differentially Expressed Proteins
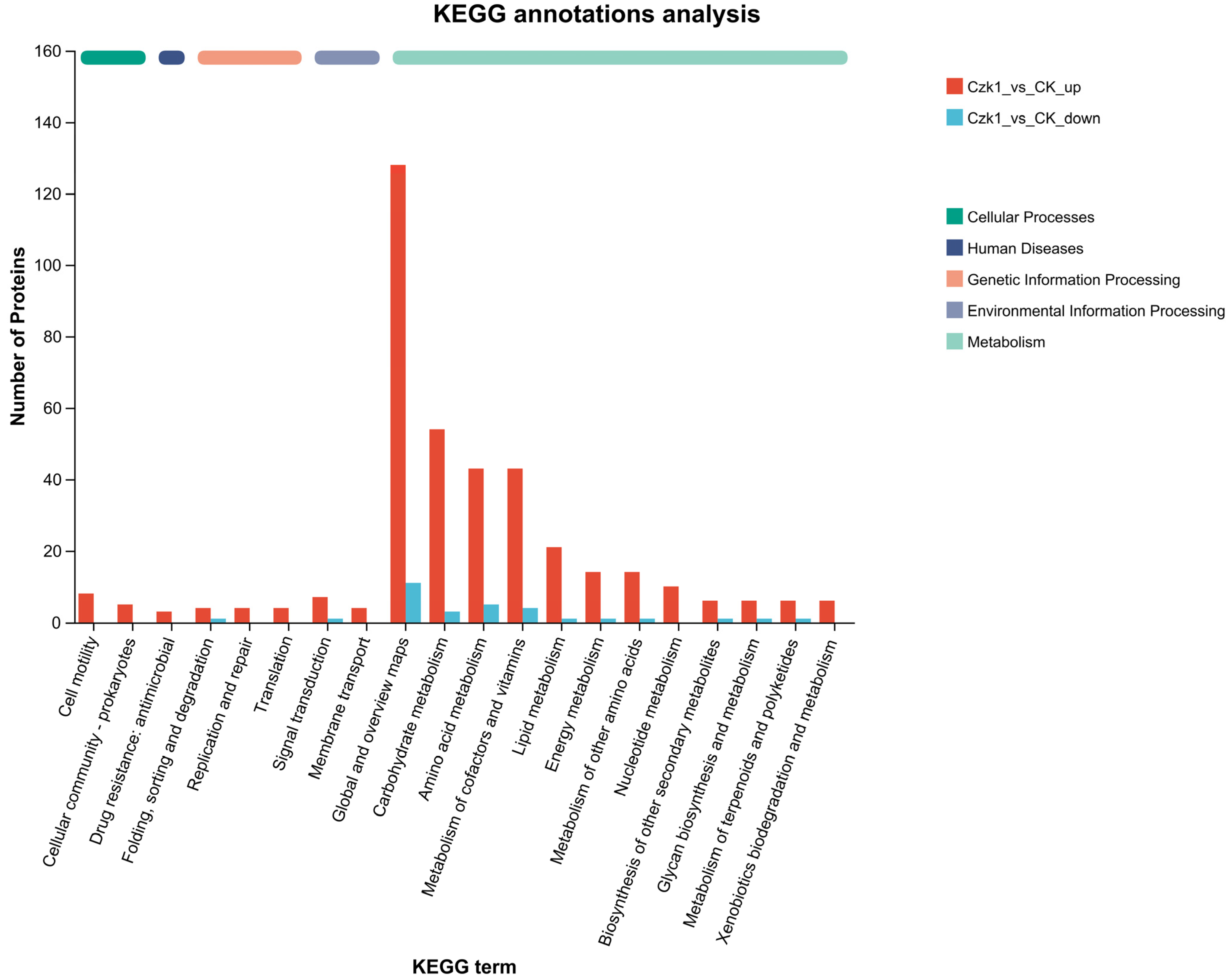
3.8. KEGG Metabolic Pathway of Differentially Expressed Proteins
3.9. EggNOG Annotation
3.10. Pfam Classification
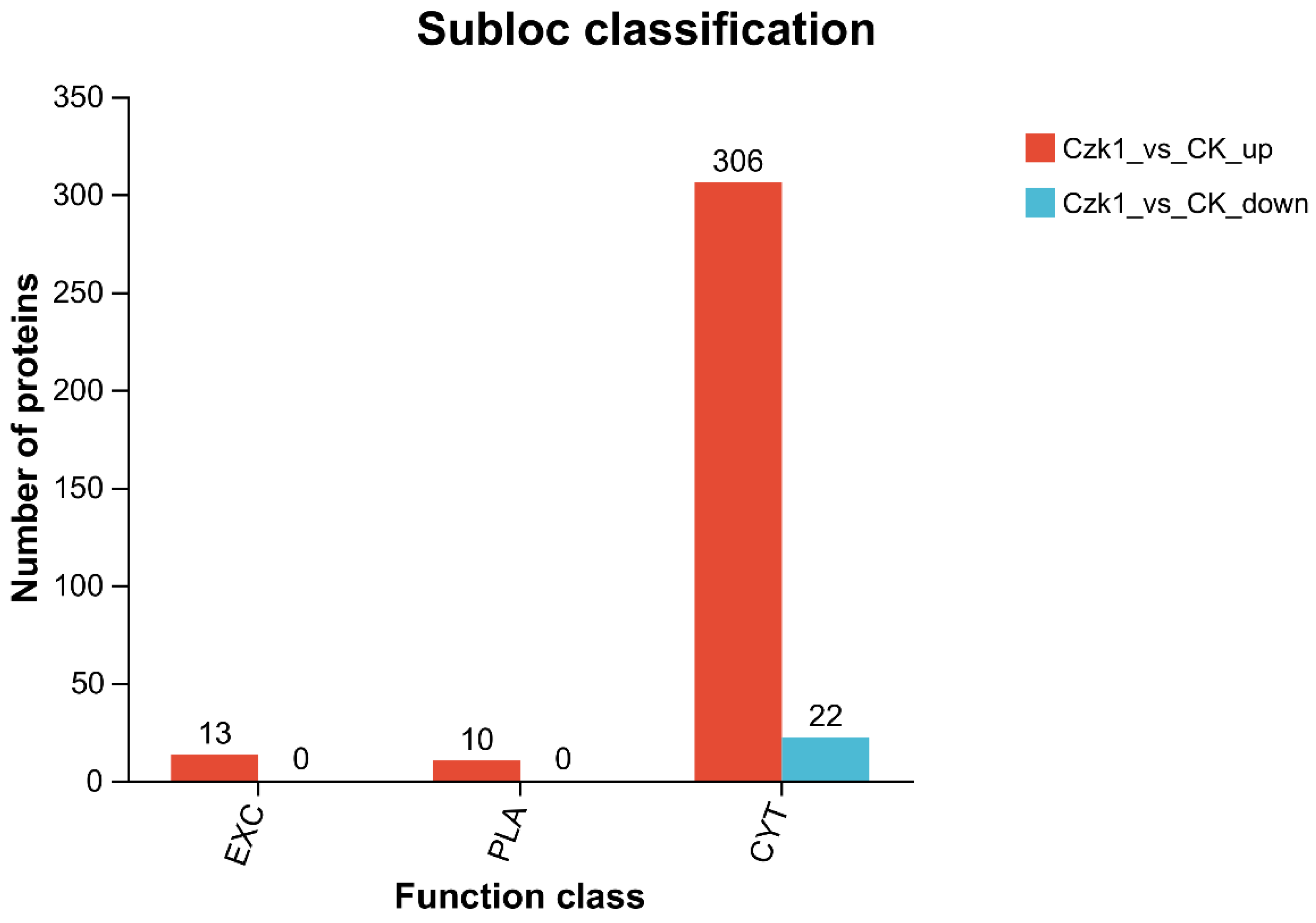
3.11. Subloc Annotation
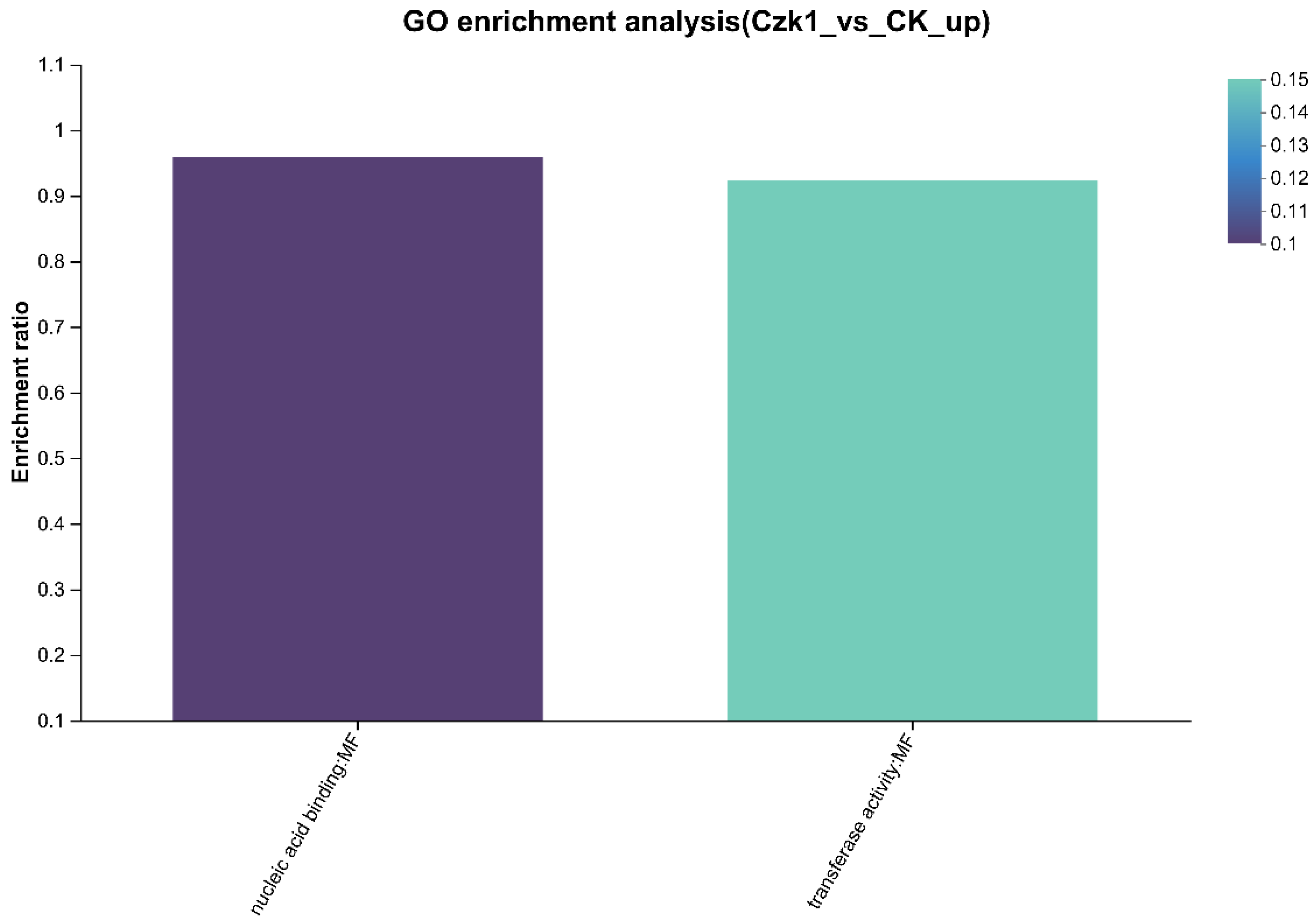
3.12. GO Enrichment
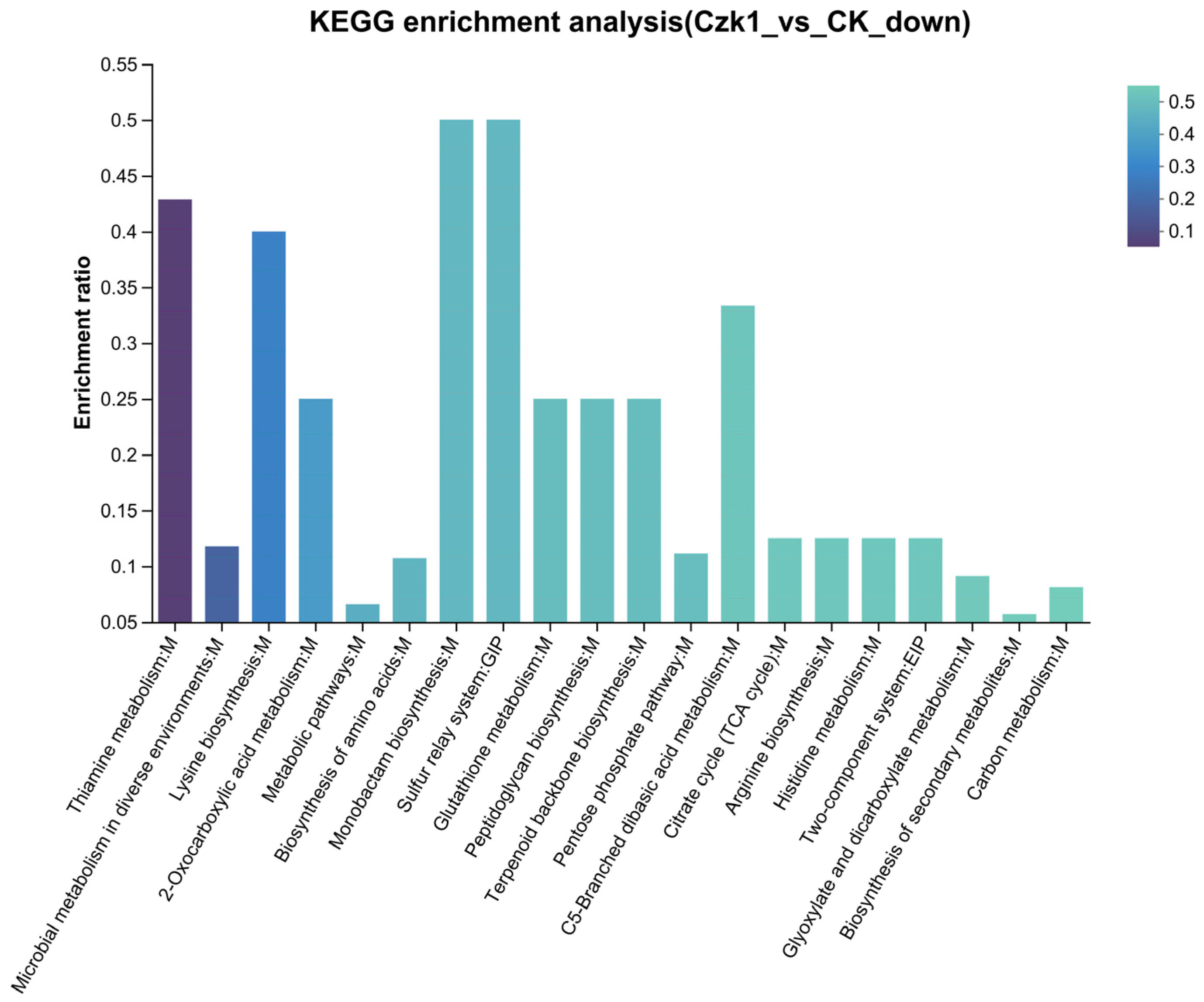
3.13. KEGG Pathway Enrichment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chao, J.; Yang, S.; Chen, Y.; Tian, W.M. Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis of the Gene Expression in Laticifers on the Basis of Latex Flow in Rubber Tree (Hevea brasiliensis Muell. Arg.). Front. Plant Sci. 2016, 7, 1149. [Google Scholar] [CrossRef]
- Jayashree, R.; Nazeem, P.A.; Rekha, K.; Sreelatha, S.; Thulaseedharan, A.; Krishnakumar, R.; Kala, R.G.; Vineetha, M.; Leda, P.; Jinu, U.; et al. Over-expression of 3-hydroxy-3- methylglutaryl-coenzyme A reductase 1 (hmgr1) gene under super-promoter for enhanced latex biosynthesis in rubber tree (Hevea brasiliensis Muell. Arg.). Plant Physiol. Biochem. 2018, 127, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Men, X.; Wang, F.; Chen, G.Q.; Zhang, H.B.; Xian, M. Biosynthesis of Natural Rubber: Current State and Perspectives. Int. J. Mol. Sci. 2019, 20, 50. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Cai, H.; Luo, Y.; Zhao, X.; Fu, Y.; Zou, L.; Zhou, Y.; Tu, M. Biocontrol Potential of Endophytic Bacillus velezensis LSR7 Against Rubber Red Root Rot Disease. J. Fungi 2024, 10, 849. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Yin, J.; Wu, H.; Liang, Y.; Wu, W.; Lu, Y.; Li, R.; Tan, S.; He, C.; Yi, K. Synergistic mechanism of Bacillus subtilis Czk1 combined with propiconazole and tebuconazole mixtures against Pyrrhoderma noxium. Chem. Biol. Technol. Agric. 2023, 10, 113. [Google Scholar] [CrossRef]
- Liu, Y.X.; Shi, Y.P.; Dai, L.M.; Li, L.L.; Cai, Z.Y. Screening, identification and fermentation optimization of an antimicrobial actinomycete strain 17-7 to Phellinus noxius. Microbiol. China 2020, 47, 118–129. [Google Scholar] [CrossRef]
- Tu, M.; Zhu, Z.; Zhao, X.; Cai, H.; Zhang, Y.; Yan, Y.; Yin, K.; Sha, Z.; Zhou, Y.; Chen, G.; et al. The versatile plant probiotic bacterium Bacillus velezensis SF305 reduces red root rot disease severity in the rubber tree by degradingthe mycelia of Ganoderma pseudoferreum. J. Integr. Agric. 2024, 24, 3112–3126. [Google Scholar] [CrossRef]
- Qin, Y.; Tu, M.; Yang, M.; Qi, J.; Tang, C. Antagonistic Effect of Eight Strains of Trichoderma on Ganoderma pseudoferreum Causing Red Root Rot of Rubber Tree. Chin. J. Trop. Agric. 2015, 3, 41–45+53. [Google Scholar] [CrossRef]
- Liu, T.Y.; Chen, C.H.; Yang, Y.L.; Tsai, I.J.; Ho, Y.N.; Chung, C.L. The brown root rot fungus Phellinus noxius affects microbial communities in different root-associated niches of Ficus trees. Environ. Microbiol. 2022, 24, 276–297. [Google Scholar] [CrossRef]
- Liao, T.Z.; Chen, Y.H.; Tsai, J.N.; Chao, C.; Huang, T.P.; Hong, C.F.; Wu, Z.C.; Tsai, I.J.; Lee, H.H.; Klopfenstein, N.B.; et al. Translocation of Fungicides and Their Efficacy in Controlling Phellinus noxius, the Cause of Brown Root Rot Disease. Plant Dis. 2023, 107, 2039–2053. [Google Scholar] [CrossRef]
- Wang, Y.; Meng, H.; Gu, V.; Gu, J. Molecular diagnosis of the brown root rot disease agent Phellinus noxius on trees and in soil by rDNA ITS analysis. Appl. Environ. Microbiol. 2016, 1, 81–91. [Google Scholar] [CrossRef]
- Peng, Y.; Li, S.J.; Yan, J.; Tang, Y.; Cheng, J.P.; Gao, A.J.; Yao, X.; Ruan, J.J.; Xu, B.L. Research progress on phytopathogenic fungi and their role as biocontrol agents. Front. Microbiol. 2021, 12, 670135. [Google Scholar] [CrossRef]
- Qiu, D.; Ke, M.; Zhang, Q.; Zhang, F.; Lu, T.; Sun, L.; Qian, H. Response of microbial antibiotic resistance to pesticides: An emerging health threat. Sci. Tot. Environ. 2022, 850, 158057. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of Action of Microbial Biological Control Agents Against Plant Diseases: Relevance Beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as Biological Control Agents of Plant Diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef]
- Devi, S.I.; Kiesewalter, H.T.; Kovács, R.; Frisvad, J.C.; Weber, T.; Larsen, T.O.; Kovács, Á.T.; Ding, L. Depiction of secondary metabolites and antifungal activity of Bacillus velezensis DTU001. Synth. Syst. Biotechnol. 2019, 4, 142–149. [Google Scholar] [CrossRef]
- Khan, M.H.; Salman, M.; Jan, S.A.; Shinwari, Z.K. Biological control of fungal phytopathogens: A comprehensive review based on Bacillus species. MOJ Biol. Med. 2021, 6, 90–92. [Google Scholar] [CrossRef]
- Tuyến, Đ.T.; Trung, N.T.; Thảo, N.T.; Thanh, N.S.L.; Nguyen, N.P.D.; Tuyết, N.T.; Cuong, N.T.; Chan, S.S.; Khoo, K.S.; Show, P.L. Antifungal activity of secondary metabolites purified from Bacillus subtilis isolated in Vietnam and evaluated on in vitro and in vivo models. Int. Biodeter. Biodegr. 2023, 179, 105558. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, Z.; Shi, Y.; Guo, D.; Pang, B.; Chen, X.; Shao, D.; Liu, Y.; Shi, J. Bacillus subtilis inhibits Aspergillus carbonarius by producing iturin A, which disturbs the transport, energy metabolism, and osmotic pressure of fungal cells as revealed by transcriptomics analysis. Int. J. Food Microbiol. 2020, 330, 108783. [Google Scholar] [CrossRef]
- Kaspar, F.; Neubauer, P.; Gimpel, M. Bioactive Secondary Metabolites from Bacillus subtilis: A Comprehensive Review. J. Nat. Prod. 2019, 82, 2038–2053. [Google Scholar] [CrossRef]
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 2, 255–261. [Google Scholar] [CrossRef]
- Mertins, P.; Qiao, J.; Patel, J.; Udeshi, N.D.; Clauser, K.R.; Mani, D.R.; Burgess, M.; Gillette, M.A.; Jaffe, J.D.; Carr, S.A. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 2013, 10, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Nikolovski, N.; Shliaha, P.V.; Gatto, L.; Dupree, P.; Lilley, K.S. Label-Free Protein Quantification for Plant Golgi Protein Localization and Abundance. Plant Physiol. 2014, 166, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Toprakcioglu, Z.; Challa, P.K.; Xu, C.K.; Knowles, T.P.J. Label-Free Analysis of Protein Aggregation and Phase Behavior. ACS Nano 2019, 13, 13940–13948. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Dong, C.; Yao, C.; Ding, Y.; Zhou, X. Preliminary Analysis of Secreted Proteins of Phytophthora infestans Under Nitrogen Starvation Condition Based on Label Free Quantitative Proteomics. Southwest China J. Agric. Sci. 2019, 32, 2324–2329. [Google Scholar]
- Liu, C.; Niu, G.; Li, X.; Zhang, H.; Chen, H.; Hou, D.; Lan, P.; Hong, Z. Comparative Label-Free Quantitative Proteomics Analysis Reveals the Essential Roles of N-Glycans in Salt Tolerance by Modulating Protein Abundance in Arabidopsis. Front. Plant Sci. 2021, 12, 646425. [Google Scholar] [CrossRef]
- Song, C.; Zhang, Y.; Chen, R.; Zhu, F.; Wei, P.; Pan, H.; Chen, C.; Dai, J. Label-Free Quantitative Proteomics Unravel the Impacts of Salt Stress on Dendrobium huoshanense. Front. Plant Sci. 2022, 13, 874579. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Wang, Z.; Li, X.; Ge, X.; Min, L.; Wang, J.; Zhou, P. Effects of liquid nitrogen freezing, immersion freezing, and air freezing on properties of Perca fluviatilis fillets and analysis of potential protein markers based on label-free proteomics. Food Biosci. 2024, 59, 104262. [Google Scholar] [CrossRef]
- Menezes, L.A.A.; Pimentel, M.; Alves, T.D.O.; Nascimento, T.P.D.; Evaristo, J.A.M.; Nogueira, F.C.S.; Ferreira, M.S.L.; Lindner, J.D.D. Label-free quantitative proteomics to exploit the impact of sourdough fermentation on reducing wheat allergenic fractions. Food Chem. 2024, 430, 137037. [Google Scholar] [CrossRef]
- He, C.P.; Fan, L.Y.; Wu, W.H.; Liang, Y.Q.; Li, R.; Tang, W.; Zheng, X.L.; Xiao, Y.N.; Liu, Z.X.; Zheng, F.C. Identification of lipopeptides produced by Bacillus subtilis Czk1 isolated from the aerial roots of rubber trees. Genet. Mol. Res. 2017, 16, gmr16018710. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wu, W.; Li, R.; Lu, Y.; Wang, G.; Tang, S.; Chen, H.; Xi, J.; Xing, H.; He, C.; et al. Evaluation of Bacillus subtilis Czk1 Metabolites by LC–MS/MS and Their Antifungal Potential Against Pyrrhoderma noxium Causing Brow Rot Disease. Agriculture 2023, 13, 1396. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, X.; Huang, A. Proteomic analysis and food-grade enzymes of Moringa oleifer Lam. a Lam. flower. Int. J. Biol. Macromol. 2018, 115, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Luján, R.; Pont, L.; Minić, Z.; Berezovski, M.V.; Sanz-Nebot, V.; Benavente, F. Characterization and differentiation of quinoa seed proteomes by label-free mass spectrometry-based shotgun proteomics. Food Chem. 2021, 363, 130250. [Google Scholar] [CrossRef] [PubMed]
- Yeung, J.; Gadjeva, M.; Geddes-McAlister, J. Label-Free Quantitative Proteomics Distinguishes General and Site-Specific Host Responses to Pseudomonas aeruginosa Infection at the Ocular Surface. Proteomics 2020, 20, e1900290. [Google Scholar] [CrossRef]
- Singh, L.K.; Pandey, M.; Baithalu, R.K.; Fernandes, A.; Ali, S.A.; Jaiswal, L.; Pannu, S.; Neeraj; Mohanty, T.K.; Kumaresan, A.; et al. Comparative Proteome Profiling of Saliva Between Estrus and Non-Estrus Stages by Employing Label-Free Quantitation (LFQ) and Tandem Mass Tag (TMT)-LC-MS/MS Analysis: An Approach for Estrus Biomarker Identification in Bubalus bubalis. Front. Genet. 2022, 13, 867909. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Xu, J.; Xu, J.; Tong, Z.; Du, B.; Liu, B.; Mu, X.; Guo, T.; Yu, S.; Liu, S.; Gao, C.; et al. Performance of feature extraction method for classification and identification of proteins based on three-dimensional fluorescence spectrometry. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 285, 121841. [Google Scholar] [CrossRef]
- Ibrahim, K.A. AI-driven detection and analysis of label-free protein aggregates. Nat. Rev. Mol. Cell Biol. 2024, 25, 337. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Andreeva, A.; Blum, M.; Chuguransky, S.R.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Llinares-López, F.; Meng-Papaxanthos, L.; et al. The Pfam protein families database: Embracing AI/ML. Nucleic Acids Res. 2025, 53, D523–D534. [Google Scholar] [CrossRef]
- Darzi, Y.; Letunic, I.; Bork, P.; Yamada, T. iPath3.0: Interactive pathways explorer v3. Nucleic Acids Res. 2018, 46, W510–W513. [Google Scholar] [CrossRef]
- Liu, H.; Chen, B.; Hu, S.; Liang, X.; Lu, X.; Shao, Y. Quantitative Proteomic Analysis of Germination of Nosema bombycis Spores under Extremely Alkaline Conditions. Front. Microbiol. 2016, 7, 1459. [Google Scholar] [CrossRef]
- Rooden, E.J.V.; Florea, B.I.; Deng, H.; Baggelaar, M.P.; Esbroeck, A.C.M.V.; Zhou, J.; Overkleeft, H.S.; Stelt, M.V.D. Mapping in vivo target interaction profiles of covalent inhibitors using chemical proteomics with label-free quantification. Nat. Protoc. 2018, 13, 752–767. [Google Scholar] [CrossRef]
- Yu, X.; Wang, Y.; Kohnen, M.V.; Piao, M.; Tu, M.; Gao, Y.; Lin, C.; Zuo, Z.; Gu, L. Large Scale Profiling of Protein Isoforms Using Label-Free Quantitative Proteomics Revealed the Regulation of Nonsense-Mediated Decay in Moso Bamboo (Phyllostachys edulis). Cells 2019, 8, 744. [Google Scholar] [CrossRef]
- Wang, P.; Xia, Q.; Li, Y. Label free-based proteomic analysis of proteins in Bacillus cereus spores regulated by high pressure processing and slightly acidic electrolyzed water treatment. Food Control 2018, 90, 392–400. [Google Scholar] [CrossRef]
- Panda, A.; Rangani, J.; Parida, A.K. Comprehensive proteomic analysis revealing multifaceted regulatory network of the xero-halophyte Haloxylon salicornicum involved in salt tolerance. J. Biotechnol. 2020, 324, 143–161. [Google Scholar] [CrossRef]
- Yang, M.; Ma, L.; Yang, X.; Li, L.; Chen, S.; Qi, B.; Wang, Y.; Li, C.; Wei, Y.; Zhao, Y. Photosynthetic Protein-Based Edible Quality Formation in Various Porphyra dentata Harvests Determined by Label-Free Proteomics Analysis. Cells 2022, 11, 1136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liang, X.; Zhu, Q.; Liu, A.; Bi, J.; Quan, Z.S.; Luo, X.; Zheng, Y.; Yang, N.; Yue, X.; et al. Label-free quantitative proteomic analysis of milk fat globule membrane proteins in porcine colostrum and mature milk. Food Chem. 2023, 426, 136447. [Google Scholar] [CrossRef]
- Jia, Z.; Zhou, J.; Han, J.; Liu, D.; Lv, R. Proteomics-based analysis of the stress response of Bacillus cereus spores under ultrasound and electrolyzed water treatment. Ultrason. Sonochem. 2023, 98, 106523. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Lai, H.; Zhao, W.; Cui, M.; Zhang, X. Antibacterial mechanism of bacteriocin from Lactobacillus plantarum YP36 based on proteomics technology. China Brew. 2024, 43, 98–104. [Google Scholar]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Tan, S.; Lu, Y.; Chen, H.; Huang, X.; Yi, K.; He, C.; Wu, W. Molecular Insights into Ammonium Sulfate-Induced Secretome Reprogramming of Bacillus subtilis Czk1 for Enhanced Biocontrol Against Rubber Tree Root Rot. Microorganisms 2025, 13, 2212. https://doi.org/10.3390/microorganisms13092212
Liang Y, Tan S, Lu Y, Chen H, Huang X, Yi K, He C, Wu W. Molecular Insights into Ammonium Sulfate-Induced Secretome Reprogramming of Bacillus subtilis Czk1 for Enhanced Biocontrol Against Rubber Tree Root Rot. Microorganisms. 2025; 13(9):2212. https://doi.org/10.3390/microorganisms13092212
Chicago/Turabian StyleLiang, Yanqiong, Shibei Tan, Ying Lu, Helong Chen, Xing Huang, Kexian Yi, Chunping He, and Weihuai Wu. 2025. "Molecular Insights into Ammonium Sulfate-Induced Secretome Reprogramming of Bacillus subtilis Czk1 for Enhanced Biocontrol Against Rubber Tree Root Rot" Microorganisms 13, no. 9: 2212. https://doi.org/10.3390/microorganisms13092212
APA StyleLiang, Y., Tan, S., Lu, Y., Chen, H., Huang, X., Yi, K., He, C., & Wu, W. (2025). Molecular Insights into Ammonium Sulfate-Induced Secretome Reprogramming of Bacillus subtilis Czk1 for Enhanced Biocontrol Against Rubber Tree Root Rot. Microorganisms, 13(9), 2212. https://doi.org/10.3390/microorganisms13092212





