Development of an RPA-CRISPR/LbaCas12a-Lateral Flow Assay for the Visual Detection of Chrysotila dentata (Haptophyta)
Abstract
1. Introduction
2. Materials and Methods
2.1. Algal Species Culturing
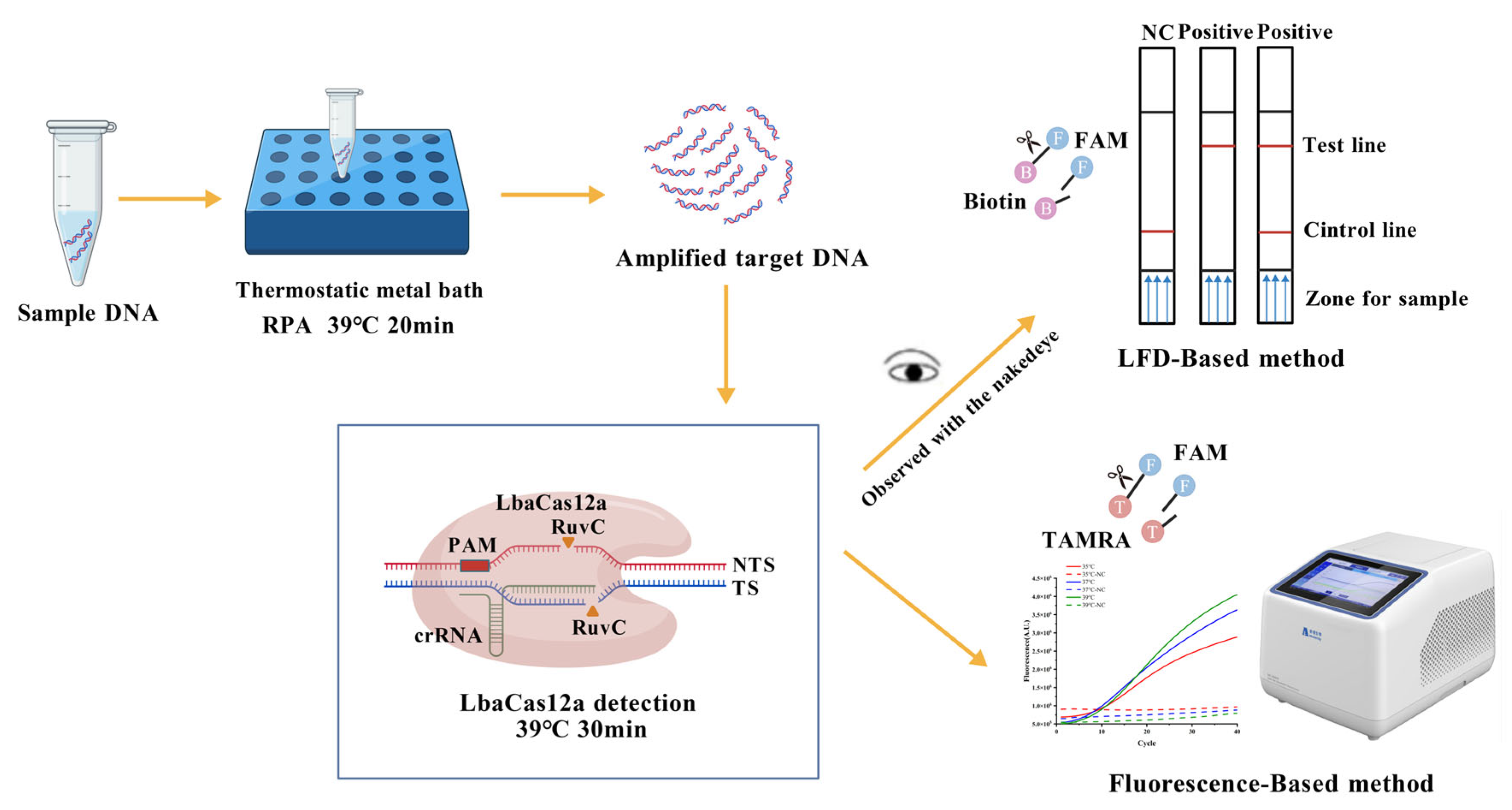
2.2. Construction of RPA-CRISPR/LbaCas12a-LFD System
2.3. Design and Screening of crRNA and RPA Primers
2.4. Optimization of RPA-CRISPR/LbaCas12a-LFD Reaction System
2.4.1. Optimization of ssDNA Reporter Concentration
2.4.2. Optimization of Reaction Time
2.4.3. Optimization of Reaction Temperature
2.5. Specificity and Sensitivity Evaluation
2.6. Environmental Water Samples Analysis
3. Results
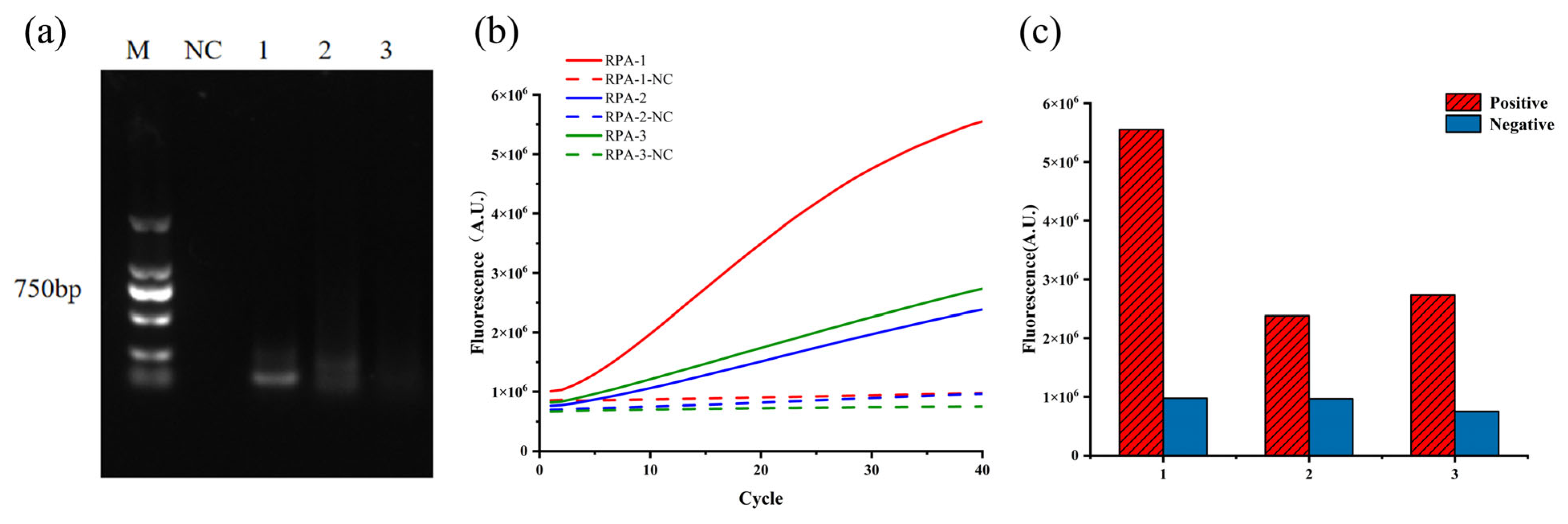
3.1. Optimal crRNA and RPA Primers
3.2. Optimizing the Reaction Conditions for the RPA-CRISPR/LbaCas12a-LFD Platform
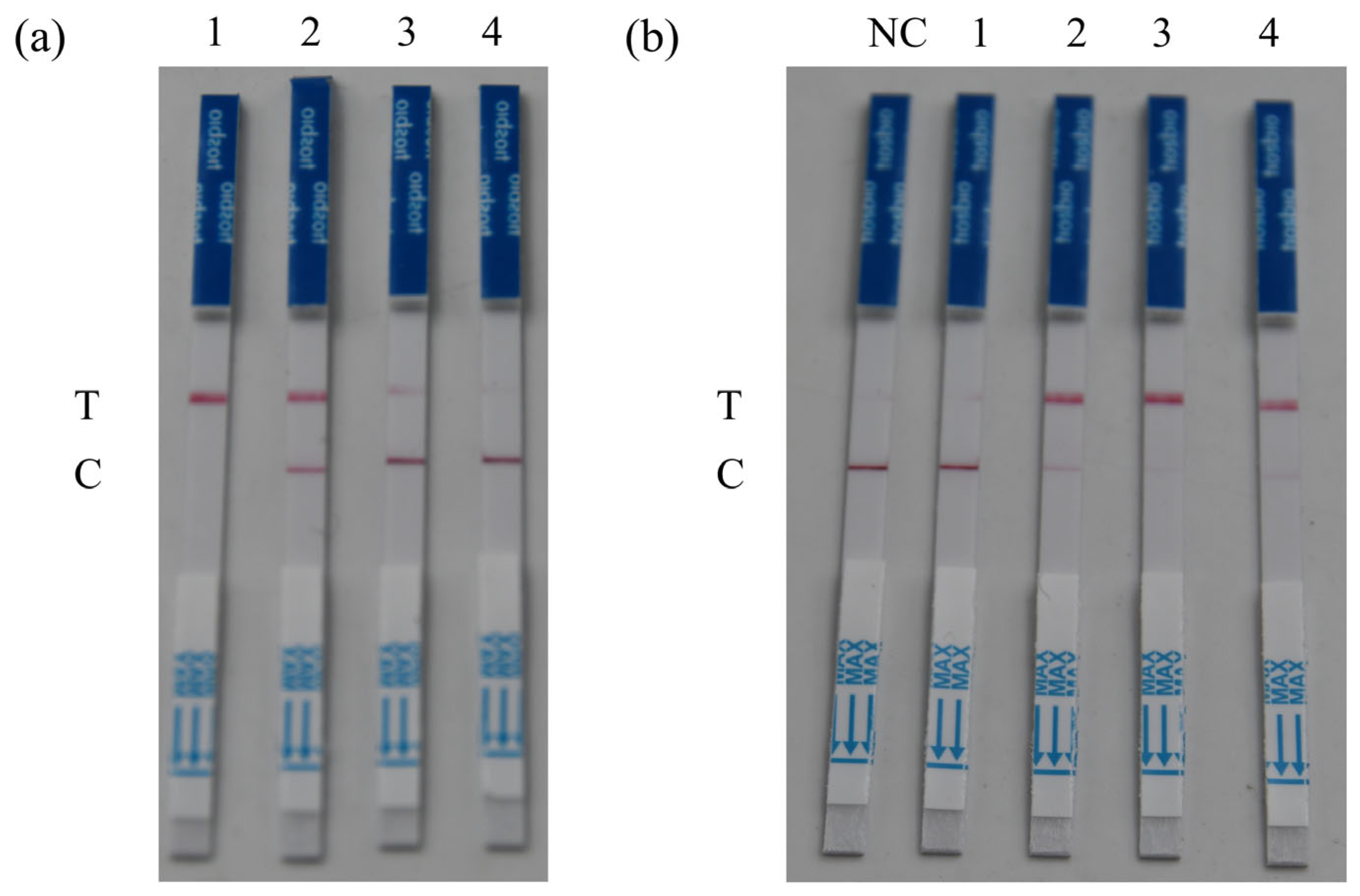
3.2.1. Optimal ssDNA Reporter Concentration
3.2.2. Optimal Reaction Time
3.2.3. Optimal Reaction Temperature
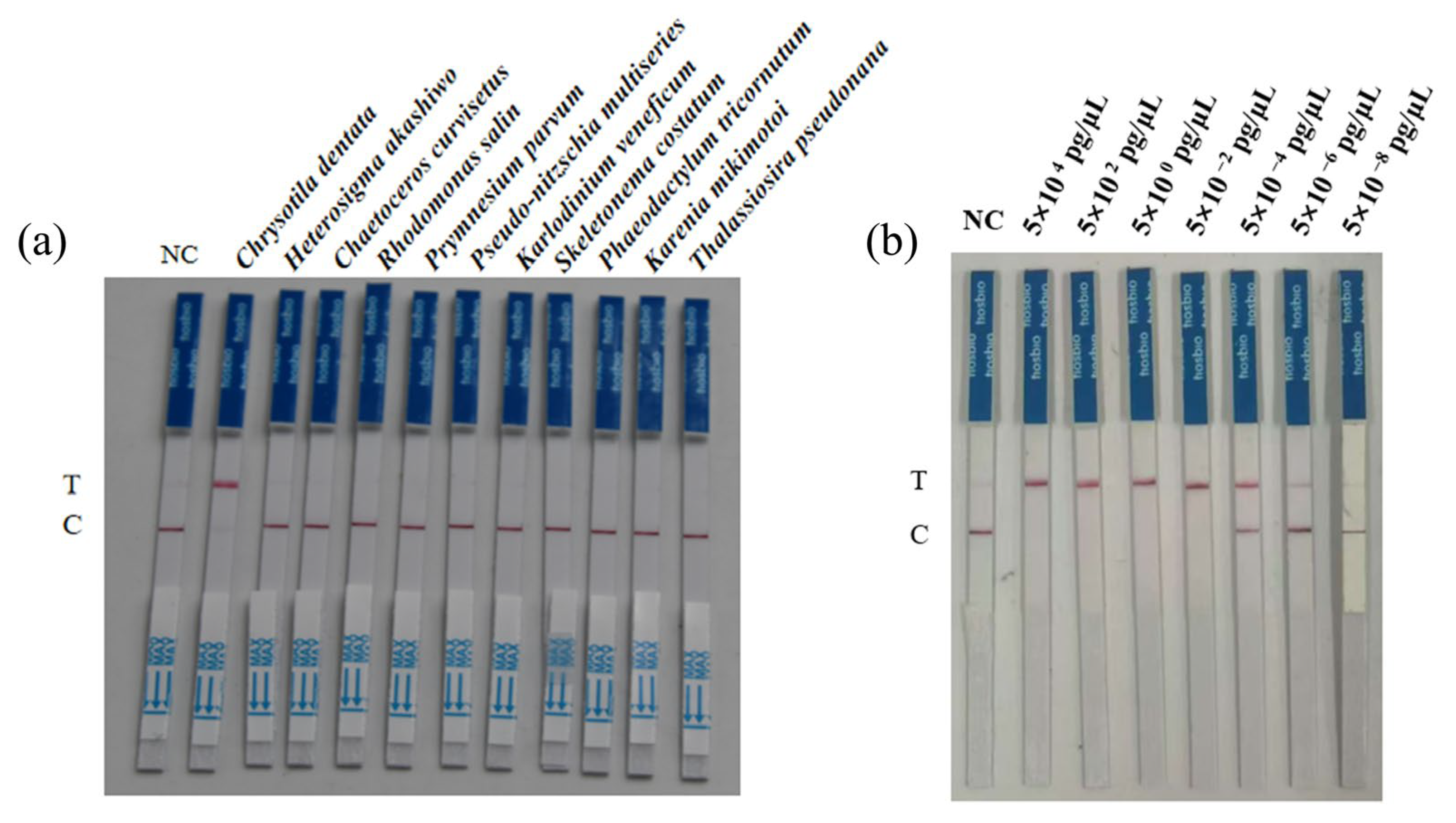
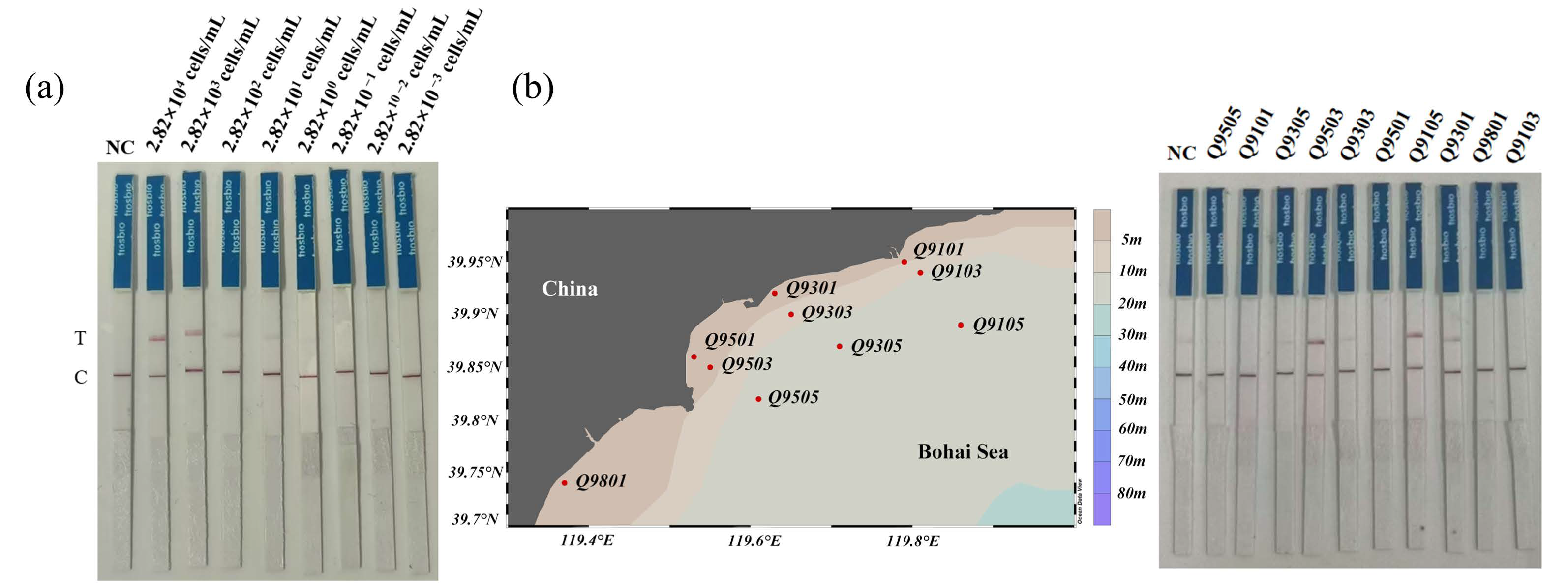
3.3. Evaluation of Detection Specificity and Sensitivity
3.4. Testing of Environmental Water Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thangaraj, S.; Liu, H.; Guo, Y.; Ding, C.; Kim, I.N.; Sun, J. Transitional traits determine the acclimation characteristics of the coccolithophore Chrysotila dentata to ocean warming and acidification. Environ. Microbiol. 2023, 25, 1099–1117. [Google Scholar] [CrossRef]
- Brown, C.W.; Yoder, J.A. Distribution pattern of coccolithophorid blooms in the western north Atlantic Ocean. Cont. Shelf Res. 1994, 14, 175–197. [Google Scholar] [CrossRef]
- Taylor, A.R.; Brownlee, C.; Wheeler, G. Coccolithophore cell biology: Chalking up progress. Annu. Rev. Mar. Sci. 2017, 9, 283–310. [Google Scholar] [CrossRef]
- Sáez, A.G.; Probert, I.; Geisen, M.; Quinn, P.; Young, J.R.; Medlin, L.K. Pseudo-cryptic speciation in coccolithophores. Proc. Natl. Acad. Sci. USA 2003, 100, 7163–7168. [Google Scholar] [CrossRef]
- An, X.L.; Yao, Q.; Pan, J. Coastal Red Tides in Hebei Province; China Environmental Science Press: Beijing, China, 2011. [Google Scholar]
- Endo, H.; Umezawa, Y.; Takeda, S.; Suzuki, K. Haptophyte communities along the Kuroshio current reveal their geographical sources and ecological traits. Mol. Ecol. 2023, 32, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Fon, M.; Chen, A.; Ghesquière, R.; Uhlig, S.; Edvardsen, B.; Solhaug, A. The cytotoxicity of the haptophyte Chrysochromulina leadbeateri towards the Atlantic salmon gill cell line ASG-10. Harmful Algae 2025, 142, 102797. [Google Scholar] [CrossRef] [PubMed]
- Paudel, Y.P.; Hu, Z.; Khatiwada, J.R.; Fan, L.; Pradhan, S.; Liu, B.; Qin, W. Chloroplast genome analysis of Chrysotila dentata. Gene 2021, 804, 145871. [Google Scholar] [CrossRef] [PubMed]
- Penna, A.; Bertozzini, E.; Battocchi, C.; Galluzzi, L.; Giacobbe, M.G.; Vila, M.; Garces, E.; Lugliè, A.; Magnani, M. Monitoring of HAB species in the Mediterranean Sea through molecular methods. J. Plankton Res. 2007, 29, 19–38. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, C.; Wang, Y.; Chen, G. A review of the current and emerging detection methods of marine harmful microalgae. Sci. Total Environ. 2022, 815, 152913. [Google Scholar] [CrossRef]
- McQuillan, J.S.; Alrefaey, A.; Turner, A.D.; Morrell, N.; Stoner, O.; Brown, R.; Kay, S.; Cooke, S.; Bage, T. Quantitative Polymerase Chain Reaction for the estimation of toxigenic microalgae abundance in shellfish production waters. Harmful Algae 2023, 128, 102497. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR/Cas technologies and applications. Nat. Rev. Mol. Cell Bio. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Delaney, J.A.; Ulrich, R.M.; Paul, J.H. Detection of the toxic marine diatom Pseudo-nitzschia multiseries using the RuBisCO small subunit (rbcS) gene in two real-time RNA amplification formats. Harmful Algae 2011, 11, 54–64. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, C.; Wang, Y.; Chen, G. Advances in multiplex molecular detection technologies for harmful algae. Environ. Sci. Pollut. Res. 2022, 29, 43745–43757. [Google Scholar] [CrossRef]
- de Oliveira, K.G.; Borba, J.C.; Bailao, A.M.; de Almeida Soares, C.M.; Carrilho, E.; Duarte, G.R.M. Loop-mediated isothermal amplification in disposable polyester-toner microdevices. Anal. Biochem. 2017, 534, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.F.; Zhu, P.; Huang, H.L. Recombinase Polymerase Amplification: A New DNA/RNA Amplification Strategy. Chin. J. Biochem. Mol. Biol. 2016, 32, 627–634. [Google Scholar]
- Yang, C.; Zhen, Y.; Hou, J.; Mi, T. Development of a rapid detection method to Prorocentrum lima by loop-mediated isothermal amplification with hydroxy naphthol blue. Mar. Biotechnol. 2024, 26, 475–487. [Google Scholar] [CrossRef]
- Huang, H.L.; Zhu, P.; Zhou, C.X.; He, S.; Yan, X.J. The development of loop-mediated isothermal amplification combined with lateral flow dipstick for detection of Karlodinium veneficum. Harmful Algae 2017, 62, 20–29. [Google Scholar] [CrossRef]
- Luo, N.J.; Huang, H.L.; Jiang, H.B. Establishment of methods for rapid detection of Prymnesium parvum by recombinase polymerase amplification combined with a lateral flow dipstick. Front. Mar. Sci. 2022, 9, 1032847. [Google Scholar] [CrossRef]
- Anbazhagan, P.; Parameswari, B.; Anitha, K.; Chaitra, G.V.; Bajaru, B.; Rajashree, A.; Mangrauthia, S.K.; Yousuf, F.; Chalam, V.C.; Singh, G.P. Advances in plant pathogen detection: Integrating recombinase polymerase amplification with CRISPR/Cas systems. 3 Biotech 2024, 14, 214. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trac-Trend Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Durán-Vinet, B.; Araya-Castro, K.; Chao, T.C.; Wood, S.A.; Gallardo, V.; Godoy, K.; Abanto, M. Potential applications of CRISPR/Cas for next-generation biomonitoring of harmful algae blooms: A review. Harmful Algae 2021, 103, 102027. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Y.; Pan, S.; Pan, F.; Chen, J. Rapid and sensitive detection of Karlodinium veneficum using RAA and CRISPR-Cas12a technologies. Harmful Algae 2025, 146, 102864. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shi, Y.; Chu, J.; Zhu, S.; Luo, X.; Shen, W.; Chen, X. Efficient biosynthesis of acidic/lactonic sophorolipids and their application in the remediation of cyanobacterial harmful algal blooms. Int. J. Mol. Sci. 2023, 24, 12389. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Anand, U.; Saha, S.C.; Sundaramurthy, S.; Okeke, E.S.; Kumar, M.; Radha Bontempi, E.; Albertini, E.; Dey, A.; Di Maria, F. Novel CRISPR/Cas technology in the realm of algal bloom biomonitoring: Recent trends and future perspectives. Environ. Res. 2023, 231, 115989. [Google Scholar] [CrossRef] [PubMed]
- Assefa, A.; Wamae, K.; Hennelly, C.M.; Ngasala, B.; Muller, M.; Kalonji, A.; Phanzu, F.; Cunningham, C.H.; Lin, J.T.; Parr, J.B. Detection of P. malariae using a new rapid isothermal amplification lateral flow assay. Malar. J. 2024, 23, 104. [Google Scholar] [CrossRef] [PubMed]
- Kiatpathomchai, W.; Jaroenram, W.; Arunrut, N.; Jitrapakdee, S.; Flegel, T.W. Shrimp Taura syndrome virus detection by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Virol. Methods 2008, 153, 214–217. [Google Scholar] [CrossRef]
- Sajid, M.; Kawde, A.N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi. Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef]
- Weng, Z.; You, Z.; Yang, J.; Mohammad, N.; Lin, M.; Wei, Q.; Gao, X.; Zhang, Y. CRISPR-Cas biochemistry and CRISPR-based molecular diagnostics. Angew. Chem. Int. Edit. 2023, 62, e202214987. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, T.; Lv, X.; Shi, L.; Bai, W.; Ye, L. An RPA-based CRISPR/Cas12a assay in combination with a lateral flow assay for the rapid detection of shigella flexneri in food samples. Foods 2024, 13, 3200. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Pan, F.; Yao, G.; Chen, J. Development of a rapid detection method for Karenia mikimotoi by using CRISPR-Cas12a. Front. Microbiol. 2023, 14, 1205765. [Google Scholar] [CrossRef]
- Huang, H.L.; Luo, N.J.; Chen, W.Z.; Wang, X.W.; Zhou, C.X.; Jiang, H.B. A highly specific and ultrasensitive approach to detect Prymnesium parvum based on RPA-CRISPR-LbaCas12a-LFD system. Anal. Chim. Acta 2024, 1315, 342797. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zhang, L.; Mu, W.; Zhang, Y.; Chen, T.; Wu, J.; Tang, H.; Zheng, S.; Liu, Y.; et al. Generic Diagramming Platform (GDP): A comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 2025, 53, D1670–D1676. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.W.; Pushparaj, S.S.C.; Muthu, M.; Gopal, J. Review of harmful algal blooms (HABs) causing marine fish kills: Toxicity and mitigation. Plants 2023, 12, 3936. [Google Scholar] [CrossRef]
- Xi, Y.; Ding, R.; Zha, D.; Li, Y.; Yan, G.; Zhang, Y.; Wu, H.; Jiang, T. Phytoplankton community dynamics during Alexandrium blooms in 2019 off the Qinhuangdao coast, Bohai Sea, China. J. Oceanol. Limnol. 2022, 40, 2416–2429. [Google Scholar] [CrossRef]
- Eliason, O.; Segev, E. Coccolith Sr/Ca is a robust temperature and growth rate indicator that withstands dynamic microbial interactions. Geobiology 2022, 20, 435–443. [Google Scholar] [CrossRef]
- Deng, H.; Gao, Z. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal. Chim. Acta 2025, 853, 30–45. [Google Scholar] [CrossRef]
- Nalefski, E.A.; Patel, N.; Leung, P.J.; Islam, Z.; Kooistra, R.M.; Parikh, I.; Marion, E.; Kontt, G.J.; Doudna, J.A.; Madan, D. Kinetic analysis of Cas12a and Cas13a RNA-Guided nucleases for development of improved CRISPR-Based diagnostics. iScience 2021, 24, 102996. [Google Scholar]
- Qiu, X.; Xu, S.; Liu, X.; Ren, H.; Han, L.; Li, Z. CRISPR/Cas12a-based diagnostic platform accurately detects Nocardia farcinica targeting a novel species-specific gene. Front. Cell Infect. Mi. 2022, 12, 884411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Xie, K. Evaluation of CRISPR/Cas12a-based DNA detection for fast pathogen diagnosis and GMO test in rice. Mol. Breeding 2020, 40, 11. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Yue, H.; Tian, T.; Xiong, E.; Zhu, D.; Jiang, Y.; Zhou, X. Glycerol additive boosts 100-fold sensitivity enhancement for one-pot RPA-CRISPR/Cas12a assay. Anal. Chem. 2022, 94, 8277–8284. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ke, Y.; Liu, W.; Sun, Y.; Ding, X. A one-pot toolbox based on Cas12a/crRNA enables rapid foodborne pathogen detection at attomolar level. ACS Sensors 2020, 5, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Wang, J.C.; Liu, L.B.; Han, Q.A.; Wang, J.F.; Yuan, W.Z. An exo probe-based recombinase polymerase amplification assay for the rapid detection of porcine parvovirus. J. Virol. Methods 2017, 248, 145–147. [Google Scholar] [CrossRef]
- Fallon, T.; Shende, V.; Wierzbicki, I.; Pendleton, L.; Watervoort, F.; Auber, P.; Gonzalez, J.; Wisecaver, H.; Moore, S. Giant polyketide synthase enzymes in the biosynthesis of giant marine polyether toxins. Science 2024, 385, 671–678. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Shen, Y.; Zhang, Q.; Luo, X.; Zong, Y.; Zhou, C.; Huang, H.; Jiang, H. Development of an RPA-CRISPR/LbaCas12a-Lateral Flow Assay for the Visual Detection of Chrysotila dentata (Haptophyta). Microorganisms 2025, 13, 2203. https://doi.org/10.3390/microorganisms13092203
Yu J, Shen Y, Zhang Q, Luo X, Zong Y, Zhou C, Huang H, Jiang H. Development of an RPA-CRISPR/LbaCas12a-Lateral Flow Assay for the Visual Detection of Chrysotila dentata (Haptophyta). Microorganisms. 2025; 13(9):2203. https://doi.org/10.3390/microorganisms13092203
Chicago/Turabian StyleYu, Jiating, Yun Shen, Qinfei Zhang, Xuxu Luo, Yujie Zong, Chengxu Zhou, Hailong Huang, and Haibo Jiang. 2025. "Development of an RPA-CRISPR/LbaCas12a-Lateral Flow Assay for the Visual Detection of Chrysotila dentata (Haptophyta)" Microorganisms 13, no. 9: 2203. https://doi.org/10.3390/microorganisms13092203
APA StyleYu, J., Shen, Y., Zhang, Q., Luo, X., Zong, Y., Zhou, C., Huang, H., & Jiang, H. (2025). Development of an RPA-CRISPR/LbaCas12a-Lateral Flow Assay for the Visual Detection of Chrysotila dentata (Haptophyta). Microorganisms, 13(9), 2203. https://doi.org/10.3390/microorganisms13092203





