Rhizobium-Enhanced Drought Tolerance in Red Kidney Beans Through Modification of Transcriptome and Microbial Communities
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growing Conditions
2.2. PV-6 Isolation and Assessment of Strain Nodulation Efficiency
2.3. Sample Collection
2.4. Measurement of Enzyme Activity of Superoxide Dismutase (Sod), Peroxidase (POD), Catalase (CAT), and the Content of H2O2
2.5. DNA Extraction and 16S rRNA Amplicon Sequencing
2.6. qRT-PCR Gene Expression and Transcriptome Analysis
2.7. Statistical Analyses
3. Results
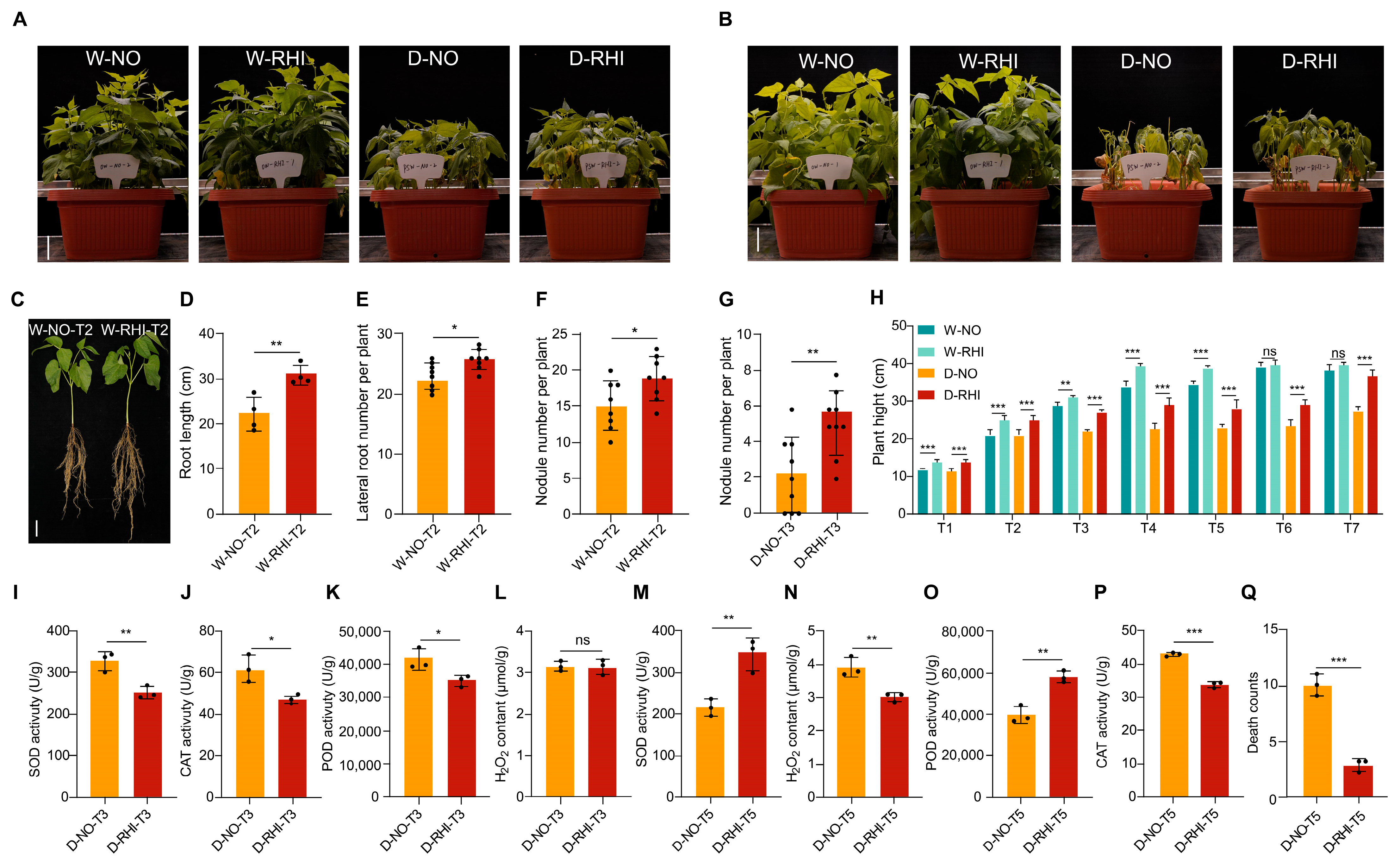
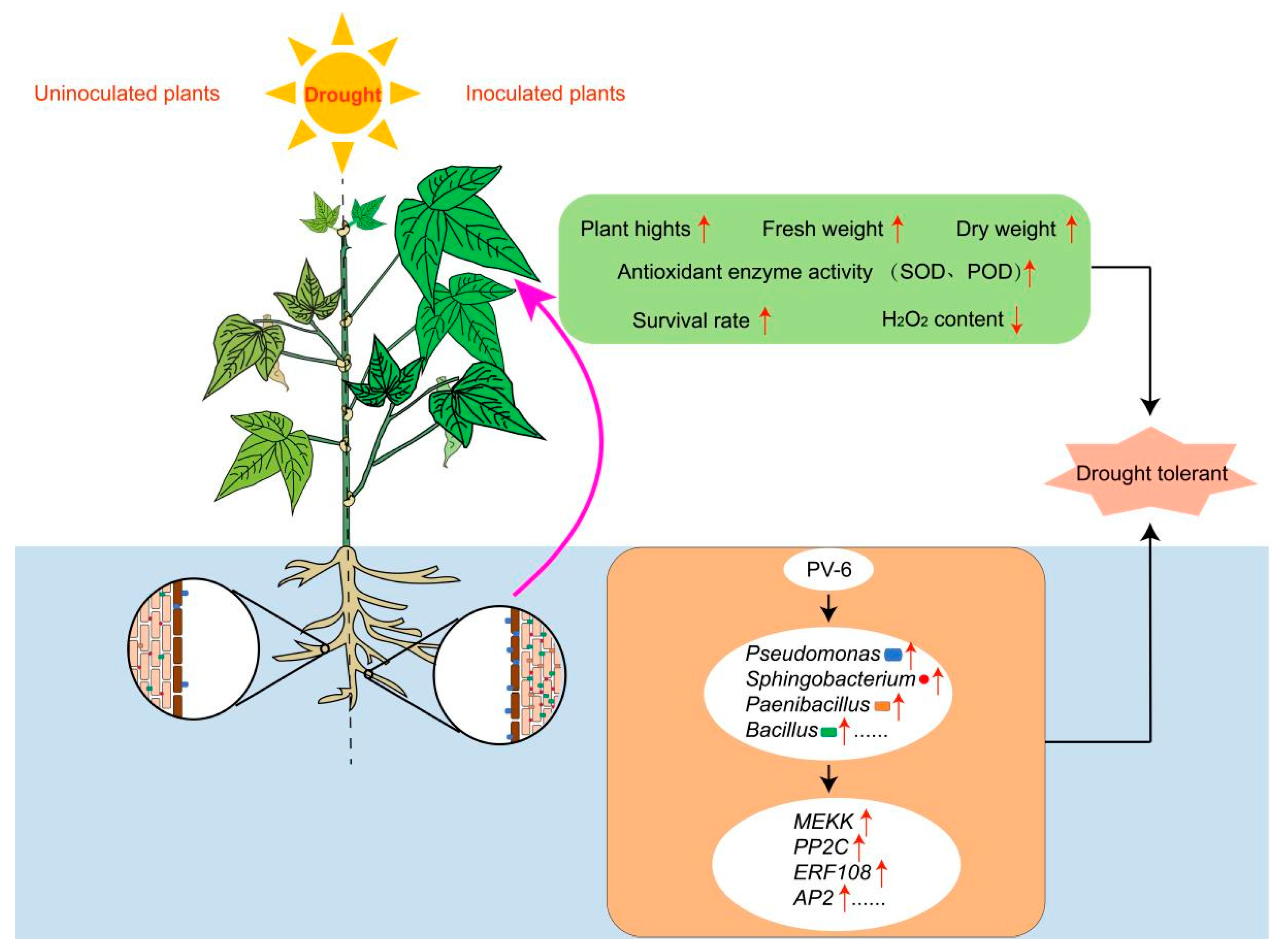
3.1. PV-6 Enhances Plant Growth and the Activity of Antioxidant Enzymes of Red Kidney Beans
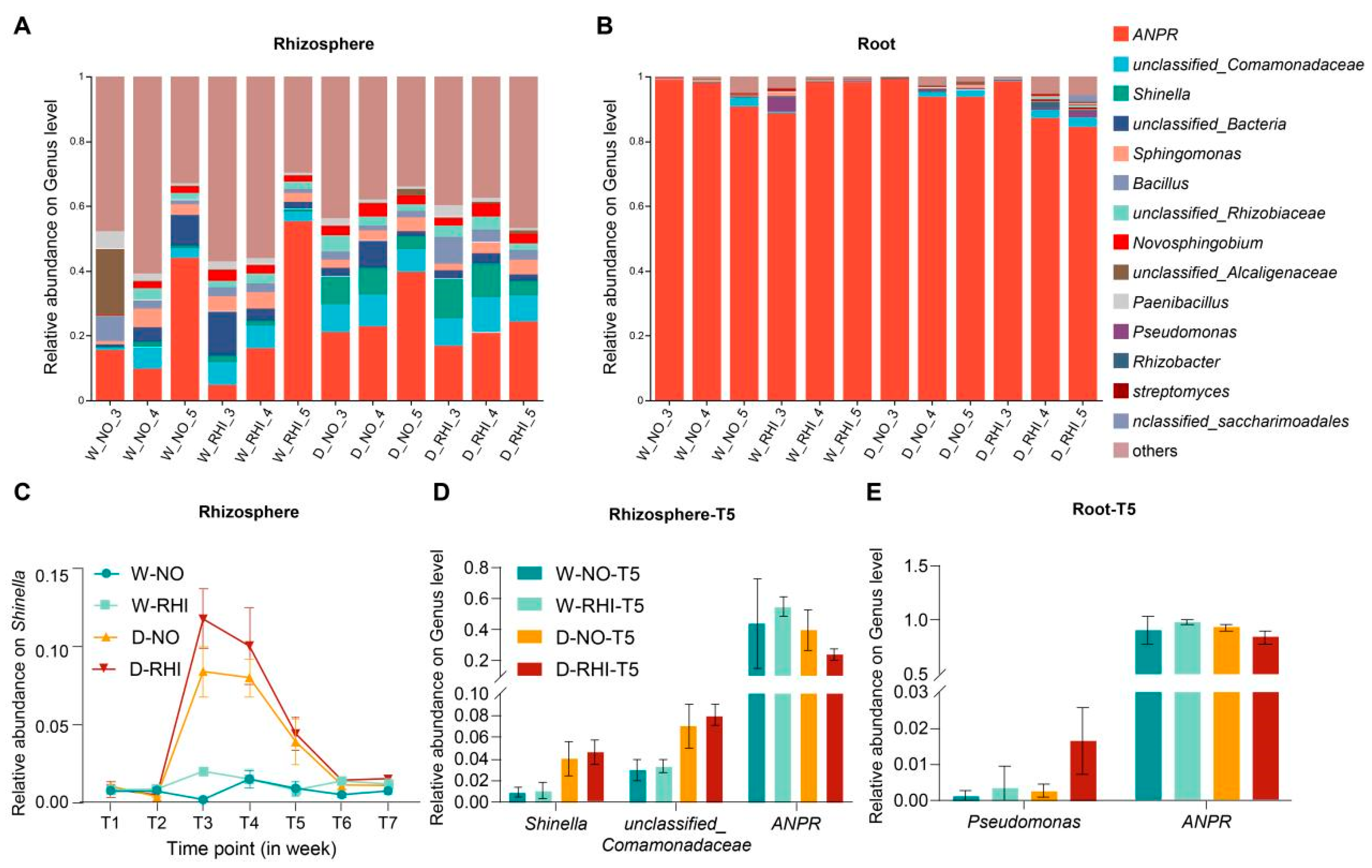
3.2. Dynamic Shifts in the Dominant Taxa of Red Kidney Beans When Subjected to Different Treatments
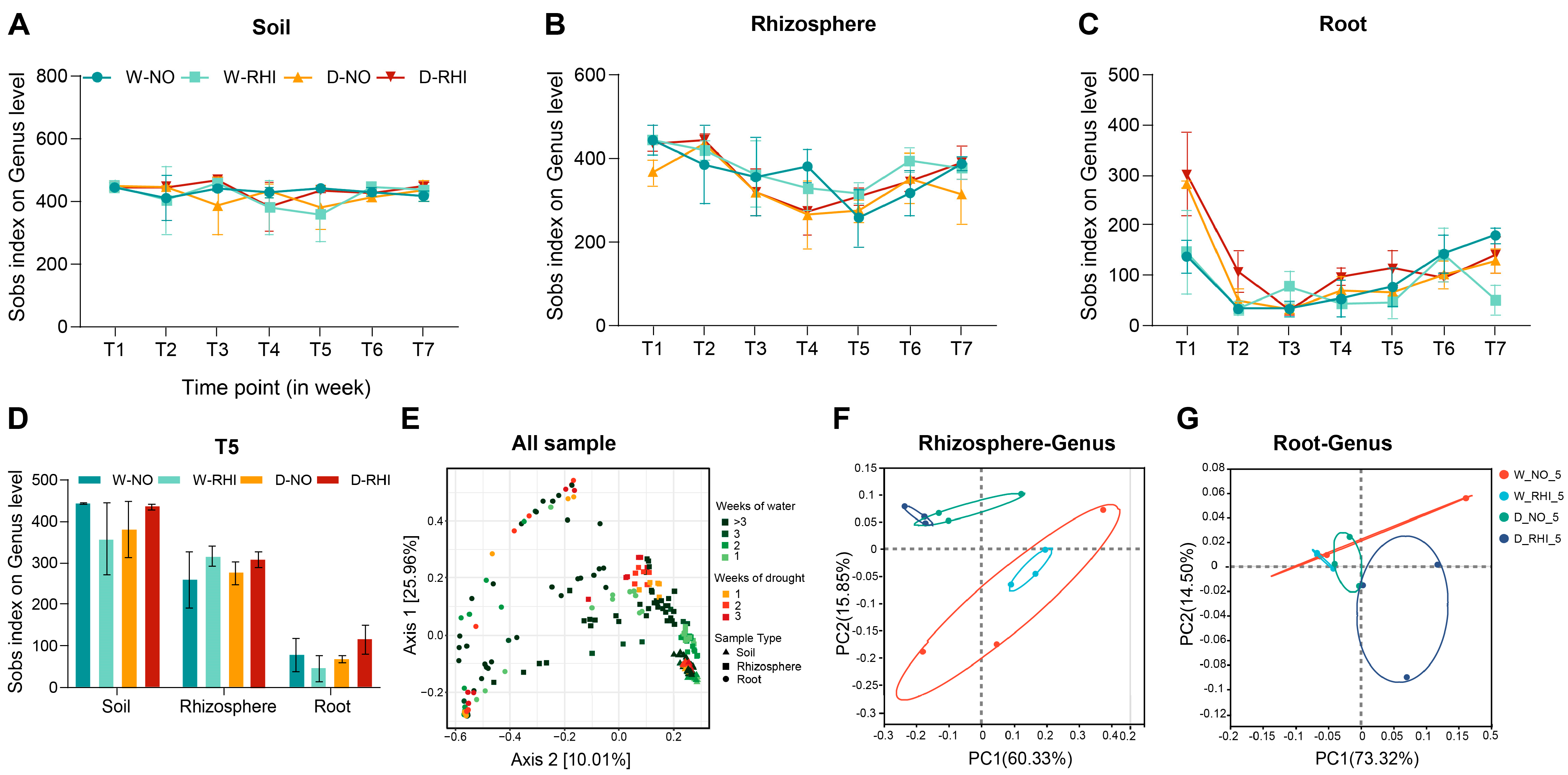
3.3. Drought Stress Alters the Microbial Community Diversity of Red Kidney Beans
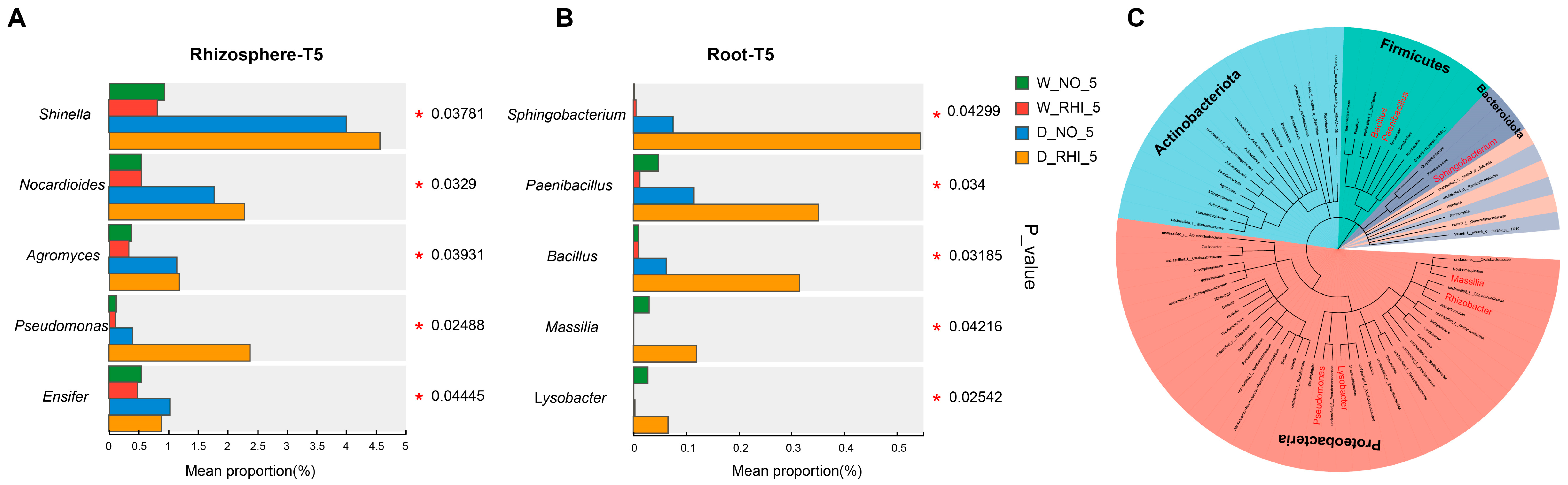
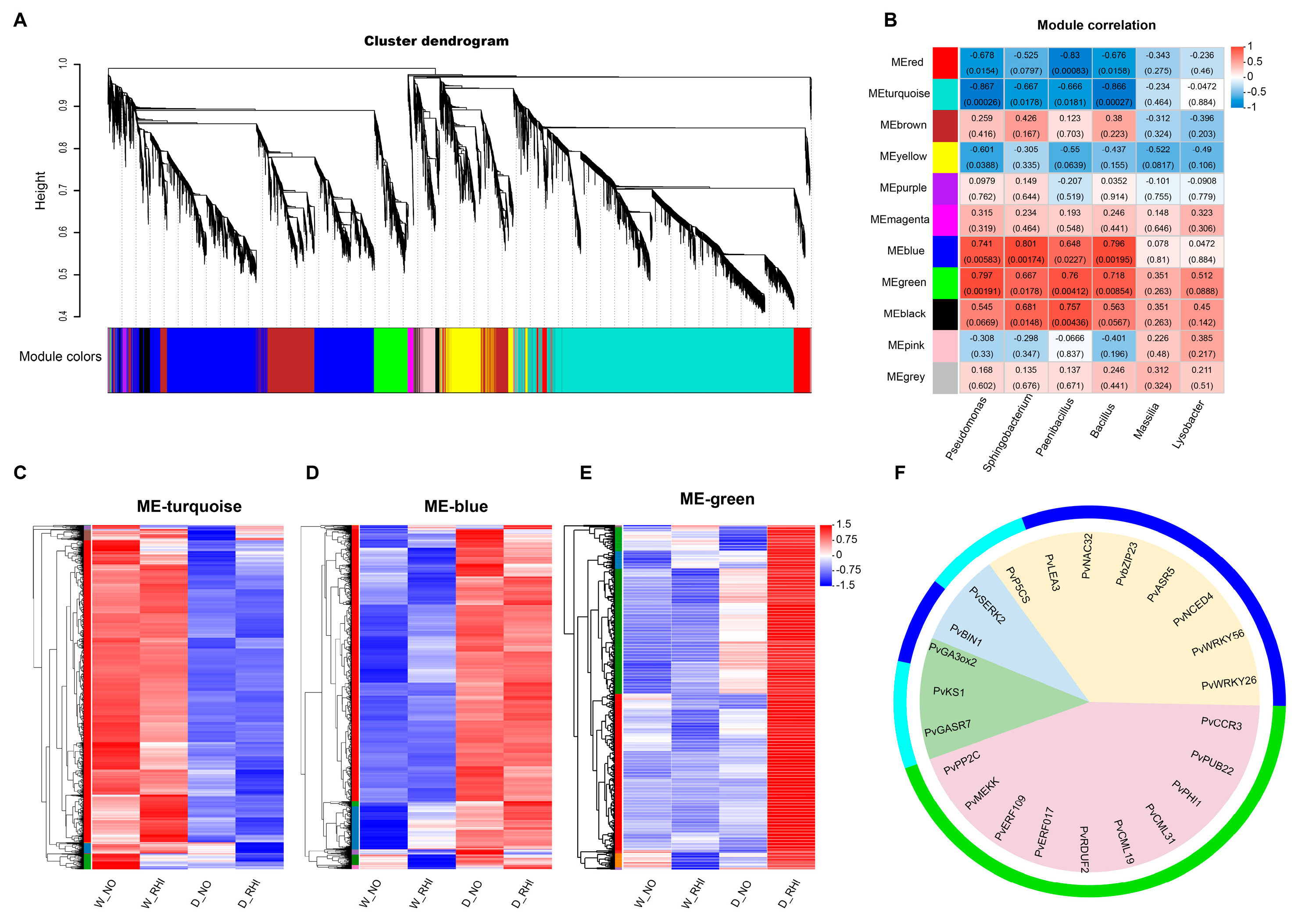
3.4. PV-6 Inoculation Reshaped the Root Bacterial Community
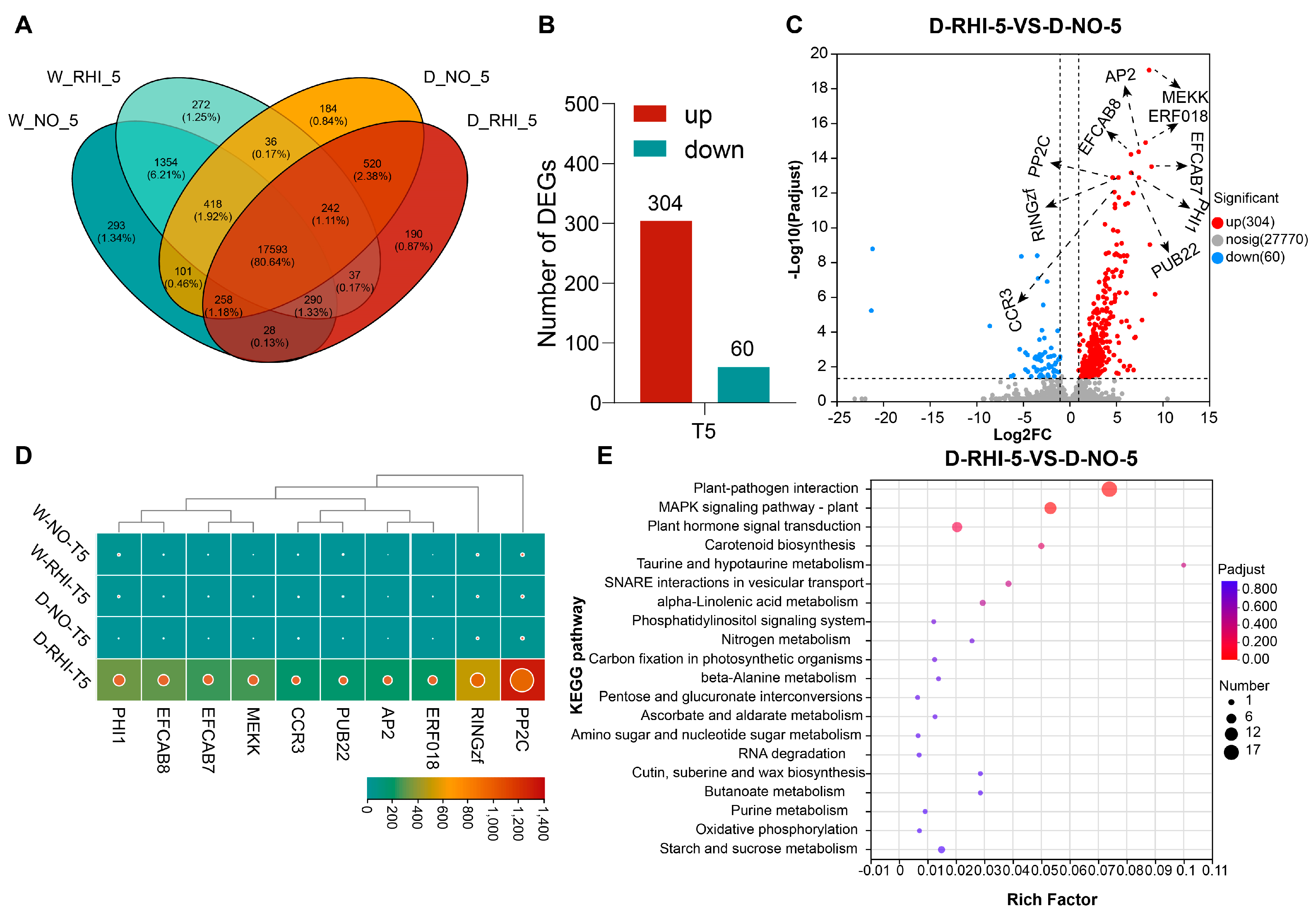
3.5. Drought-Inoculation Treatment Altered the Expression of Drought-Related Genes in Red Kidney Beans
3.6. Differentially Enriched Specific Bacteria and the Differentially Expressed Gene Identified by Integrated Association Analysis
4. Discussion
4.1. PV-6 Inoculation Positively Regulate Red Kidney Beans Growth
4.2. PV-6 Inoculation Reshapes the Root Endophytic Bacterial Community in Red Kidney Beans Under Drought Stress
4.3. PV-6 Inoculation Altered the Expression of Drought-Related Genes in Red Kidney Beans Under Drought Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carbas, B.; Machado, N.; Oppolzer, D.; Ferreira, L.; Queiroz, M.; Brites, C.; Rosa, E.A.; Barros, A.I. Nutrients, Antinutrients, Phenolic Composition, and Antioxidant Activity of Common Bean Cultivars and their Potential for Food Applications. Antioxidants 2020, 9, 186. [Google Scholar] [CrossRef]
- Ghallab, D.S.; Ghareeb, D.A.; Goda, D.A. Integrative metabolomics and chemometrics depict the metabolic alterations of differently processed red kidney beans (Phaseolus vulgaris L.) and in relation to in-vitro anti-diabetic efficacy. Food Res. Int. 2024, 192, 114786. [Google Scholar] [CrossRef]
- Los, F.G.B.; Zielinski, A.A.F.; Wojeicchowski, J.P.; Nogueira, A.; Demiate, I.M. Beans (Phaseolus vulgaris L.): Whole seeds with complex chemical composition. Curr. Opin. Food Sci. 2018, 19, 63–71. [Google Scholar] [CrossRef]
- Kan, L.; Nie, S.; Hu, J.; Wang, S.; Bai, Z.; Wang, J.; Zhou, Y.; Jiang, J.; Zeng, Q.; Song, K. Comparative study on the chemical composition, anthocyanins, tocopherols and carotenoids of selected legumes. Food Chem. 2018, 260, 317–326. [Google Scholar] [CrossRef]
- Fan, X.; Li, Y.; Cui, S. Ultrasound-assisted structural characterization and properties of glycosylation-modified products of the British red kidney bean protein antioxidant peptide fractions. Front. Nutr. 2025, 12, 1577660. [Google Scholar] [CrossRef]
- Bai, Z.; Huang, X.; Wu, G.; Ye, H.; Huang, W.; Nie, Q.; Chen, H.; Yin, J.; Chen, Y.; Nie, S. Polysaccharides from red kidney bean alleviating hyperglycemia and hyperlipidemia in type 2 diabetic rats via gut microbiota and lipid metabolic modulation. Food Chem. 2023, 404, 134598. [Google Scholar] [CrossRef]
- Une, S.; Nakata, R.; Nonaka, K.; Akiyama, J. Antiproliferative and anti-inflammatory effects of fractionated crude lectins from boiled kidney beans (Phaseolus vulgaris). J. Food Sci. 2024, 89, 671–683. [Google Scholar] [CrossRef]
- Labastida, D.; Ingvarsson, P.K.; Rendón-Anaya, M. Dissecting the genetic basis of drought responses in common bean using natural variation. Front. Plant Sci. 2023, 14, 1143873. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Woodward, A.; Smith, K.R.; Campbell-Lendrum, D.; Chadee, D.D.; Honda, Y.; Liu, Q.; Olwoch, J.; Revich, B.; Sauerborn, R.; Chafe, Z.; et al. Climate change and health: On the latest IPCC report. Lancet 2014, 383, 1185–1189. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Cheng, M.; Jiang, H.; Zhang, X.; Peng, C.; Lu, X.; Zhang, M.; Jin, J. Effect of Drought on Agronomic Traits of Rice and Wheat: A Meta-Analysis. Int. J. Environ. Res. Public Health 2018, 15, 839. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Kocsy, G.; Tari, I.; Vanková, R.; Zechmann, B.; Gulyás, Z.; Poór, P.; Galiba, G. Redox control of plant growth and development. Plant Sci. 2013, 211, 77–91. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosché, M.; Kangasjärvi, J. ROS signaling loops—Production, perception, regulation. Curr. Opin. Plant Biol. 2013, 16, 575–582. [Google Scholar] [CrossRef]
- Dat, J.; Vandenabeele, S.; Vranová, E.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef]
- Gechev, T.S.; Hille, J. Hydrogen peroxide as a signal controlling plant programmed cell death. J. Cell Biol. 2005, 168, 17–20. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef]
- Dietz, K.J.; Jacob, S.; Oelze, M.L.; Laxa, M.; Tognetti, V.; de Miranda, S.M.; Baier, M.; Finkemeier, I. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 2006, 57, 1697–1709. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Takahashi, M.; Asada, K. Superoxide production in aprotic interior of chloroplast thylakoids. Arch. Biochem. Biophys. 1988, 267, 714–722. [Google Scholar] [CrossRef]
- Kalladan, R.; Lasky, J.R.; Chang, T.Z.; Sharma, S.; Juenger, T.E.; Verslues, P.E. Natural variation identifies genes affecting drought-induced abscisic acid accumulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 11536–11541. [Google Scholar] [CrossRef]
- McAdam, S.A.M.; Brodribb, T.J. Mesophyll Cells Are the Main Site of Abscisic Acid Biosynthesis in Water-Stressed Leaves. Plant Physiol. 2018, 177, 911–917. [Google Scholar] [CrossRef]
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA Transport and Plant Water Stress Responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef]
- Park, S.Y.; Peterson, F.C.; Mosquna, A.; Yao, J.; Volkman, B.F.; Cutler, S.R. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 2015, 520, 545–548. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef]
- Mega, R.; Abe, F.; Kim, J.S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Okamoto, M.; Peterson, F.C.; Defries, A.; Park, S.Y.; Endo, A.; Nambara, E.; Volkman, B.F.; Cutler, S.R. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc. Natl. Acad. Sci. USA 2013, 110, 12132–12137. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef]
- Wang, H.; Tang, J.; Liu, J.; Hu, J.; Liu, J.; Chen, Y.; Cai, Z.; Wang, X. Abscisic Acid Signaling Inhibits Brassinosteroid Signaling through Dampening the Dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 2018, 11, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Liu, J.; Wang, H.; Yang, C.; Chen, Y.; Li, Y.; Pan, S.; Dong, R.; Tang, G.; Barajas-Lopez Jde, D.; et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 9651–9656. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.; Jiang, H.; Tang, B.; Zhang, M.; Li, Z.; Yin, Y. The AP2/ERF Transcription Factor TINY Modulates Brassinosteroid-Regulated Plant Growth and Drought Responses in Arabidopsis. Plant Cell 2019, 31, 1788–1806. [Google Scholar] [CrossRef]
- Dinneny, J.R. Developmental Responses to Water and Salinity in Root Systems. Annu. Rev. Cell Dev. Biol. 2019, 35, 239–257. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Santos-Medellín, C.; Liechty, Z.; Edwards, J.; Nguyen, B.; Huang, B.; Weimer, B.C.; Sundaresan, V. Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nat. Plants 2021, 7, 1065–1077. [Google Scholar] [CrossRef]
- Xu, L.; Naylor, D.; Dong, Z.; Simmons, T.; Pierroz, G.; Hixson, K.K.; Kim, Y.M.; Zink, E.M.; Engbrecht, K.M.; Wang, Y.; et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E4284–E4293. [Google Scholar] [CrossRef]
- Timmusk, S.; Paalme, V.; Pavlicek, T.; Bergquist, J.; Vangala, A.; Danilas, T.; Nevo, E. Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS ONE 2011, 6, e17968. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef]
- Islam, F.; Yasmeen, T.; Arif, M.S.; Ali, S.; Ali, B.; Hameed, S.; Zhou, W. Plant growth promoting bacteria confer salt tolerance in Vigna radiata by up-regulating antioxidant defense and biological soil fertility. Plant Growth Regul. 2016, 80, 23–36. [Google Scholar] [CrossRef]
- Kang, S.M.; Radhakrishnan, R.; Khan, A.L.; Kim, M.J.; Park, J.M.; Kim, B.R.; Shin, D.H.; Lee, I.J. Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 2014, 84, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Liu, N.; Fan, Z.; Liu, M.; Zhang, X.; Tian, J.; Yu, Y.; Lin, H.; Huang, Y.; Kong, Z. A novel PGPR strain, Streptomyces lasalocidi JCM 3373(T), alleviates salt stress and shapes root architecture in soybean by secreting indole-3-carboxaldehyde. Plant Cell Environ. 2024, 47, 1941–1956. [Google Scholar] [CrossRef]
- Parray, J.A.; Jan, S.; Kamili, A.N.; Qadri, R.A.; Egamberdieva, D.; Ahmad, P. Current Perspectives on Plant Growth-Promoting Rhizobacteria. J. Plant Growth Regul. 2016, 35, 877–902. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Ngumbi, E.; Kloepper, J. Bacterial-mediated drought tolerance: Current and future prospects. Appl. Soil Ecol. 2016, 105, 109–125. [Google Scholar] [CrossRef]
- Yu, W.; Luo, L.; Qi, X.; Cao, Y.; An, J.; Xie, Z.; Hu, T.; Yang, P. Insights into the Impact of Trans-Zeatin Overproduction-Engineered Sinorhizobium meliloti on Alfalfa (Medicago sativa L.) Tolerance to Drought Stress. J. Agric. Food Chem. 2024, 72, 8650–8663. [Google Scholar] [CrossRef] [PubMed]
- Lagunas, B.; Achom, M.; Bonyadi-Pour, R.; Pardal, A.J.; Richmond, B.L.; Sergaki, C.; Vázquez, S.; Schäfer, P.; Ott, S.; Hammond, J.; et al. Regulation of Resource Partitioning Coordinates Nitrogen and Rhizobia Responses and Autoregulation of Nodulation in Medicago truncatula. Mol. Plant 2019, 12, 833–846. [Google Scholar] [CrossRef]
- Wang, X.; Qiu, Z.; Zhu, W.; Wang, N.; Bai, M.; Kuang, H.; Cai, C.; Zhong, X.; Kong, F.; Lü, P.; et al. The NAC transcription factors SNAP1/2/3/4 are central regulators mediating high nitrogen responses in mature nodules of soybean. Nat. Commun. 2023, 14, 4711. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Q.; Wu, J.; Qi, M.; Zhang, C.; Huang, Y.; Wang, G.; Wang, H.; Tian, J.; Yu, Y.; et al. A legume kinesin controls vacuole morphogenesis for rhizobia endosymbiosis. Nat. Plants 2022, 8, 1275–1288. [Google Scholar] [CrossRef]
- Xu, J.; Li, X.L.; Luo, L. Effects of engineered Sinorhizobium meliloti on cytokinin synthesis and tolerance of alfalfa to extreme drought stress. Appl. Environ. Microbiol. 2012, 78, 8056–8061. [Google Scholar] [CrossRef]
- Vogel, C.M.; Potthoff, D.B.; Schäfer, M.; Barandun, N.; Vorholt, J.A. Protective role of the Arabidopsis leaf microbiota against a bacterial pathogen. Nat. Microbiol. 2021, 6, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Igiehon, O.N.; Babalola, O.O. Rhizobium and Mycorrhizal Fungal Species Improved Soybean Yield Under Drought Stress Conditions. Curr. Microbiol. 2021, 78, 1615–1627. [Google Scholar] [CrossRef]
- Tulumello, J.; Chabert, N.; Rodriguez, J.; Long, J.; Nalin, R.; Achouak, W.; Heulin, T. Rhizobium alamii improves water stress tolerance in a non-legume. Sci. Total Environ. 2021, 797, 148895. [Google Scholar] [CrossRef]
- Tak, N.; Bissa, G.; Gehlot, H.S. Methods for Isolation and Characterization of Nitrogen-Fixing Legume-Nodulating Bacteria. Methods Mol. Biol. 2020, 2057, 119–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Zhou, T.; Xu, K.; Zhao, F.; Liu, W.; Li, L.; Hua, Z.; Zhou, X. itol.toolkit accelerates working with iTOL (Interactive Tree of Life) by an automated generation of annotation files. Bioinformatics 2023, 39, btad339. [Google Scholar] [CrossRef]
- Young, J.P.W.; Moeskjær, S.; Afonin, A.; Rahi, P.; Maluk, M.; James, E.K.; Cavassim, M.I.A.; Rashid, M.H.; Aserse, A.A.; Perry, B.J.; et al. Defining the Rhizobium leguminosarum Species Complex. Genes 2021, 12, 111. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, Y.; Ren, Z.; Zhang, X.; Ren, J.; Su, J.; Zhang, C.; Tian, J.; Yu, Y.; Gao, G.F.; et al. Transfer cells mediate nitrate uptake to control root nodule symbiosis. Nat. Plants 2020, 6, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.T.; Zhang, L.; He, S.Y. Plant-Microbe Interactions Facing Environmental Challenge. Cell Host Microbe 2019, 26, 183–192. [Google Scholar] [CrossRef]
- Yue, H.; Sun, X.; Wang, T.; Zhang, A.; Han, D.; Wei, G.; Song, W.; Shu, D. Host genotype-specific rhizosphere fungus enhances drought resistance in wheat. Microbiome 2024, 12, 44. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, N.; Liu, Y.X.; Zhang, X.; Hu, B.; Qin, Y.; Xu, H.; Wang, H.; Guo, X.; Qian, J.; et al. Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci. China Life Sci. 2018, 61, 613–621. [Google Scholar] [CrossRef]
- Zhang, C.; van der Heijden, M.G.A.; Dodds, B.K.; Nguyen, T.B.; Spooren, J.; Valzano-Held, A.; Cosme, M.; Berendsen, R.L. A tripartite bacterial-fungal-plant symbiosis in the mycorrhiza-shaped microbiome drives plant growth and mycorrhization. Microbiome 2024, 12, 13. [Google Scholar] [CrossRef]
- Saleem, M.; He, J.; Alexandre, J. More Than the Sum of Its Parts: Microbiome Biodiversity as a Driver of Plant Growth and Soil Health. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 145–168. [Google Scholar] [CrossRef]
- Wang, X.; Wei, Z.; Yang, K.; Wang, J.; Jousset, A.; Xu, Y.; Shen, Q.; Friman, V.P. Phage combination therapies for bacterial wilt disease in tomato. Nat. Biotechnol. 2019, 37, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.L.; Yu, X.; Lane, D.; Sun, W.N.; Tang, Z.C.; Su, W.A. Upland rice and lowland rice exhibited different PIP expression under water deficit and ABA treatment. Cell Res. 2006, 16, 651–660. [Google Scholar] [CrossRef]
- Sripinyowanich, S.; Klomsakul, P.; Boonburapong, B.; Bangyeekhun, T.; Asami, T.; Gu, H.; Buaboocha, T.; Chadchawan, S. Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): The role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ. Exp. Bot. 2013, 86, 94–105. [Google Scholar] [CrossRef]
- Sun, J.Y.; Liu, X.S.; Khan, I.U.; Wu, X.C.; Yang, Z.M. OsPIP2;3 as an aquaporin contributes to rice resistance to water deficit but not to salt stress. Environ. Exp. Bot. 2021, 183, 104342. [Google Scholar] [CrossRef]
- Yang, L.; Chen, Y.; Xu, L.; Wang, J.; Qi, H.; Guo, J.; Zhang, L.; Shen, J.; Wang, H.; Zhang, F.; et al. The OsFTIP6-OsHB22-OsMYBR57 module regulates drought response in rice. Mol. Plant 2022, 15, 1227–1242. [Google Scholar] [CrossRef]
- Zhang, F.; Pan, Z.; Han, C.; Dong, H.; Lin, L.; Qiao, Q.; Zhao, K.; Wu, J.; Tao, S.; Zhang, S.; et al. Pyrus betulaefolia ERF3 interacts with HsfC1a to coordinately regulate aquaporin PIP1;4 and NCED4 for drought tolerance. Hortic. Res. 2024, 11, uhae090. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, X.; Ren, Z.; Abou-Elwafa, S.F.; Pu, X.; Zhu, Y.; Dou, D.; Su, H.; Cheng, H.; Liu, Z.; et al. ZmERF21 directly regulates hormone signaling and stress-responsive gene expression to influence drought tolerance in maize seedlings. Plant Cell Environ. 2022, 45, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.; Li, L.; Gao, L.; Hu, G.; Zhang, Y.; Reynolds, M.P.; Zhang, X.; Jia, J.; Mao, X.; et al. DIW1 encoding a clade I PP2C phosphatase negatively regulates drought tolerance by de-phosphorylating TaSnRK1.1 in wheat. J. Integr. Plant Biol. 2023, 65, 1918–1936. [Google Scholar] [CrossRef]
- Jeong, S.; Lim, C.W.; Lee, S.C. Pepper SnRK2.6-activated MEKK protein CaMEKK23 is directly and indirectly modulated by clade A PP2Cs in response to drought stress. New Phytol. 2023, 238, 237–251. [Google Scholar] [CrossRef]
- Meng, Y.; Lv, Q.; Li, L.; Wang, B.; Chen, L.; Yang, W.; Lei, Y.; Xie, Y.; Li, X. E3 ubiquitin ligase TaSDIR1-4A activates membrane-bound transcription factor TaWRKY29 to positively regulate drought resistance. Plant Biotechnol. J. 2024, 22, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Reinhold-Hurek, B.; Hurek, T. Life in grasses: Diazotrophic endophytes. Trends Microbiol. 1998, 6, 139–144. [Google Scholar] [CrossRef]
- van Overbeek, L.S.; Saikkonen, K. Impact of Bacterial-Fungal Interactions on the Colonization of the Endosphere. Trends Plant Sci. 2016, 21, 230–242. [Google Scholar] [CrossRef]
- Singh, T.; Bisht, N.; Ansari, M.M.; Mishra, S.K.; Chauhan, P.S. Paenibacillus lentimorbus alleviates nutrient deficiency-induced stress in Zea mays by modulating root system architecture, auxin signaling, and metabolic pathways. Plant Cell Rep. 2024, 43, 49. [Google Scholar] [CrossRef]
- Pang, Z.; Mao, X.; Zhou, S.; Yu, S.; Liu, G.; Lu, C.; Wan, J.; Hu, L.; Xu, P. Microbiota-mediated nitrogen fixation and microhabitat homeostasis in aerial root-mucilage. Microbiome 2023, 11, 85. [Google Scholar] [CrossRef]
- Cao, Y.H.; Zhao, X.W.; Nie, G.; Wang, Z.Y.; Song, X.; Zhang, M.X.; Hu, J.P.; Zhao, Q.; Jiang, Y.; Zhang, J.L. The salt-tolerance of perennial ryegrass is linked with root exudate profiles and microflora recruitment. Sci. Total Environ. 2024, 916, 170205. [Google Scholar] [CrossRef]
- Fan, X.; Matsumoto, H.; Xu, H.; Fang, H.; Pan, Q.; Lv, T.; Zhan, C.; Feng, X.; Liu, X.; Su, D.; et al. Aspergillus cvjetkovicii protects against phytopathogens through interspecies chemical signalling in the phyllosphere. Nat. Microbiol. 2024, 9, 2862–2876. [Google Scholar] [CrossRef]
- Chi, F.; Shen, S.H.; Cheng, H.P.; Jing, Y.X.; Yanni, Y.G.; Dazzo, F.B. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; Khan, M.Y.; Asghar, H.N.; Akhtar, M.J.; Zahir, Z.A. Differential response of single and co-inoculation of Rhizobium leguminosarum and Mesorhizobium ciceri for inducing water deficit stress tolerance in wheat. Ann. Microbiol. 2017, 67, 739–749. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Wang, J.; Gao, W.; Sun, X.; Xiong, Q.; Shu, X.; Miao, Y.; Shen, Q.; Xun, W.; et al. Nonpathogenic Pseudomonas syringae derivatives and its metabolites trigger the plant “cry for help” response to assemble disease suppressing and growth promoting rhizomicrobiome. Nat. Commun. 2024, 15, 1907. [Google Scholar] [CrossRef]
- Bakker, P.; Pieterse, C.M.J.; de Jonge, R.; Berendsen, R.L. The Soil-Borne Legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef]
- He, D.; Singh, S.K.; Peng, L.; Kaushal, R.; Vílchez, J.I.; Shao, C.; Wu, X.; Zheng, S.; Morcillo, R.J.L.; Paré, P.W.; et al. Flavonoid-attracted Aeromonas sp. from the Arabidopsis root microbiome enhances plant dehydration resistance. ISME J. 2022, 16, 2622–2632. [Google Scholar] [CrossRef]
- Hagaggi, N.S.A.; Abdul-Raouf, U.M. Drought-tolerant Sphingobacterium changzhouense Alv associated with Aloe vera mediates drought tolerance in maize (Zea mays). World J. Microbiol. Biotechnol. 2022, 38, 248. [Google Scholar] [CrossRef]
- Yang, N.; Nesme, J.; Røder, H.L.; Li, X.; Zuo, Z.; Petersen, M.; Burmølle, M.; Sørensen, S.J. Emergent bacterial community properties induce enhanced drought tolerance in Arabidopsis. npj Biofilm Microbiomes 2021, 7, 82. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, Z.; Lang, D.; Chu, Y.; Cui, G.; Jia, X. Bacillus pumilus improved drought tolerance in Glycyrrhiza uralensis G5 seedlings through enhancing primary and secondary metabolisms. Physiol. Plant 2021, 171, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Sikora, E.; Park, S.W. Plant growth-promoting rhizobacterium, Paenibacillus polymyxa CR1, upregulates dehydration-responsive genes, RD29A and RD29B, during priming drought tolerance in arabidopsis. Plant Physiol. Biochem. 2020, 156, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Rashid, U.; Yasmin, H.; Hassan, M.N.; Naz, R.; Nosheen, A.; Sajjad, M.; Ilyas, N.; Keyani, R.; Jabeen, Z.; Mumtaz, S.; et al. Drought-tolerant Bacillus megaterium isolated from semi-arid conditions induces systemic tolerance of wheat under drought conditions. Plant Cell Rep. 2022, 41, 549–569. [Google Scholar] [CrossRef] [PubMed]
- Sukweenadhi, J.; Kim, Y.J.; Choi, E.S.; Koh, S.C.; Lee, S.W.; Kim, Y.J.; Yang, D.C. Paenibacillus yonginensis DCY84(T) induces changes in Arabidopsis thaliana gene expression against aluminum, drought, and salt stress. Microbiol. Res. 2015, 172, 7–15. [Google Scholar] [CrossRef]
- Vaishnav, A.; Singh, J.; Singh, P.; Rajput, R.S.; Singh, H.B.; Sarma, B.K. Sphingobacterium sp. BHU-AV3 Induces Salt Tolerance in Tomato by Enhancing Antioxidant Activities and Energy Metabolism. Front. Microbiol. 2020, 11, 443. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Carvalhais, L.C.; Percy, C.D.; Prakash Verma, J.; Schenk, P.M.; Singh, B.K. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 2021, 229, 2873–2885. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Y.; Li, M.; Zhang, K.; Ma, W.; Zheng, L.; Xu, H.; Cui, B.; Liu, R.; Yang, Y.; et al. Functional assembly of root-associated microbial consortia improves nutrient efficiency and yield in soybean. J. Integr. Plant Biol. 2021, 63, 1021–1035. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Huang, C.; You, Q.; Jia, G.; Tan, Y.; Wu, S.; Kong, Z.; Wang, L. Rhizobium-Enhanced Drought Tolerance in Red Kidney Beans Through Modification of Transcriptome and Microbial Communities. Microorganisms 2025, 13, 2153. https://doi.org/10.3390/microorganisms13092153
Li X, Huang C, You Q, Jia G, Tan Y, Wu S, Kong Z, Wang L. Rhizobium-Enhanced Drought Tolerance in Red Kidney Beans Through Modification of Transcriptome and Microbial Communities. Microorganisms. 2025; 13(9):2153. https://doi.org/10.3390/microorganisms13092153
Chicago/Turabian StyleLi, Xiaoliang, Chunguo Huang, Qian You, Gaiya Jia, Yongjunlin Tan, Shenjie Wu, Zhaosheng Kong, and Lixiang Wang. 2025. "Rhizobium-Enhanced Drought Tolerance in Red Kidney Beans Through Modification of Transcriptome and Microbial Communities" Microorganisms 13, no. 9: 2153. https://doi.org/10.3390/microorganisms13092153
APA StyleLi, X., Huang, C., You, Q., Jia, G., Tan, Y., Wu, S., Kong, Z., & Wang, L. (2025). Rhizobium-Enhanced Drought Tolerance in Red Kidney Beans Through Modification of Transcriptome and Microbial Communities. Microorganisms, 13(9), 2153. https://doi.org/10.3390/microorganisms13092153





