Canine Parvovirus and Vaccine-Origin Feline Panleukopenia Virus in Wastewater, Arizona, USA: July 2022–June 2023
Abstract
1. Introduction
2. Methods
2.1. Sample Collection and Processing
2.2. Extraction and Nested Polymerase Chain Reaction (nPCR) Assay
2.3. Sequencing
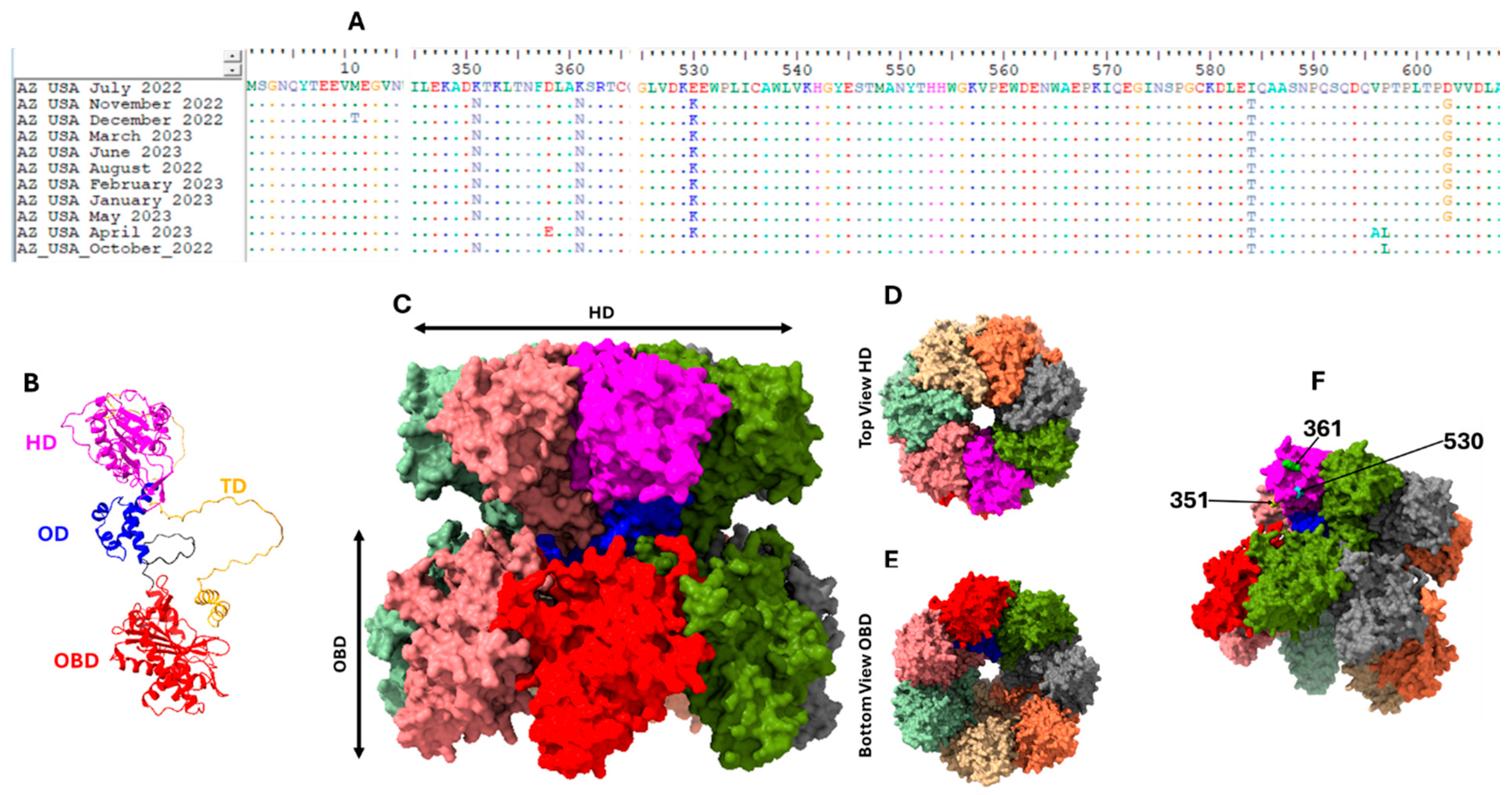
2.4. Phylogenetics, Recombination Analysis, B-Cell Epitope and Structure Prediction
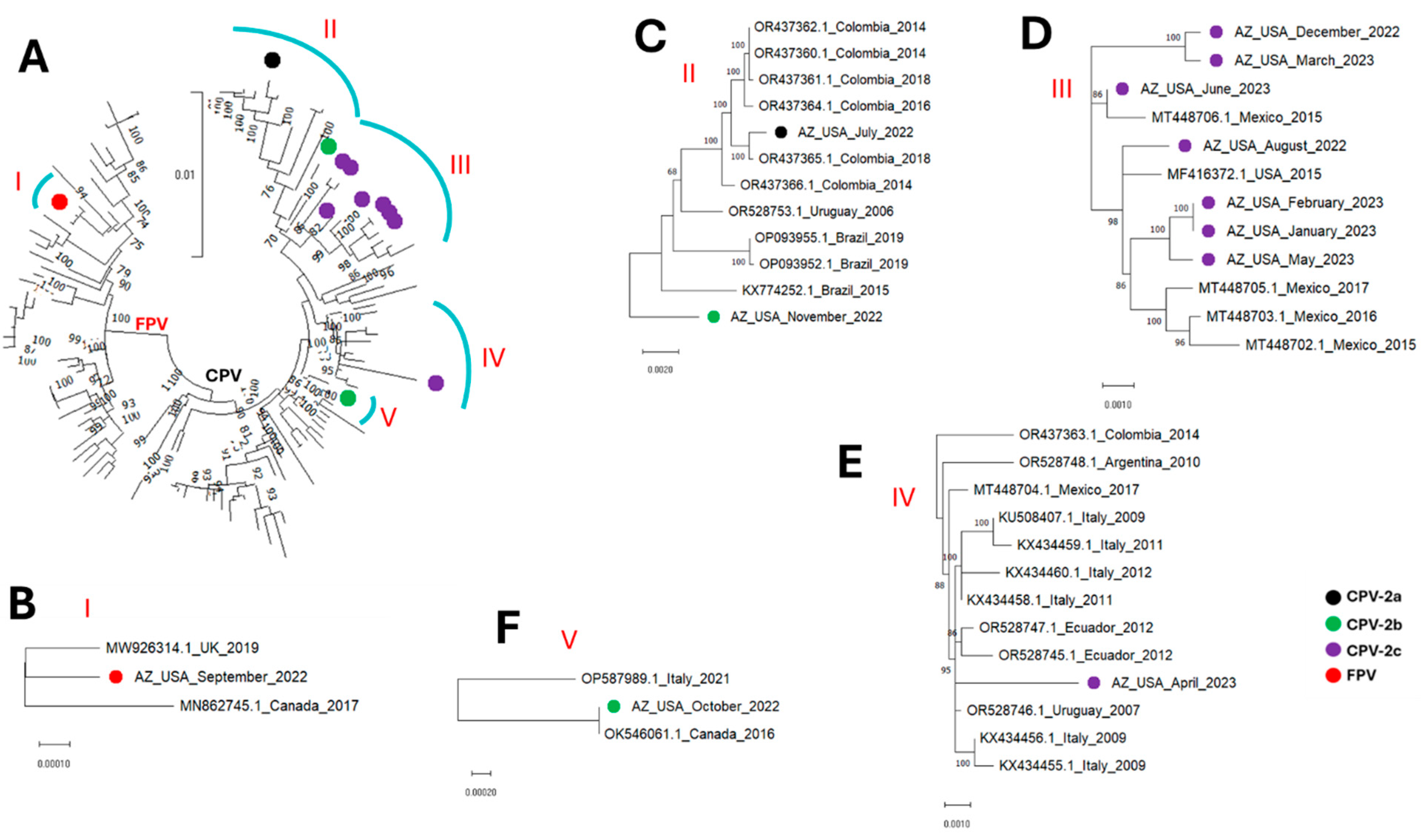
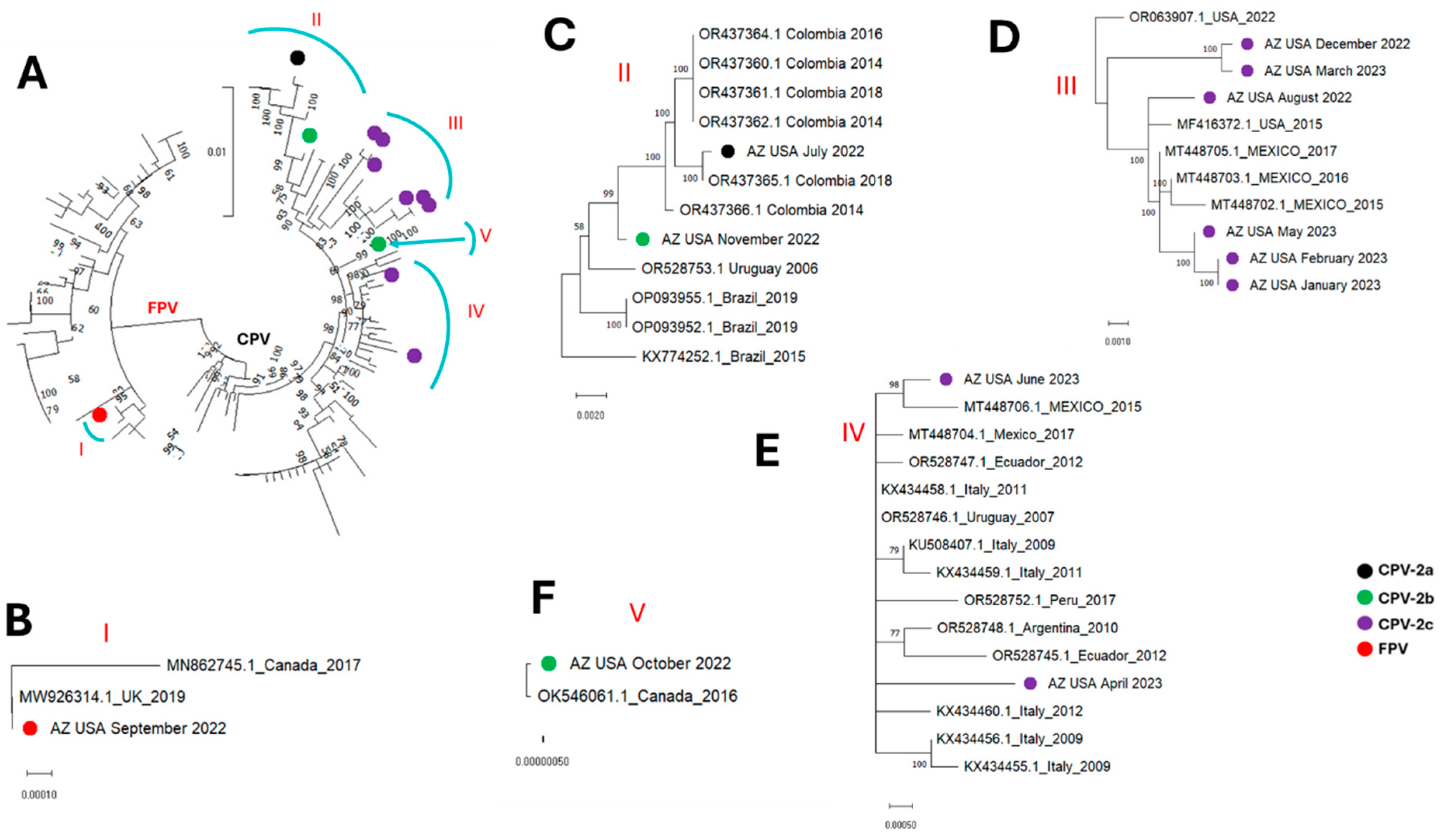
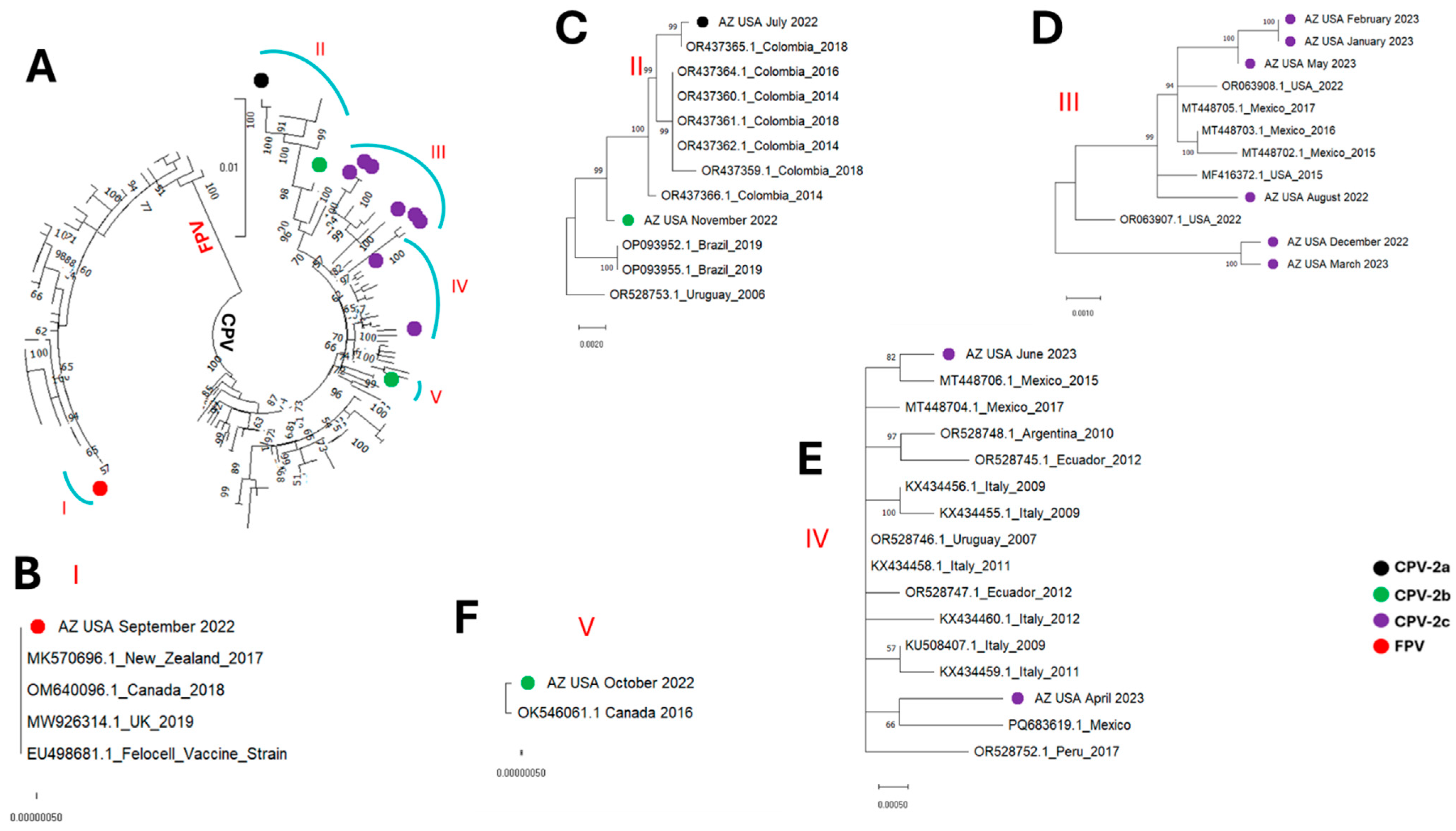
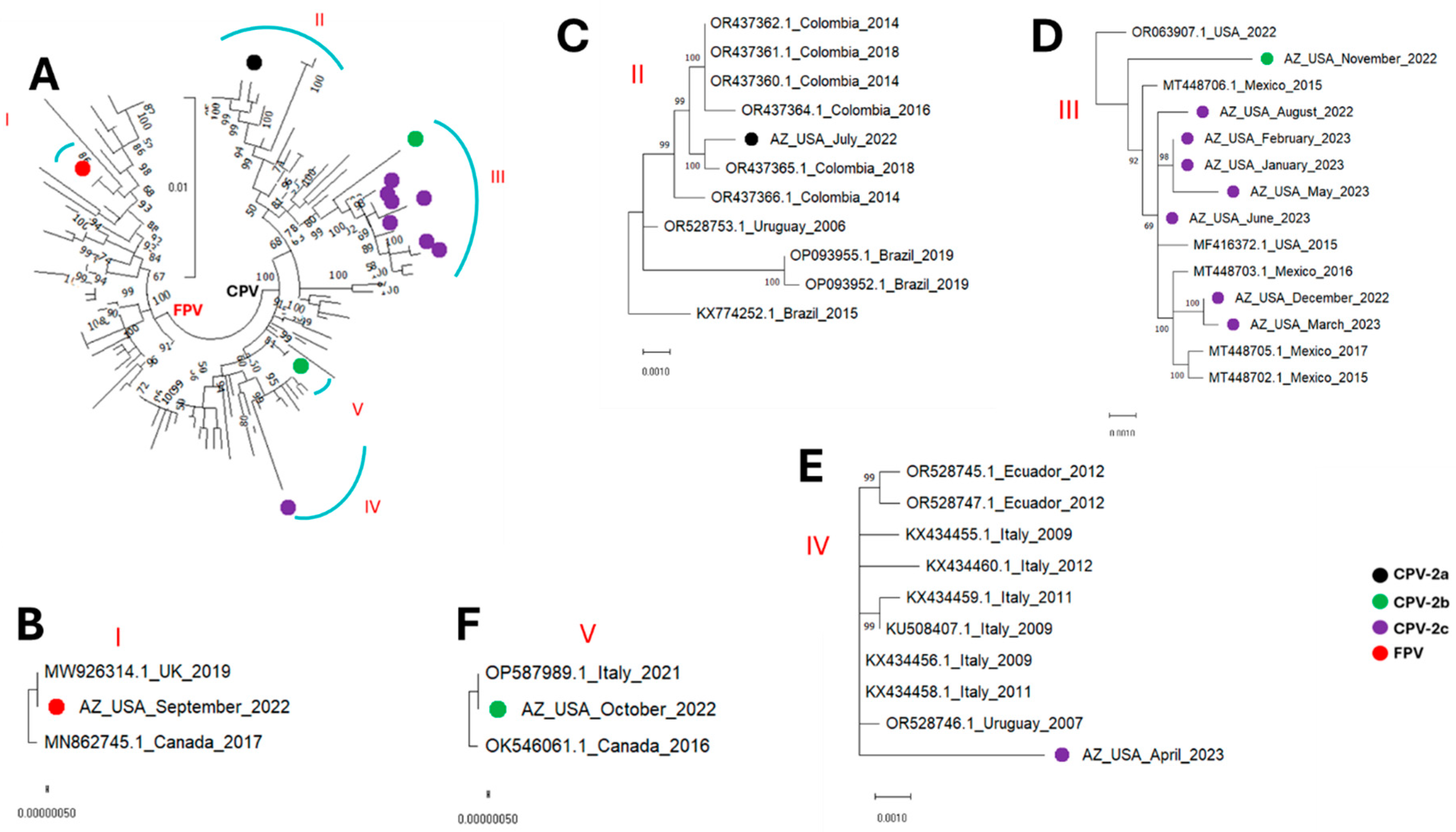
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miranda, C.; Thompson, G. Canine parvovirus: The worldwide occurrence of antigenic variants. J. Gen. Virol. 2016, 97, 2043–2057. [Google Scholar] [CrossRef]
- Adu, O.F.; Lee, H.; Fruh, S.P.; Schoenle, M.V.; Weichert, W.S.; Flyak, A.I.; Hafenstein, S.L.; Parrish, C.R. Structures and functions of the limited natural polyclonal antibody response to parvovirus infection. Proc. Natl. Acad. Sci. USA 2025, 122, e2423460122. [Google Scholar] [CrossRef]
- Faleye, T.O.C.; Driver, E.M.; Bowes, D.A.; Smith, A.; Kaiser, N.A.; Wright, J.M.; Chapman, A.R.; Halden, R.U.; Varsani, A.; Scotch, M.; et al. Canine Parvovirus 2C Identified in Dog Feces from Poop Bags Collected from Outdoor Waste Bins in Arizona USA, June 2022. Transbound Emerg Dis. 2023, 2023, 5596886. [Google Scholar] [CrossRef]
- Grassly, N.C.; Shaw, A.G.; Owusu, M. Global wastewater surveillance for pathogens with pandemic potential: Opportunities and challenges. Lancet Microbe. 2025, 6, 100939. [Google Scholar] [CrossRef]
- Kilaru, P.; Hill, D.; Anderson, K.; Collins, M.B.; Green, H.; Kmush, B.L.; Larsen, D.A. Wastewater Surveillance for Infectious Disease: A Systematic Review. Am. J. Epidemiol. 2023, 192, 305–322. [Google Scholar] [CrossRef]
- Singh, S.; Ahmed, A.I.; Almansoori, S.; Alameri, S.; Adlan, A.; Odivilas, G.; Chattaway, M.A.; Bin Salem, S.; Brudecki, G.; Elamin, W. A narrative review of wastewater surveillance: Pathogens of concern, applications, detection methods, and challenges. Front. Public Health 2024, 12, 1445961. [Google Scholar] [CrossRef] [PubMed]
- Faleye, T.O.C.; Adewumi, O.M.; Olayinka, O.A.; Donbraye, E.; Oluremi, B.; George, U.E.; Arowolo, O.A.; Omoruyi, E.C.; Ifeorah, M.I.; Oyero, A.O.; et al. Draft Genome Sequence of a Bovine Enterovirus Isolate Recovered from Sewage in Nigeria. Microbiol. Resour. Announc. 2018, 7, e01466-18. [Google Scholar] [CrossRef] [PubMed]
- Faleye, T.O.C.; Driver, E.M.; Bowes, D.A.; Holm, R.H.; Talley, D.; Yeager, R.; Bhatnagar, A.; Smith, T.; Varsani, A.; Halden, R.U.; et al. Detection of human, porcine and canine picornaviruses in municipal sewage sludge using pan-enterovirus amplicon-based long-read Illumina sequencing. Emerg. Microbes Infect. 2022, 11, 1339–1342. [Google Scholar] [CrossRef]
- Faleye, T.O.C.; Driver, E.M.; Wright, J.M.; Halden, R.U.; Varsani, A.; Scotch, M. Direct detection of canine picornavirus complete coding sequence in wastewater using long-range reverse-transcriptase polymerase chain reaction and long-read sequencing. Infect. Genet. Evol. 2024, 118, 105550. [Google Scholar] [CrossRef] [PubMed]
- Faleye, T.O.C.; Ifeorah, M.I.; Olayinka, O.A.; Oluremi, B.; George, U.E.; Arowolo, O.A.; Omoruyi, E.C.; Donbraye, E.; Oyero, A.O.; Adewumi, O.M.; et al. Near-Complete Genome Sequence of an Enterovirus Species F Isolate Recovered from Sewage in Nigeria. Microbiol. Resour. Announc. 2020, 9, e00094-20. [Google Scholar] [CrossRef]
- Ley, V.; Higgins, J.; Fayer, R. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 2002, 68, 3455–3461. [Google Scholar] [CrossRef]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef]
- Hofmann-Lehmann, R.; Fehr, D.; Grob, M.; Elgizoli, M.; Packer, C.; Martenson, J.S.; O’BRien, S.J.; Lutz, H. Prevalence of antibodies to feline parvovirus, calicivirus, herpesvirus, coronavirus, and immunodeficiency virus and of feline leukemia virus antigen and the interrelationship of these viral infections in free-ranging lions in east Africa. Clin. Diagn. Lab. Immunol. 1996, 3, 554–562. [Google Scholar] [CrossRef]
- Mira, F.; Purpari, G.; Lorusso, E.; Di Bella, S.; Gucciardi, F.; Desario, C.; Macaluso, G.; Decaro, N.; Guercio, A. Introduction of Asian canine parvovirus in Europe through dog importation. Transbound. Emerg. Dis. 2018, 65, 16–21. [Google Scholar] [CrossRef]
- Perez, R.; Calleros, L.; Marandino, A.; Sarute, N.; Iraola, G.; Grecco, S.; Blanc, H.; Vignuzzi, M.; Isakov, O.; Shomron, N.; et al. Phylogenetic and genome-wide deep-sequencing analyses of canine parvovirus reveal co-infection with field variants and emergence of a recent recombinant strain. PLoS ONE 2014, 9, e111779. [Google Scholar] [CrossRef] [PubMed]
- de Koning, W.; Miladi, M.; Hiltemann, S.; Heikema, A.; Hays, J.P.; Flemming, S.; Beek, M.v.D.; Mustafa, D.A.; Backofen, R.; Grüning, B.; et al. NanoGalaxy: Nanopore long-read sequencing data analysis in Galaxy. GigaScience 2020, 9, giaa105. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Callaway, H.M.; Cifuente, J.O.; Bator, C.M.; Parrish, C.R.; Hafenstein, S.L. Transferrin receptor binds virus capsid with dynamic motion. Proc. Natl. Acad. Sci. USA 2019, 116, 20462–20471. [Google Scholar] [CrossRef]
- Xie, Q.; Wang, J.; Gu, C.; Wu, J.; Liu, W. Structure and function of the parvoviral NS1 protein: A review. Virus Genes 2023, 59, 195–203. [Google Scholar] [CrossRef]
- Bergmann, M.; Schwertler, S.; Speck, S.; Truyen, U.; Reese, S.; Hartmann, K. Faecal shedding of parvovirus deoxyribonucleic acid following modified live feline panleucopenia virus vaccination in healthy cats. Vet. Rec. 2019, 185, 83. [Google Scholar] [CrossRef]
- Decaro, N.; Crescenzo, G.; Desario, C.; Cavalli, A.; Losurdo, M.; Colaianni, M.L.; Ventrella, G.; Rizzi, S.; Aulicino, S.; Lucente, M.S.; et al. Long-term viremia and fecal shedding in pups after modified-live canine parvovirus vaccination. Vaccine 2014, 32, 3850–3853. [Google Scholar] [CrossRef]
- Freisl, M.; Speck, S.; Truyen, U.; Reese, S.; Proksch, A.L.; Hartmann, K. Faecal shedding of canine parvovirus after modified-live vaccination in healthy adult dogs. Vet. J. 2017, 219, 15–21. [Google Scholar] [CrossRef]
- Castro, C.J.; Oderinde, B.S.; Poston, K.D.; Mawashi, K.Y.; Bullard, K.; Akinola, M.; Meade, C.; Liu, H.; Hu, F.; Bullows, J.E.; et al. Complete genome sequences of nine double recombinant vaccine-derived novel oral poliovirus type 2 genomes from Nigeria 2023–2024. Microbiol. Resour. Announc. 2024, 13, e0088124. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, M.; Ohshima, T.; Une, Y.; Yachi, A. Recombination between vaccine and field strains of canine parvovirus is revealed by isolation of virus in canine and feline cell cultures. J. Vet. Med. Sci. 2008, 70, 1305–1314. [Google Scholar] [CrossRef]
- Pan, S.; Man, Y.; Xu, X.; Ji, J.; Zhang, S.; Huang, H.; Li, Y.; Bi, Y.; Yao, L. Genetic Diversity and Recombination Analysis of Canine Parvoviruses Prevalent in Central and Eastern China, from 2020 to 2023. Microorganisms 2024, 12, 2173. [Google Scholar] [CrossRef]
- Adeniji, J.A.; Faleye, T.O. Enterovirus C strains circulating in Nigeria and their contribution to the emergence of recombinant circulating vaccine-derived polioviruses. Arch. Virol. 2015, 160, 675–683. [Google Scholar] [CrossRef]
- Burns, C.C.; Diop, O.M.; Sutter, R.W.; Kew, O.M. Vaccine-derived polioviruses. J. Infect. Dis. 2014, 210 (Suppl. 1), S283–S293. [Google Scholar] [CrossRef]
- Hao, X.; Li, Y.; Xiao, X.; Chen, B.; Zhou, P.; Li, S. The Changes in Canine Parvovirus Variants over the Years. Int. J. Mol. Sci. 2022, 23, 11540. [Google Scholar] [CrossRef]
- Lopez-Astacio, R.A.; Adu, O.F.; Goetschius, D.J.; Lee, H.; Weichert, W.S.; Wasik, B.R.; Frueh, S.P.; Alford, B.K.; Voorhees, I.E.H.; Flint, J.F.; et al. Viral Capsid, Antibody, and Receptor Interactions: Experimental Analysis of the Antibody Escape Evolution of Canine Parvovirus. J. Virol. 2023, 97, e0009023. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, S.R.; Charnesky, A.J.; Fruh, S.P.; Lopez-Astacio, R.A.; Weichert, W.S.; DiNunno, N.; Cho, S.H.; Bator, C.M.; Parrish, C.R.; Hafenstein, S.L. Cryo EM structures map a post vaccination polyclonal antibody response to canine parvovirus. Commun. Biol. 2023, 6, 955. [Google Scholar] [CrossRef] [PubMed]
- Boros, A.; Albert, M.; Urban, P.; Herczeg, R.; Gaspar, G.; Balazs, B.; Cságola, A.; Pankovics, P.; Gyenesei, A.; Reuter, G. Unusual “Asian-origin” 2c to 2b point mutant canine parvovirus (Parvoviridae) and canine astrovirus (Astroviridae) co-infection detected in vaccinated dogs with an outbreak of severe haemorrhagic gastroenteritis with high mortality rate in Hungary. Vet. Res. Commun. 2022, 46, 1355–1361. [Google Scholar] [CrossRef]
- Sarabandi, S.; Pourtaghi, H. Whole genome sequence analysis of CPV-2 isolates from 1998 to 2020. Virol. J. 2023, 20, 138. [Google Scholar] [CrossRef]

| Amino Acid Residue at Major Antigenic Sites in VP2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus Type | 80 | 87 | 93 | 101 | 103 | 300 | 305 | 323 | 426 | 555 | 564 | 568 |
| FPV | K | M | K | I | V | A | D | D | N | V | N | A |
| CPV-2 | R | M | N | I | A | A | D | N | N | V | S | G |
| CPV-2a | R | L | N | T | A | G | Y | N | N | I | S | G |
| CPV-2b | R | L | N | T | A | G | Y | N | D | V | S | G |
| CPV-2c | R | L | N | T | A | G | Y | N | E | V | S | G |
| Amino Acid in Select Positions in VP2 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | # Reads | # Size-Selected Reads Length [4.4 K–4.9 K] | Contig Length (GC%) | Closest GenBank Hit (% Similarity, Year, Country) | 80 | 87 | 93 | 101 | 103 | 300 | 305 | 323 | 426 | 555 | 564 | 568 | Type |
| AZ-USA-July-2022 | 20,334 | 16,183 | 4610 nt (36%) | OR437365.1, (99.85%, 2018, Colombia,) | R | L | N | T | A | G | Y | N | N | V | S | G | CPV-2a |
| AZ-USA-August-2022 | 14,583 | 12,467 | 4574 nt (36%) | MF416372.1 (99.71%, 2015, USA) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| AZ-USA-September-2022 | 8000 | 6786 | 4555 nt (35%) | OR528756.1 (99.21%, 2009, Argentina) | K | M | K | T | V | A | D | D | N | V | N | A | FPV |
| AZ-USA-October-2022 | 13,940 | 9828 | 4579 nt (36%) | OP587989.1 (99.74%, 2021, Italy) | R | L | N | T | A | G | Y | N | D | V | S | G | CPV-2b |
| AZ-USA-November-2022 | 11,247 | 9260 | 4675 nt (36%) | KX434458.1 (99.36%, 2011, Italy) | R | L | N | T | A | G | Y | N | D | V | S | G | CPV-2b |
| AZ-USA-December-2022 | 11,743 | 8353 | 4566 nt (36%) | MT448706.1 (99.47%, 2015, Mexico) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| AZ-USA-January-2023 | 9167 | 7369 | 4673 nt (36%) | MF416372.1 (99.67%, 2015, USA) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| AZ-USA-February-2023 | 13,072 | 3346 | 4626 nt (36%) | MF416372.1 (99.67%, 2015, USA) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| AZ-USA-March-2023 | 4298 | 2040 | 4656 nt (36%) | MT448706.1 (99.47%, 2015, Mexico) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| AZ-USA-April-2023 | 5941 | 3186 | 4574 nt (36%) | KX434458.1 (99.54%, 2011, Italy) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| AZ-USA-May-2023 | 6741 | 4892 | 4622 nt (36%) | MF416372.1 (99.67%, 2015, USA) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| AZ-USA-June-2023 | 5195 | 3148 | 4663 nt (36%) | MT448706.1 (99.87%, 2015, Mexico) | R | L | N | T | A | G | Y | N | E | V | S | G | CPV-2c |
| Total | 124,261 | 86,858 | |||||||||||||||
| First Sequence | Second Sequence | Similarity (%) |
|---|---|---|
| EU498681.1_Felocell_Vaccine_Strain | MK570696.1_New_Zealand_2017 | 100 |
| EU498681.1_Felocell_Vaccine_Strain | AZ_USA_September_2022 | 100 |
| EU498681.1_Felocell_Vaccine_Strain | MW926314.1_UK_2019 | 100 |
| EU498681.1_Felocell_Vaccine_Strain | OM640096.1_Canada_2018 | 100 |
| AZ_USA_September_2022 | OM640096.1_Canada_2018 | 100 |
| MW926314.1_UK_2019 | AZ_USA_September_2022 | 100 |
| MK570696.1_New_Zealand_2017 | OM640096.1_Canada_2018 | 100 |
| MW926314.1_UK_2019 | MK570696.1_New_Zealand_2017 | 100 |
| MW926314.1_UK_2019 | OM640096.1_Canada_2018 | 100 |
| MK570696.1_New_Zealand_2017 | AZ_USA_September_2022 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas, J.; Bermudez-Rivera, B.; Block, I.; Shaffer, G.; Estrada, L.; Dadd, T.; Dickerson, T.; Curtis, C.; Woods, C.; Driver, E.M.; et al. Canine Parvovirus and Vaccine-Origin Feline Panleukopenia Virus in Wastewater, Arizona, USA: July 2022–June 2023. Microorganisms 2025, 13, 2124. https://doi.org/10.3390/microorganisms13092124
Vargas J, Bermudez-Rivera B, Block I, Shaffer G, Estrada L, Dadd T, Dickerson T, Curtis C, Woods C, Driver EM, et al. Canine Parvovirus and Vaccine-Origin Feline Panleukopenia Virus in Wastewater, Arizona, USA: July 2022–June 2023. Microorganisms. 2025; 13(9):2124. https://doi.org/10.3390/microorganisms13092124
Chicago/Turabian StyleVargas, Jacqueline, Brenda Bermudez-Rivera, Izabella Block, Gray Shaffer, Lesley Estrada, Tegan Dadd, Tanner Dickerson, Courtney Curtis, Craig Woods, Erin M. Driver, and et al. 2025. "Canine Parvovirus and Vaccine-Origin Feline Panleukopenia Virus in Wastewater, Arizona, USA: July 2022–June 2023" Microorganisms 13, no. 9: 2124. https://doi.org/10.3390/microorganisms13092124
APA StyleVargas, J., Bermudez-Rivera, B., Block, I., Shaffer, G., Estrada, L., Dadd, T., Dickerson, T., Curtis, C., Woods, C., Driver, E. M., Halden, R. U., Varsani, A., Scotch, M., & Faleye, T. O. C. (2025). Canine Parvovirus and Vaccine-Origin Feline Panleukopenia Virus in Wastewater, Arizona, USA: July 2022–June 2023. Microorganisms, 13(9), 2124. https://doi.org/10.3390/microorganisms13092124








