Flagellimonas algicida sp. Nov.: A Novel Broad-Spectrum Algicidal Bacterium Targeting Harmful Algal Bloom Species and Genomic Insights into Its Secondary Metabolites
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Algal Strains and Culture Conditions
2.3. Analysis of Algicidal Characteristics of Strain SN16T
2.4. Determination of Algicidal Mode
2.5. Dose-Dependent Algicidal Activity Against P. globosa Morphotypes
2.6. Algicidal Spectrum of Strain SN16T
2.7. Polyphasic Taxonomic Characterization of Strain SN16T
2.7.1. Phylogenetic and Genomic Analysis
2.7.2. Phenotypic, Physiological, and Biochemical Characterization
2.7.3. Chemotaxonomic Characterization
2.8. Statistical Analysis
3. Results
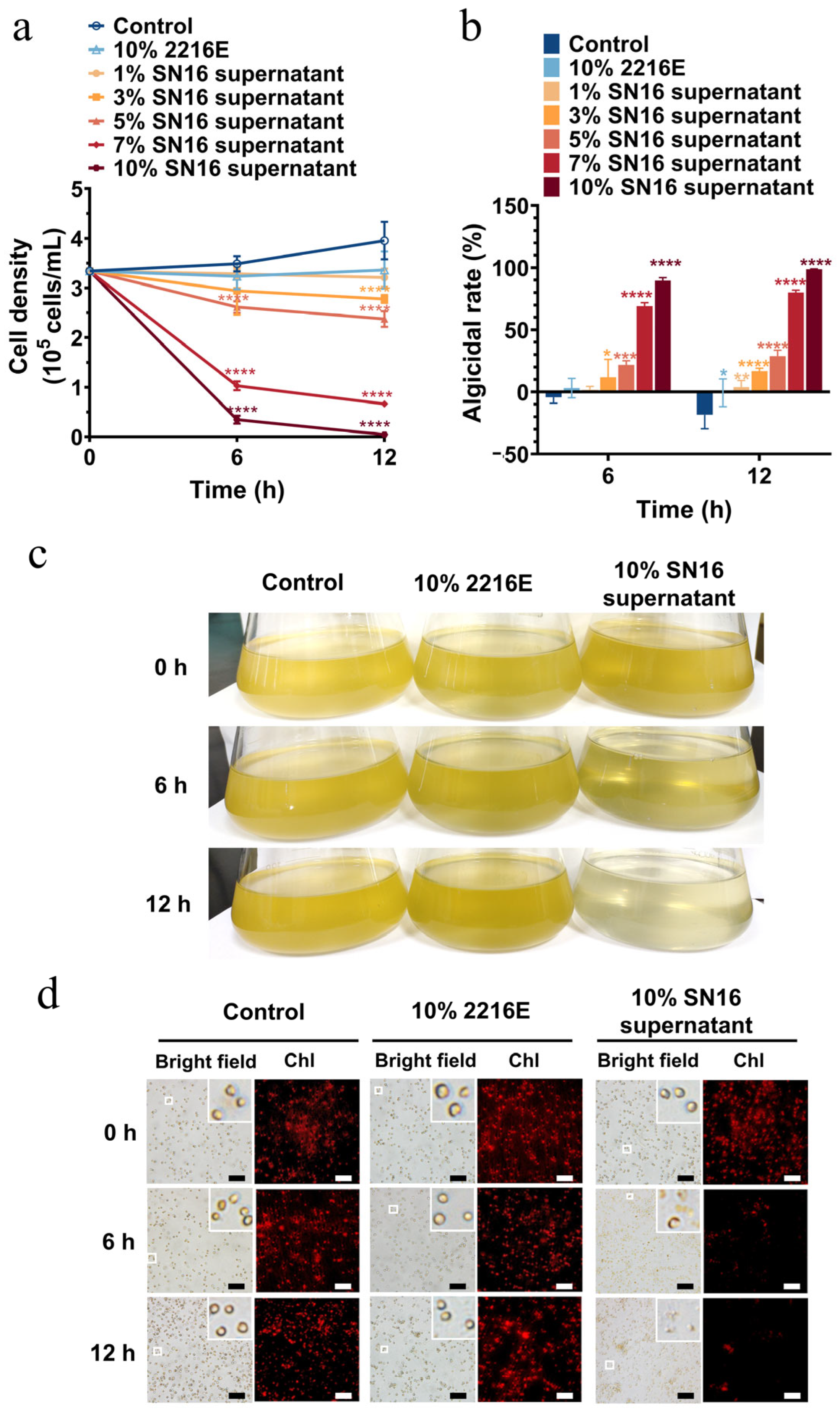
3.1. Growth and Algicidal Characteristics of Strain SN16T
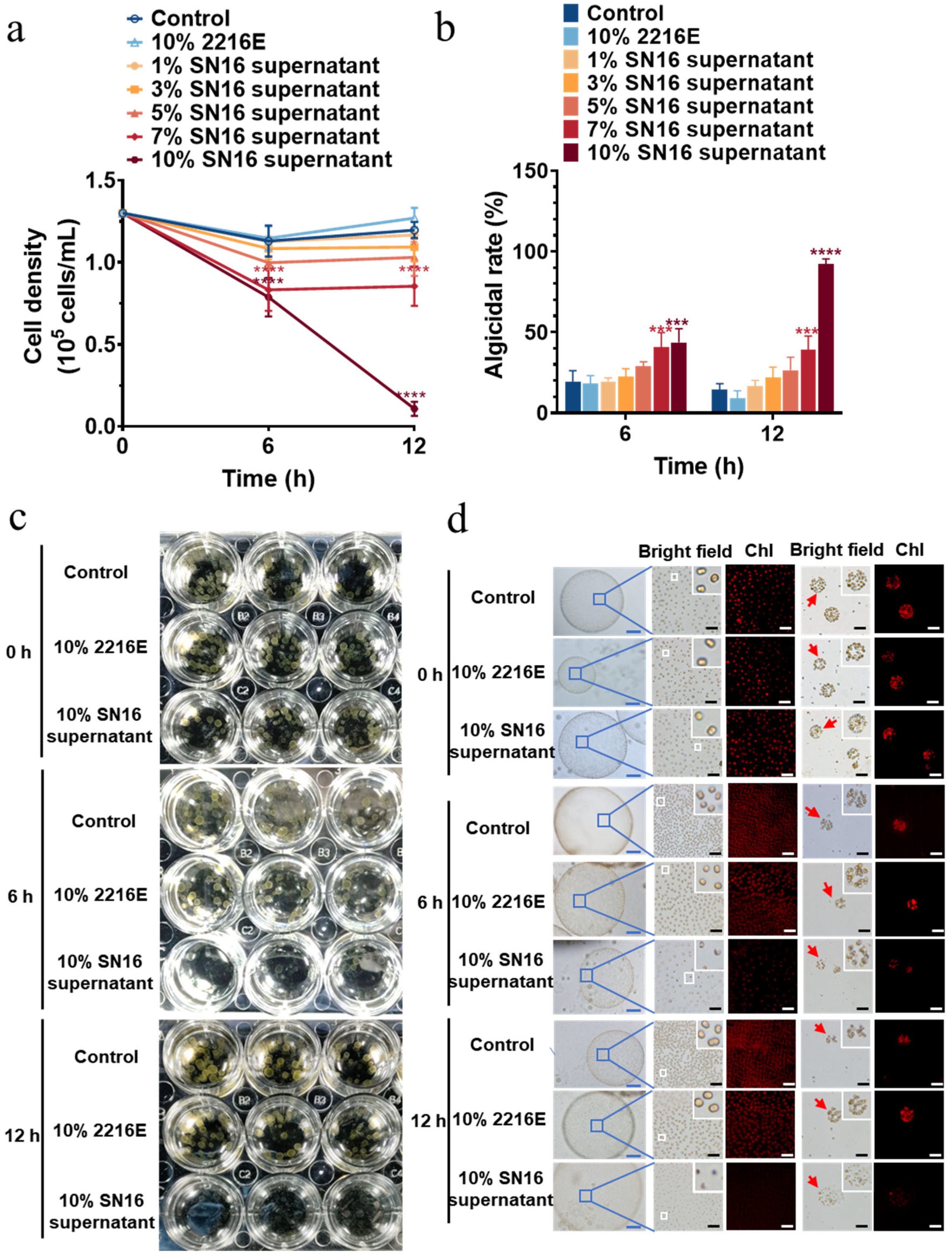
3.2. Dose-Dependent Algicidal Effects on P. globosa Morphotypes
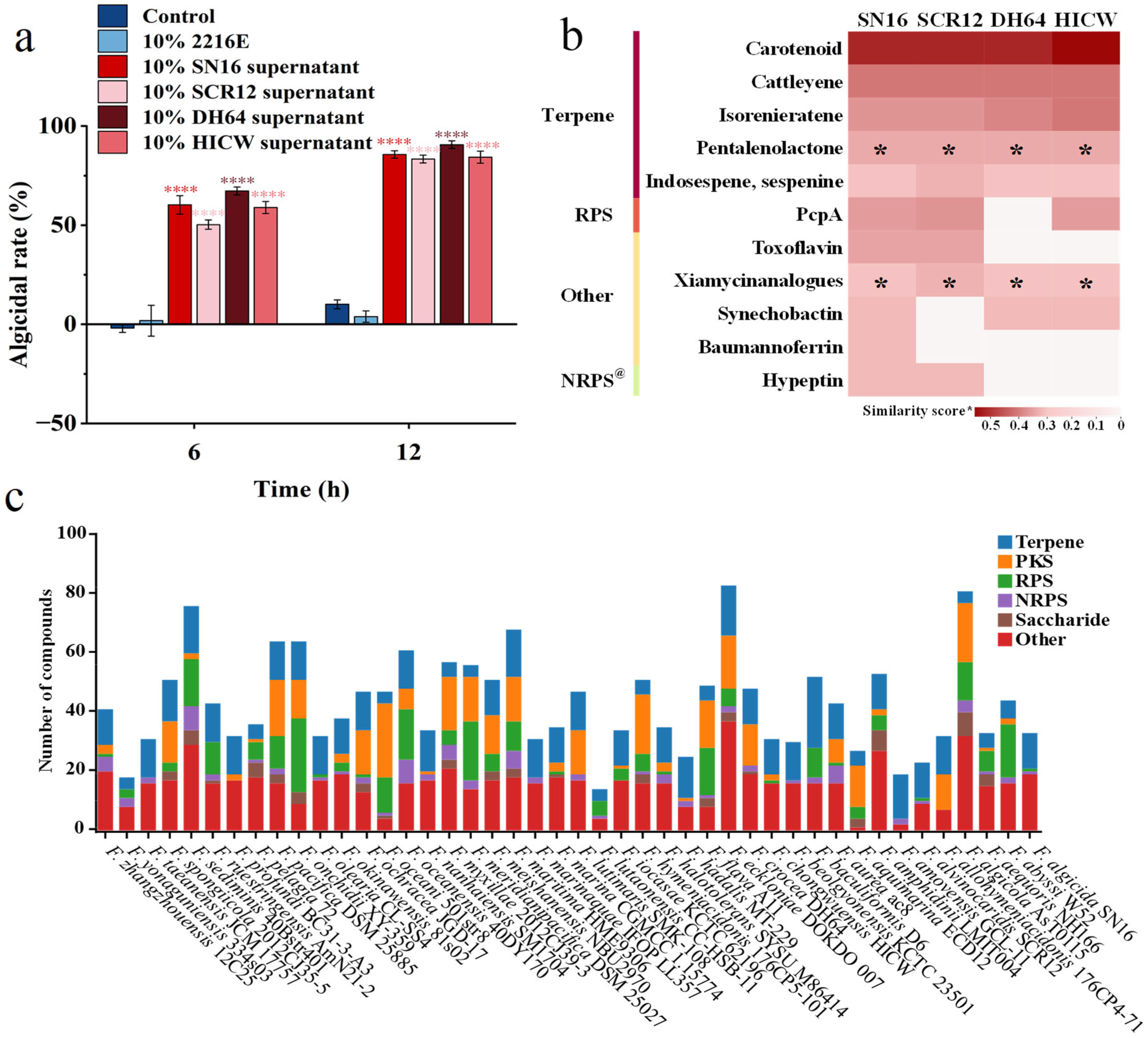
3.3. Broad-Spectrum Algicidal Activity of Strain SN16T
3.4. Phylogenetic and Genomic Characterization of Strain SN16T
3.4.1. 16 S rRNA Gene-Based Phylogenetic Analysis
3.4.2. Genome Properties and Phylogenomic Analysis
3.4.3. Genomic Evidence for a Novel Species
3.5. Physiological and Biochemical Characterization
3.6. Chemotaxonomic Characteristics
3.7. Algicidal Activities and Assessment of the Secondary Metabolic Potential of the Genus Flagellimonas
4. Discussion
4.1. Proposal of Flagellimonas Algicida sp. Nov.
4.2. Description of Flagellimonas algicida sp. Nov.
4.3. Algicidal Property of Flagellimonas algicida sp. Nov.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, X.; Song, H.; Wang, Y.; Chen, N. Research on the biology and ecology of the harmful algal bloom species Phaeocystis globosa in China: Progresses in the last 20 years. Harmful Algae 2021, 107, 102057. [Google Scholar] [CrossRef]
- Zhu, J.; Yu, Z.; He, L.; Yuan, Y.; Wang, W.; Cao, X.; Chen, N.; Wang, W.; Song, X. Mechanisms of Phaeocystis globosa blooms in the Beibu Gulf revealed by metatranscriptome analysis. Harmful Algae 2023, 124, 102407. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.O.; Trimborn, S. Phaeocystis: A Global Enigma. Annu. Rev. Mar. Sci. 2024, 16, 417–441. [Google Scholar] [CrossRef] [PubMed]
- Peperzak, L.; van Wezel, R. Human fatalities related to a Phaeocystis harmful algal bloom in the North Sea. Harmful Algae 2023, 130, 102545. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, T.; Zhou, J. Historical Occurrence of Algal Blooms in the Northern Beibu Gulf of China and Implications for Future Trends. Front. Microbiol. 2019, 10, 451. [Google Scholar] [CrossRef]
- Xu, S.; Lyu, P.; Zheng, X.; Yang, H.; Xia, B.; Li, H.; Zhang, H.; Ma, S. Monitoring and control methods of harmful algal blooms in Chinese freshwater system: A review. Environ. Sci. Pollut. Res. Int. 2022, 29, 56908–56927. [Google Scholar] [CrossRef]
- Che, M.; Shan, C.; Huang, R.; Cui, M.; Qi, W.; Klemes, J.J.; Su, R. A rapid removal of Phaeocystis globosa from seawater by peroxymonosulfate enhanced cellulose nanocrystals coagulation. Ecotoxicol. Environ. Saf. 2023, 262, 115318. [Google Scholar] [CrossRef]
- Qiao, L.; Che, M.; Shan, C.; Su, R.; Huang, R. Removal of Phaeocystis globosa from seawater via polyvinylpyrrolidone-modified pyrite activated persulfate. Chem.—Eur. J. 2025, 31, e202501261. [Google Scholar] [CrossRef]
- Pal, M.; Yesankar, P.J.; Dwivedi, A.; Qureshi, A. Biotic control of harmful algal blooms (HABs): A brief review. J. Environ. Manag. 2020, 268, 110687. [Google Scholar] [CrossRef]
- Guan, C.; Guo, X.; Li, Y.; Zhang, H.; Lei, X.; Cai, G.; Guo, J.; Yu, Z.; Zheng, T. Photoinhibition of Phaeocystis globosa resulting from oxidative stress induced by a marine algicidal bacterium Bacillus sp. LP-10. Sci. Rep. 2015, 5, 17002. [Google Scholar] [CrossRef]
- Tan, S.; Hu, X.; Yin, P.; Zhao, L. Photosynthetic inhibition and oxidative stress to the toxic Phaeocystis globosa caused by a diketopiperazine isolated from products of algicidal bacterium metabolism. J. Microbiol. 2016, 54, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Yang, X.; Lai, Q.; Yu, X.; Zhang, H.; Li, Y.; Chen, Z.; Lei, X.; Zheng, W.; Xu, H.; et al. Lysing bloom-causing alga Phaeocystis globosa with microbial algicide: An efficient process that decreases the toxicity of algal exudates. Sci. Rep. 2016, 6, 20081. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, B.; Cai, G.; Wang, H.; Li, D.; Yang, X.; An, X.; Zheng, X.; Tian, Y.; Zheng, W.; Zheng, T. Streptomyces alboflavus RPS and its novel and high algicidal activity against harmful algal bloom species Phaeocystis globosa. PLoS ONE 2014, 9, e92907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, S.; Peng, Y.; Li, Y.; Cai, G.; Chen, Z.; Zheng, W.; Tian, Y.; Xu, H.; Zheng, T. Effectiveness and toxicity of a novel isolated actinomycete strain Streptomyces sp. JS01 on a harmful alga Phaeocystis globosa. Appl. Microbiol. Biotechnol. 2015, 99, 4807–4814. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, B.; Zhang, J.; Huang, L.; Lin, J.; Li, X.; Zhou, Y.; Wang, H.; Yang, X.; Su, J.; et al. A marine algicidal actinomycete and its active substance against the harmful algal bloom species Phaeocystis globosa. Appl. Microbiol. Biotechnol. 2013, 97, 9207–9215. [Google Scholar] [CrossRef]
- Wang, L.; Xiang, W.; Wei, H.; Lv, J.; Wu, H.; Wu, H. Study on the Algicidal Characteristics and Physiological Response of Microbacterium sp. CBA01 to Phaeocystis globosa. Biotechnol. Bull. 2021, 37, 91–99. [Google Scholar]
- Zhang, H.; Wang, H.; Zheng, W.; Yao, Z.; Peng, Y.; Zhang, S.; Hu, Z.; Tao, Z.; Zheng, T. Toxic effects of prodigiosin Secreted by Hahella sp. KA22 on harmful alga Phaeocystis globosa. Front. Microbiol. 2017, 8, 999. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, S.; Luo, G.; Zheng, W.; Tian, Y.; Lei, X.; Yao, L.; Wu, C.; Xu, H. A novel algicidal bacterium, Microbulbifer sp. YX04, triggered oxidative damage and autophagic cell death in Phaeocystis globosa, which causes harmful algal blooms. Microbiol. Spectr. 2022, 10, e00934-21. [Google Scholar] [CrossRef]
- Bae, S.S.; Kwon, K.K.; Yang, S.H.; Lee, H.-S.; Kim, S.-J.; Lee, J.-H. Flagellimonas eckloniae gen. nov., sp nov., a mesophilic marine bacterium of the family Flavobacteriaceae, isolated from the rhizosphere of Ecklonia kurome. Int. J. Syst. Evol. Microbiol. 2007, 57, 1050–1054. [Google Scholar] [CrossRef]
- Yoon, B.-J.; Oh, D.-C. Spongiibacterium flavum gen. nov., sp nov., a member of the family Flavobacteriaceae isolated from the marine sponge Halichondria oshoro, and emended descriptions of the genera Croceitalea and Flagellimonas. Int. J. Syst. Evol. Microbiol. 2012, 62, 1158–1164. [Google Scholar] [CrossRef]
- Choi, S.; Lee, J.H.; Kang, J.W.; Choe, H.N.; Seong, C.N. Flagellimonas aquimarina sp. nov., and transfer of Spongiibacterium flavum Yoon and Oh 2012 and S-pacificum Gao et al. 2015 to the genus Flagellimonas Bae et al. 2007 as Flagellimonas flava comb. nov and F.pacifica comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2018, 68, 3266–3272. [Google Scholar] [CrossRef] [PubMed]
- Novoa, E.A.M.; Deshmukh, U.B.; Oren, A. Reclassification of Allomuricauda and Muricauda species as members of the genus Flagellimonas Bae et al. 2007 and emended description of the genus Flagellimonas. Int. J. Syst. Evol. Microbiol. 2024, 74, 6286. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Goeker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Kang, S.-J.; Jung, Y.-T.; Oh, T.-K. Muricauda lutimaris sp. nov., isolated from a tidal flat of the Yellow Sea. Int. J. Syst. Evol. Microbiol. 2008, 58, 1603–1607. [Google Scholar] [CrossRef][Green Version]
- Dang, Y.-R.; Sun, Y.-Y.; Sun, L.-L.; Yuan, X.-X.; Li, Y.; Qin, Q.-L.; Chen, X.-L.; Zhang, Y.-Z.; Shi, M.; Zhang, X.-Y. Muricauda nanhaiensis sp. nov., isolated from seawater of the South China Sea. Int. J. Syst. Evol. Microbiol. 2019, 69, 2089–2094. [Google Scholar] [CrossRef]
- Guo, L.-L.; Wu, D.; Sun, C.; Cheng, H.; Xu, X.-W.; Wu, M.; Wu, Y.-H. Muricauda maritima sp. nov., Muricauda aequoris sp. nov. and Muricauda oceanensis sp. nov., three marine bacteria isolated from seawater. Int. J. Syst. Evol. Microbiol. 2020, 70, 6240–6250. [Google Scholar] [CrossRef]
- Kim, J.; Kim, K.H.; Chun, B.H.; Khan, S.A.; Jeon, C.O. Flagellimonas algicola sp. nov., Isolated from a Marine Red Alga, Asparagopsis taxiformis. Curr. Microbiol. 2020, 77, 294–299. [Google Scholar] [CrossRef]
- Park, J.-S. Muricauda hymeniacidonis sp. nov., isolated from sponge of Hymeniacidon sinapium. Int. J. Syst. Evol. Microbiol. 2019, 69, 3800–3805. [Google Scholar] [CrossRef]
- Liu, L.; Yu, M.; Zhou, S.; Fu, T.; Sun, W.; Wang, L.; Zhang, X.-H. Muricauda alvinocaridis sp. nov., isolated from shrimp gill from the Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2020, 70, 1666–1671. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Y.; Pei, J.; Cao, J.; Xie, Z.; Liu, R.; Wang, L.; Wei, Y.; Fang, J. Muricauda hadalis sp. nov., a novel piezophile isolated from hadopelagic water of the Mariana Trench and reclassification of Muricauda antarctica as a later heterotypic synonym of Muricauda teanensis. Int. J. Syst. Evol. Microbiol. 2020, 70, 4315–4320. [Google Scholar] [CrossRef]
- Chen, M.-X.; He, X.-Y.; Li, H.-Y. Muricauda chongwuensis sp. nov., isolated from coastal seawater of China. Arch. Microbiol. 2021, 203, 6245–6252. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.-G.; Park, J.-S. Flagellimonas hymeniacidonis sp. nov., Isolated from the Sponge Hymeniacidon sinapium. Curr. Microbiol. 2021, 78, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.-L.; Kim, K.-H.; Park, J.-S. Muricauda myxillae sp. nov., isolated from a marine sponge (Myxilla rosacea), and reclassification of Flagellimonas hymeniacidonis as Muricauda symbiotica nom. nov. Int. J. Syst. Evol. Microbiol. 2023, 73, 6040. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, H.; Cha, I.; Joh, K. Flagellimonas maritima sp. nov., isolated from surface seawater. Int. J. Syst. Evol. Microbiol. 2020, 70, 187–192. [Google Scholar] [CrossRef]
- Yoon, J.; Yasumoto-Hirose, M.; Kasai, H. Flagellimonas algarum sp. nov., isolated from dense mats of filamentous algae. Folia Microbiol. 2025, 70, 455–462. [Google Scholar] [CrossRef]
- Hameed, A.; Arun, A.B.; Ho, H.-P.; Chang, C.-M.J.; Rekha, P.D.; Lee, M.-R.; Singh, S.; Young, C.-C. Supercritical Carbon Dioxide Micronization of Zeaxanthin from Moderately Thermophilic Bacteria Muricauda lutaonensis CC-HSB-11T. J. Agric. Food Chem. 2011, 59, 4119–4124. [Google Scholar] [CrossRef]
- Prabhu, S.; Rekha, P.D.; Arun, A.B. Zeaxanthin Biosynthesis by Members of the Genus Muricauda. Pol. J. Microbiol. 2014, 63, 115–119. [Google Scholar] [CrossRef]
- Prabhu, S.; Rekha, P.D.; Young, C.-C.; Hameed, A.; Lin, S.-Y.; Arun, A.B. Zeaxanthin production by novel Marine isolates from coastal sand of India and its antioxidant properties. Appl. Biochem. Biotechnol. 2013, 171, 817–831. [Google Scholar] [CrossRef]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.-H. MomL, a Novel Marine-Derived N-Acyl Homoserine Lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef]
- Tang, K.; Zhang, X.-H. A culture-dependent method for the identification of quorum quenching enzymes of microbial origin. Methods Mol. Biol. 2018, 1673, 297–309. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Cui, X.; Zhang, X.-H. A novel stress response mechanism, triggered by indole, involved in quorum quenching enzyme MomL and iron-sulfur cluster in Muricauda olearia Th120. Sci. Rep. 2017, 7, 4245. [Google Scholar] [CrossRef]
- Tran, V.H.N.; Nguyen, T.T.; Meier, S.; Holck, J.; Cao, H.T.T.; Van, T.T.T.; Meyer, A.S.; Mikkelsen, M.D. The endo-α(1,3)-fucoidanase Mef2 releases uniquely branched oligosaccharides from Saccharina latissima fucoidans. Mar. Drugs 2022, 20, 305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ye, Q.; Chen, Q.; Yang, K.; Zhang, D.; Chen, Z.; Lu, S.; Shao, X.; Fan, Y.; Yao, L.; et al. Algicidal activity of novel marine bacterium Paracoccus sp. strain Y42 against a harmful algal-bloom-causing dinoflagellate, Prorocentrum donghaiense. Appl. Environ. Microbiol. 2018, 84, e01015-18. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhou, Y.; Sun, R.; Wei, H.; Li, Y.; Yin, L.; Pu, Y. Biodegradation of microcystin-LR and-RR by a novel microcystin-degrading bacterium isolated from Lake Taihu. Biodegradation 2014, 25, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.-S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A genome- driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 6421. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Connor, R.; Feldgarden, M.; Fine, A.M.; Funk, K.; Hoffman, J.; et al. Database resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res. 2024, 53, D20–D29. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Goeker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-m.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Vader, L.; Szenei, J.; Reitz, Z.L.; Augustijn, H.E.; Cediel-Becerra, J.D.D.; de Crecy-Lagard, V.; Koetsier, R.A.; Williams, S.E.; et al. antiSMASH 8.0: Extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation. Nucleic Acids Res. 2025, 53, W32–W38. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Yoon, J.; Lee, K.; Kho, Y.; Kang, K.; Kim, C.; Park, Y. Halomonas alimentaria sp nov., isolated from jeotgal, a traditional Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 2002, 52, 123–130. [Google Scholar] [CrossRef]
- Minnikin, D.; Odonnell, A.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, Y.; Lai, Q.; Oren, A.; Wang, W. Paracrocinitomix mangrovi gen. nov., sp. nov., isolated from a mangrove sediment: Proposal of two new families, Phaeocystidibacteraceae fam. nov. and Owenweeksiaceae fam. nov., and emended description of the family Schleiferiaceae. Antonie Leeuwenhoek 2023, 116, 171–184. [Google Scholar] [CrossRef]
- Riesco, R.; Trujillo, M.E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2024, 74, 6300. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Hwang, C.Y.; Kim, M.H.; Bae, G.D.; Zhang, G.I.; Kim, Y.H.; Cho, B.C. Muricauda olearia sp. nov., isolated from crude-oil-contaminated seawater, and emended description of the genus Muricauda. Int. J. Syst. Evol. Microbiol. 2009, 59, 1856–1861. [Google Scholar] [CrossRef]
- Wang, M.-l.; Fu, G.-Y.; Xu, X.-W. Flagellimonas baculiformis sp. nov. and Flagellimonas crocea sp. nov., isolated from surface seawater of the Pacific Ocean. Int. J. Syst. Evol. Microbiol. 2024, 74, 6316. [Google Scholar] [CrossRef]



| Phylum | Species | Algicidal Activity (%) |
|---|---|---|
| Chlorophyta | Chlorella vulgaris | - |
| Tetraselmis helgolandica | - | |
| Nannochloris oculata | - | |
| Pyrrophyta | Prorocentrum donghaiense | 73.48 |
| Amphidinium carterae | 93.01 | |
| Alexandrium tamarense | 93.17 | |
| Alexandrium catenella | 84.09 | |
| Karenia mikimotoi | 89.01 | |
| Scrippsiella trochoidea | 77.67 | |
| Bacillariophyta | Entomoneis alata | - |
| Conticribra weissflogii | 87.19 | |
| Skeletonema costatum | 59.09 | |
| Cylindrotheca closterium | - | |
| Chrysophyta | Isochrysis galbana | - |
| Haptophyta | Phaeocystis globosa | 96.8% |
| Xanthophyta | Heterosigma akashiwo | - |
| Strains | GenBank Accession Number | ANI (%) | AAI (%) | dDDH (%) | |
|---|---|---|---|---|---|
| 1 | Flagellimonas olearia CL-SS4T | WELG01000074 | 91.37 | 94.80 | 44.0 |
| 2 | Flagellimonas alvinocaridis SCR12T | SNTZ01000001 | 86.17 | 90.87 | 30.3 |
| 3 | Flagellimonas beolgyonensis KCTC 23501T | RZMY01000002 | 76.18 | 80.21 | 19.3 |
| 4 | Flagellimonas yonaguniensis 334s03T | JARFVB010000001 | 76.00 | 80.08 | 19.4 |
| 5 | Flagellimonas ruestringensis DSM 13258T | CP002999 | 75.76 | 79.85 | 19.3 |
| 6 | Flagellimonas oceani 501str8T | CP049616 | 75.56 | 79.21 | 19.0 |
| 7 | Flagellimonas aurea ac8T | CP159476 | 75.48 | 79.31 | 18.9 |
| 8 | Flagellimonas chongwuensis HICWT | WYET01000004 | 75.47 | 79.47 | 18.9 |
| 9 | Flagellimonas crocea DH64T | JAUZVX010000001 | 75.38 | 79.67 | 19.3 |
| Characteristic | 1 | 2 * | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Temperature range for growth (°C) (Optimum) | 15–40 (20–30) | 15–40 (25–30) | 16–40 (37) @ | 10–40 # | 15–40 (25–30) & |
| NaCl range for growth (%, w/v) (Optimum) | 3–8 (4) | 1–6(2–3) | 1–5 (3) @ | 0.5–8.0 # | 0.5–8 (2–3) & |
| pH range for growth (Optimum) | 6–10 (6–7) | 5.2–9.4 (6.8–7.7) | 5.5–8.5 (6.5) @ | 5.5–8.5# | 6–8 (7) & |
| Oxidase | + | + | − @ | + | − & |
| Catalase | + | + | − @ | + | + & |
| Enzyme activity (API ZYM) | |||||
| Lipase (C14) | + | − | w | + | + |
| Trypsin | + | − | + | + | + |
| α-Chymotrypsin | + | − | + | + | + |
| α-Galactosidase | − | − | + | + | + |
| β-Galactosidase | w | − | + | + | + |
| β-Glucuronidase | − | − | + | − | − |
| α-Glucosidase | + | − | + | + | + |
| β-Glucosidase | + | − | + | + | + |
| α-Mannosidase | + | − | + | + | + |
| α-Fucosidase | − | − | + | − | − |
| API 20E results: | |||||
| Arginine dihydrolase | − | w | − | − | − |
| Gelatinase | − | + | − | − | − |
| Mannitol | w | − | w | w | − |
| Rhamnose | − | NM | w | w | − |
| Saccharose/Amygdalin | w | NM | + | w | + |
| Arabinose | − | − | − | w | + |
| API 20NE results: | |||||
| Reduction of nitrate to nitrite | − | − | + | − | w |
| Denitrification | − | NM | + | − | w |
| D-Glucose fermentation | + | NM | + | w | + |
| Gelatin hydrolysis | − | + | − | − | − |
| D-Glucose | − | + | − | − | − |
| D-Mannose | − | + | − | − | − |
| Major respiratory quinone | MK−6 | MK−6 | MK−6 @ | MK−6 # | MK−6 & |
| DNA G + C content (mol%) | 43.9 | 50.7 | 42.3 @ | 42.6 # | 41.4 & |
| 1 | 2 * | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| Straight-chain | |||||
| C15:0 | − | 10.7 | − | − | − |
| C16:0 | 1.09 | TR | 1.46 | 1.36 | TR |
| C18:0 | − | − | 2.81 | TR | TR |
| Branched | |||||
| iso-C13:0 | 1.19 | − | TR | 1.17 | 1.37 |
| iso-C15:0 | 42.15 | 18.7 | 38.55 | 49.06 | 46.43 |
| anteiso-C15:0 | 2.69 | 1.4 | TR | 3.12 | 1.59 |
| iso-C15:1 G | 27.97 | 17.1 | 21.28 | 24.33 | 28.76 |
| iso-C16:0 | TR | TR | TR | 1.83 | TR |
| iso-C17:1ω9c | − | 3.6 | − | − | − |
| Unsaturated | |||||
| C15:1ω6c | TR | 1.4 | TR | TR | TR |
| C17:1ω6c | TR | 1.6 | 1.34 | TR | TR |
| C20:2ω6,9c | TR | − | TR | 1.10 | TR |
| Hydroxy | |||||
| C15:0 3-OH | TR | 1.9 | − | 4.19 | TR |
| iso-C15:0 3-OH | 5.93 | 4.4 | 4.83 | − | 5.39 |
| iso-C16:0 3-OH | TR | 1.4 | TR | TR | TR |
| C17:0 3-OH | TR | 1.2 | TR | TR | TR |
| iso-C17:0 3-OH | 4.52 | 20.5 | 4.78 | 4.06 | 6.01 |
| Summed Features # | |||||
| 1 | − | 7.1 | − | − | − |
| 3 | 4.68 | − | − | 1.30 | 3.23 |
| 8 | TR | − | − | TR | TR |
| 9 | 2.88 | − | − | 2.38 | TR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Liang, Y.; Zhou, H.; Chi, Y.; Chen, L.; Lai, Q.; Xu, H. Flagellimonas algicida sp. Nov.: A Novel Broad-Spectrum Algicidal Bacterium Targeting Harmful Algal Bloom Species and Genomic Insights into Its Secondary Metabolites. Microorganisms 2025, 13, 2062. https://doi.org/10.3390/microorganisms13092062
Wang N, Liang Y, Zhou H, Chi Y, Chen L, Lai Q, Xu H. Flagellimonas algicida sp. Nov.: A Novel Broad-Spectrum Algicidal Bacterium Targeting Harmful Algal Bloom Species and Genomic Insights into Its Secondary Metabolites. Microorganisms. 2025; 13(9):2062. https://doi.org/10.3390/microorganisms13092062
Chicago/Turabian StyleWang, Ning, Yiling Liang, Hui Zhou, Yutian Chi, Lizhu Chen, Qiliang Lai, and Hong Xu. 2025. "Flagellimonas algicida sp. Nov.: A Novel Broad-Spectrum Algicidal Bacterium Targeting Harmful Algal Bloom Species and Genomic Insights into Its Secondary Metabolites" Microorganisms 13, no. 9: 2062. https://doi.org/10.3390/microorganisms13092062
APA StyleWang, N., Liang, Y., Zhou, H., Chi, Y., Chen, L., Lai, Q., & Xu, H. (2025). Flagellimonas algicida sp. Nov.: A Novel Broad-Spectrum Algicidal Bacterium Targeting Harmful Algal Bloom Species and Genomic Insights into Its Secondary Metabolites. Microorganisms, 13(9), 2062. https://doi.org/10.3390/microorganisms13092062





