Acute Toxoplasma gondii Infection Drives Gut Microbiome Dysbiosis and Functional Disruption in Mice as Revealed by Metagenomic Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. DNA Extraction, Metagenomic Library Preparation, and Sequencing
2.3. Bioinformatics Analysis of Metagenomic Sequencing Data
2.4. Differential Abundance Analysis and Statistical Assessment of Diversity Across Groups
2.5. Statistical Analysis
3. Results
3.1. Metagenomic Sequencing Quality and Data Overview
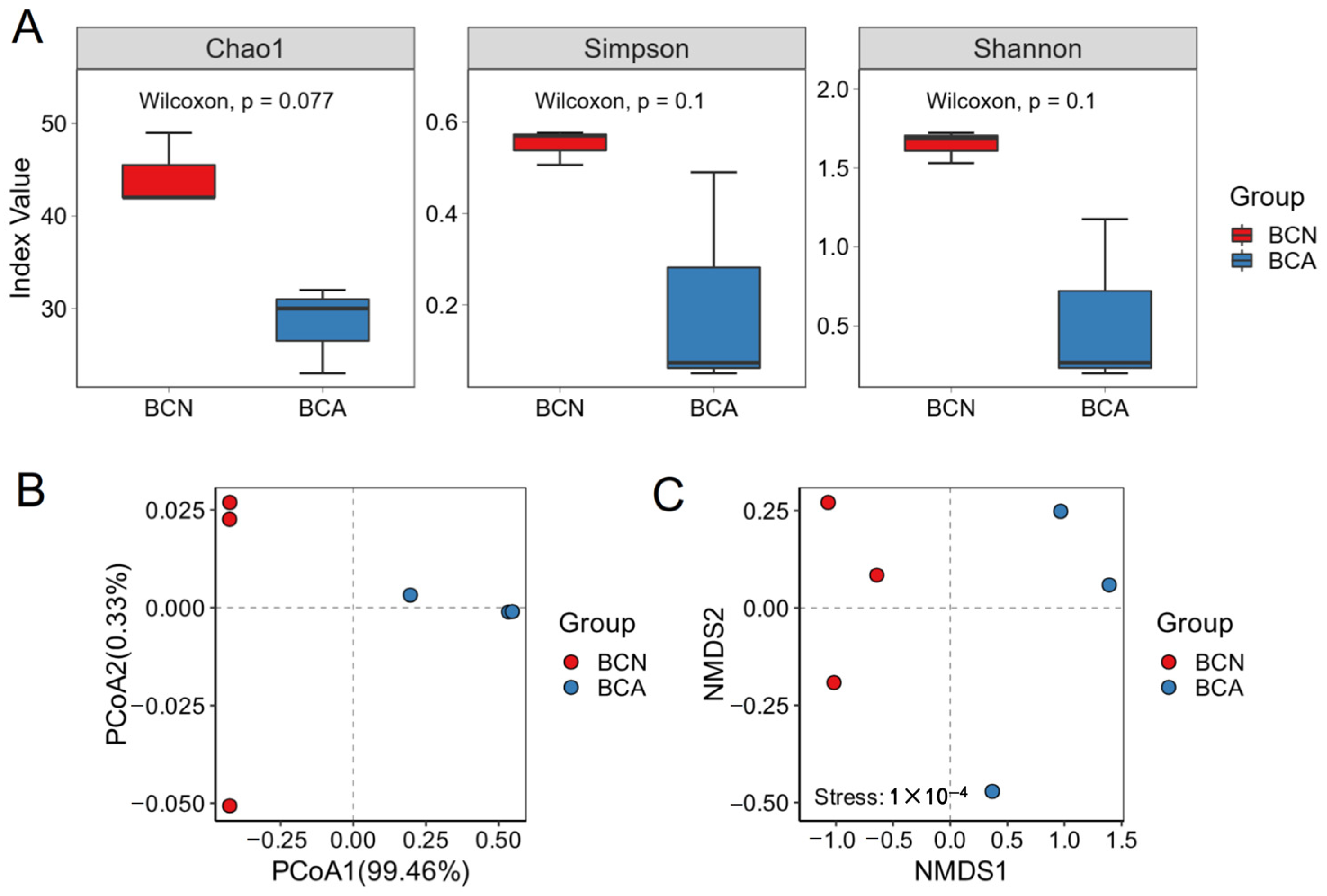
3.2. T. gondii Infection Leads to Changes in Gut Microbial Diversity in Mice
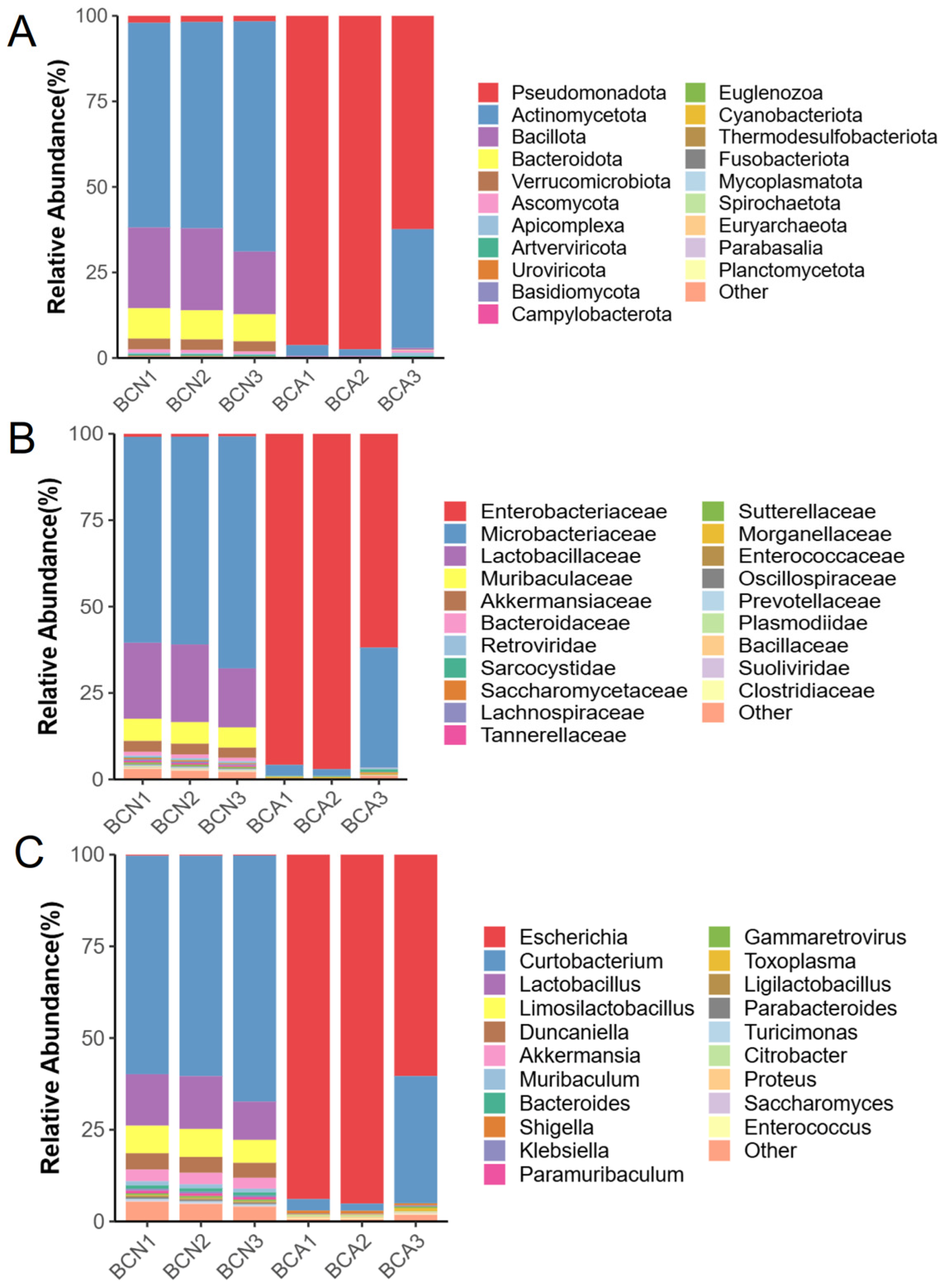
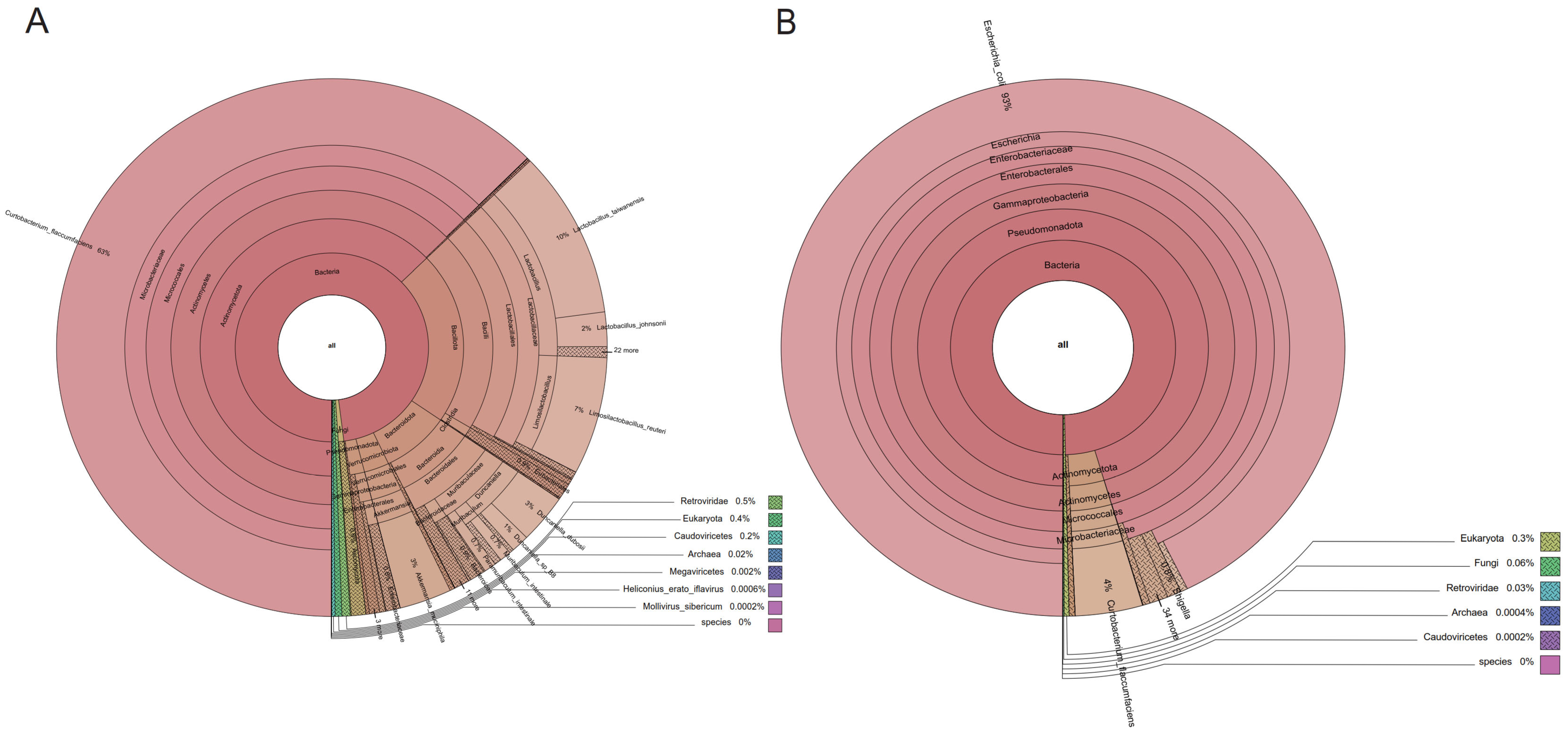
3.3. Disturbance of Gut Microbiome Composition in Diseased Mice
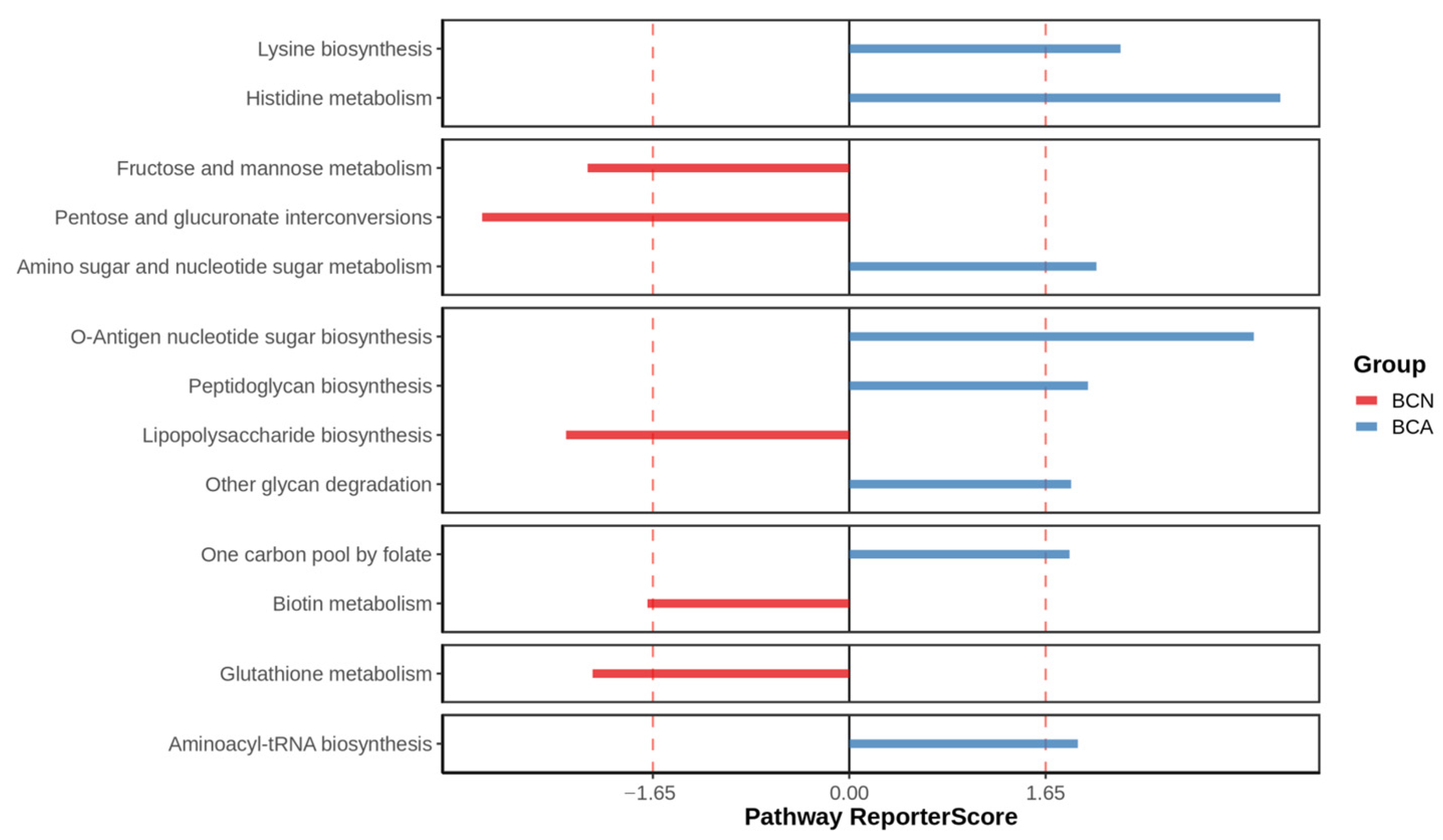
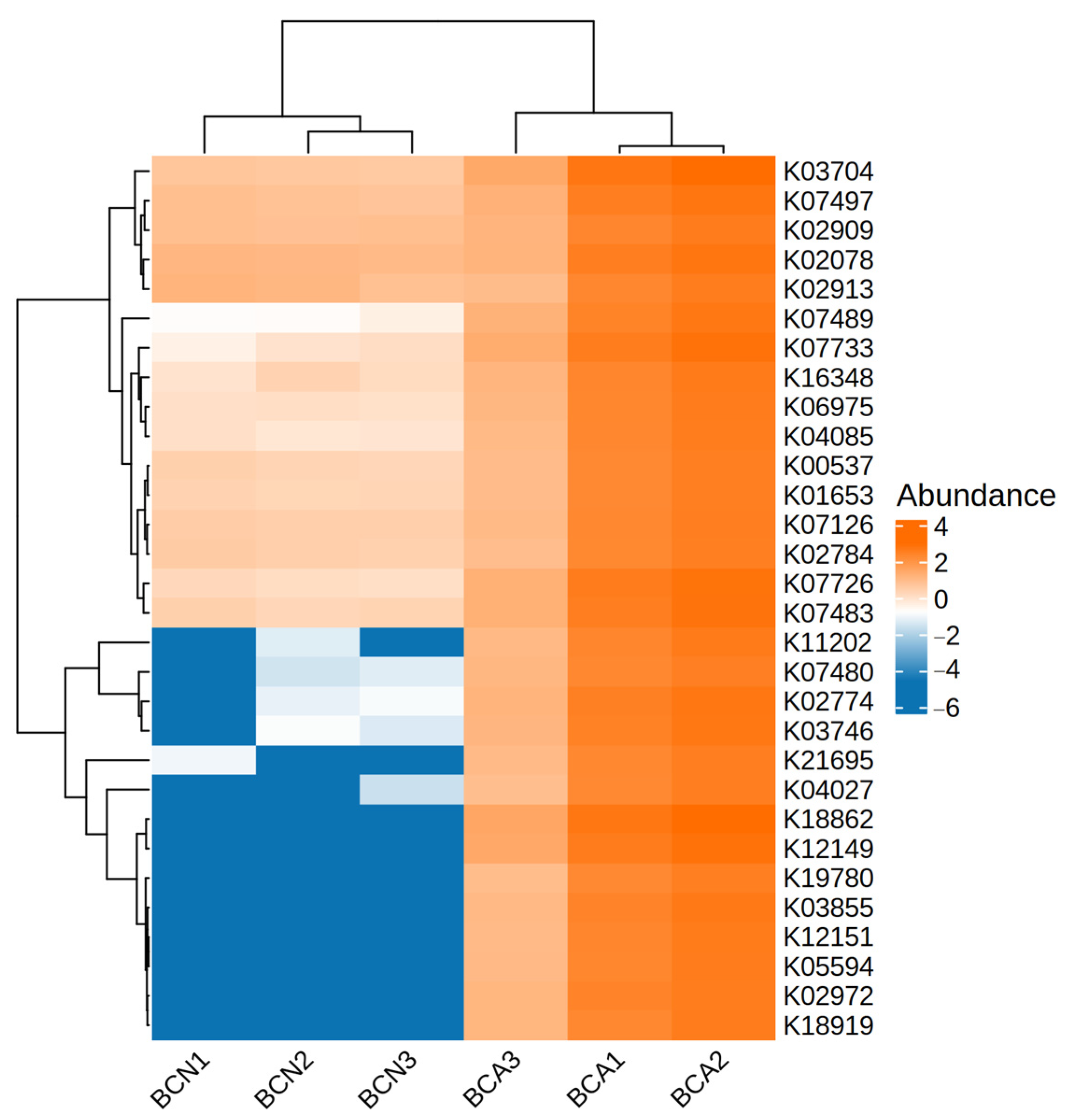
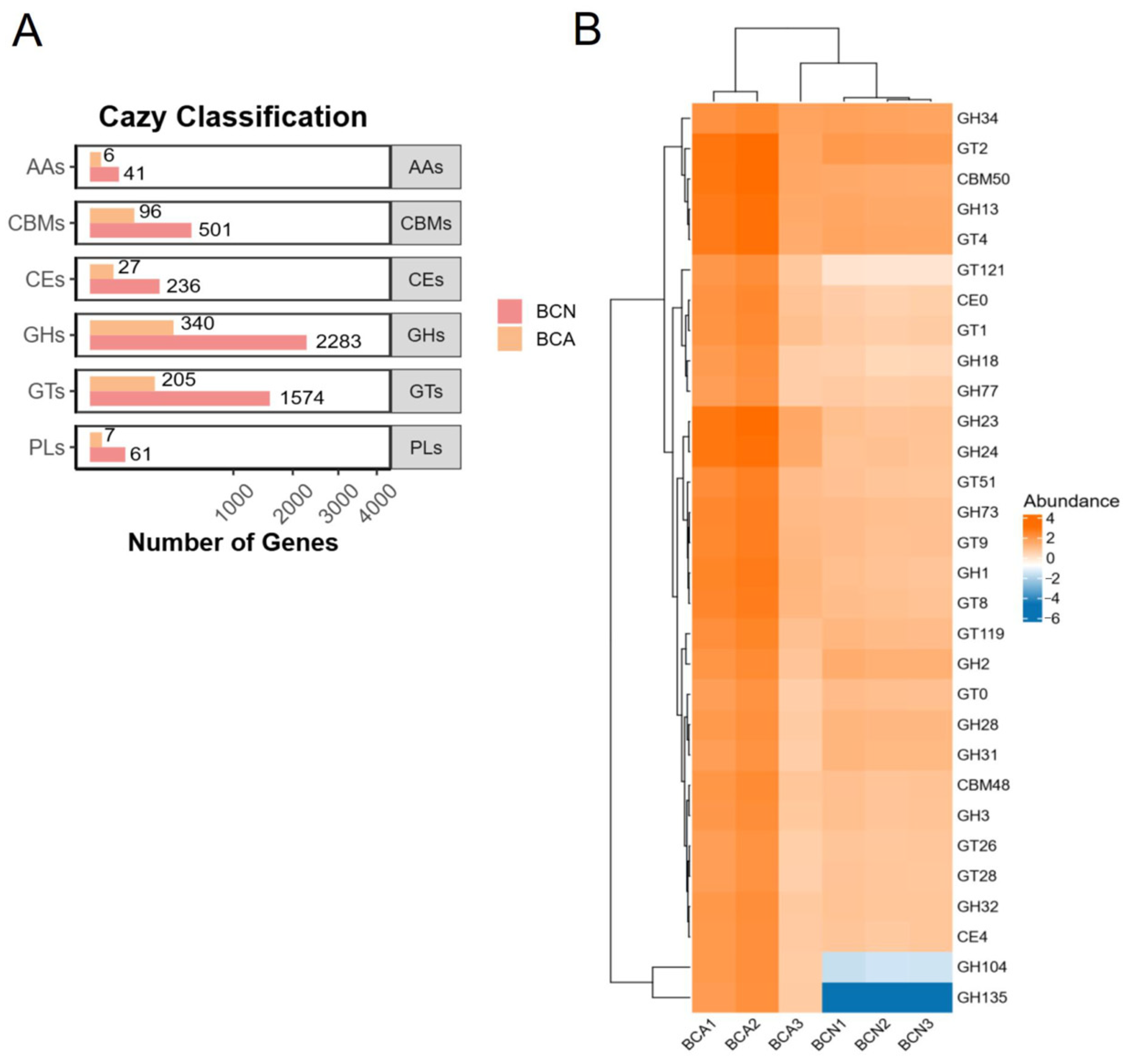
3.4. Functional Changes in the Gut Microbiome of Infected Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DNBSeq | DNA Nanoball Sequencing |
| PE150 | Paired-End 150 bp Sequencing |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CAZy | Carbohydrate-Active enZymes |
| KO | KEGG Ortholog |
| LDA | Linear Discriminant Analysis |
| LEfSe | Linear Discriminant Analysis Effect Size |
| PCoA | Principal Coordinates Analysis |
| NMDS | Non-metric Multidimensional Scaling |
| PERMANOVA | Permutational Multivariate Analysis of Variance |
| LPS | Lipopolysaccharide |
| TLR4 | Toll-Like Receptor 4 |
| IFN-γ | Interferon gamma |
| IDO | Indoleamine 2,3-dioxygenase |
| AhR | Aryl Hydrocarbon Receptor |
| GHs | Glycoside Hydrolases |
| GTs | GlycosylTransferases |
| CBMs | Carbohydrate-Binding Modules |
| CEs | Carboxylesterases |
| PLs | Polysaccharide Lyases |
| AAs | Auxiliary Activityies |
References
- Dubey, J.P.; Jones, J.L. Toxoplasma gondii infection in humans and animals in the United States. Int. J. Parasitol. 2008, 38, 1257–1278. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef]
- Israelski, D.M.; Remington, J.S. Toxoplasmic encephalitis in patients with AIDS. Infect. Dis. Clin. N. Am. 1988, 2, 429–446. [Google Scholar] [CrossRef]
- Couturier-Maillard, A.; Froux, N.; Piotet-Morin, J.; Michaudel, C.; Brault, L.; Le Bérichel, J.; Sénéchal, A.; Robinet, P.; Chenuet, P.; Jejou, S.; et al. Correction: Interleukin-22-deficiency and microbiota contribute to the exacerbation of Toxoplasma gondii-induced intestinal inflammation. Mucosal Immunol. 2019, 12, 290. [Google Scholar] [CrossRef]
- Gay, G.; Braun, L.; Brenier-Pinchart, M.-P.; Vollaire, J.; Josserand, V.; Bertini, R.-L.; Varesano, A.; Touquet, B.; De Bock, P.-J.; Coute, Y.; et al. Toxoplasma gondii TgIST Co-Opts host chromatin repressors dampening STAT1-dependent gene regulation and IFN-γ-mediated host defenses. J. Exp. Med. 2016, 213, 1779–1798. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, C.; Yang, Z.; Wu, W.; Zou, W.; Xin, Z.; Zheng, S.; Liu, R.; Yang, L.; Peng, H. Intestinal microbiota imbalance resulted by anti-Toxoplasma gondii immune responses aggravate gut and brain injury. Parasit Vectors 2024, 17, 284. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.-X.; Wei, X.-Y.; Guo, H.; Chen, Y.; Wang, W.; Geng, H.-L.; Yang, X.; Jiang, J.; Zhang, X.-X. Metagenomic insights into the composition and function of the gut microbiota of mice infected with Toxoplasma gondii. Front. Immunol. 2023, 14, 1156397. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.A.; Hoffmann, C.; Abt, M.C.; Du, Y.; Kobuley, D.; Kirn, T.J.; Bushman, F.D.; Artis, D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010, 3, 148–158. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Yang, J.; Liu, S.; Zhao, Q.; Li, X.; Jiang, K. Gut microbiota-related metabolite alpha-linolenic acid mitigates intestinal inflammation induced by oral infection with Toxoplasma gondii. Microbiome 2023, 11, 273. [Google Scholar] [CrossRef]
- Shao, D.Y.; Bai, X.; Tong, M.W.; Zhang, Y.Y.; Liu, X.L.; Zhou, Y.H.; Li, C.; Cai, W.; Gao, X.; Liu, M.; et al. Changes to the gut microbiota in mice induced by infection with Toxoplasma gondii. Acta Trop. 2020, 203, 105301. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A mapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Patil, K.R.; Nielsen, J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. USA 2005, 102, 2685–2689. [Google Scholar] [CrossRef]
- Matsouaka, R.A.; Singhal, A.B.; Betensky, R.A. An optimal wilcoxon-mann-whitney test of mortality and a continuous outcome. Stat. Methods Med. Res. 2018, 27, 2384–2400. [Google Scholar] [CrossRef]
- Green, G.H.; Diggle, P.J. On the operational characteristics of the benjamini and hochberg false discovery rate procedure. Stat. Appl. Genet. Mol. Biol. 2007, 6, Article27. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.P.; Dubey, J.P. Toxoplasmosis of animals and humans. Parasites Vectors 2010, 3, 112. [Google Scholar] [CrossRef]
- Partida-Rodríguez, O.; Serrano-Vázquez, A.; Nieves-Ramírez, M.E.; Moran, P.; Rojas, L.; Portillo, T.; González, E.; Hernández, E.; Finlay, B.B.; Ximenez, C. Human intestinal microbiota: Interaction between parasites and the host immune response. Arch. Med. Res. 2017, 48, 690–700. [Google Scholar] [CrossRef]
- Prandovszky, E.; Severance, E.G.; Splan, V.W.; Liu, H.; Xiao, J.; Yolken, R.H. Toxoplasma-induced behavior changes-is microbial dysbiosis the missing link? Front. Cell Infect. Microbiol. 2024, 14, 1415079. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashidi, H.S.; El-Wakil, E.S. Parasites and microbiota: Dual interactions and therapeutic perspectives. Microorganisms 2024, 12, 2076. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; El-Fahmawi, A.; Christian, D.A.; Fang, Q.; Radaelli, E.; Chen, L.; Sullivan, M.C.; Misic, A.M.; Ellringer, J.A.; Zhu, X.-Q.; et al. Infection-induced intestinal dysbiosis is mediated by macrophage activation and nitrate production. mBio 2019, 10, e00935–e19. [Google Scholar] [CrossRef]
- Heimesaat, M.M.; Bereswill, S.; Fischer, A.; Fuchs, D.; Struck, D.; Niebergall, J.; Jahn, H.-K.; Dunay, I.R.; Moter, A.; Gescher, D.M.; et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J. Immunol. 2006, 177, 8785–8795. [Google Scholar] [CrossRef]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Aldars-García, L.; Marin, A.C.; Chaparro, M.; Gisbert, J.P. The interplay between immune system and microbiota in inflammatory bowel disease: A narrative review. Int. J. Mol. Sci. 2021, 22, 3076. [Google Scholar] [CrossRef]
- Fingas, F.; Volke, D.; Hassert, R.; Fornefett, J.; Funk, S.; Baums, C.G.; Hoffmann, R. Sensitive and immunogen-specific serological detection of rodentibacter pneumotropicus infections in mice. BMC Microbiol. 2019, 19, 43. [Google Scholar] [CrossRef]
- Hoogland, I.C.M.; Houbolt, C.; van Westerloo, D.J.; van Gool, W.A.; van de Beek, D. Systemic inflammation and microglial activation: Systematic review of animal experiments. J. Neuroinflamm. 2015, 12, 114. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Kim, M.-S.; Kim, E.; Cheon, J.H.; Lee, Y.-S.; Kim, Y.; Lee, S.-H.; Seo, S.-U.; Shin, S.-H.; Choi, S.S.; et al. Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-β production. Immunity 2016, 44, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Dicksved, J.; Halfvarson, J.; Rosenquist, M.; Järnerot, G.; Tysk, C.; Apajalahti, J.; Engstrand, L.; Jansson, J.K. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008, 2, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Gaiani, F.; Mancabelli, L.; Milani, C.; Grandi, A.; Bolchi, A.; Santoni, A.; Lugli, G.A.; Ferrario, C.; Mangifesta, M.; et al. Elucidating the gut microbiome of ulcerative colitis: Bifidobacteria as novel microbial biomarkers. FEMS Microbiol. Ecol. 2016, 92, fiw191. [Google Scholar] [CrossRef] [PubMed]
- Sartor, R.B.; Wu, G.D. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 2017, 152, 327–339.e4. [Google Scholar] [CrossRef]
- Gophna, U.; Sommerfeld, K.; Gophna, S.; Doolittle, W.F.; Veldhuyzen van Zanten, S.J.O. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006, 44, 4136–4141. [Google Scholar] [CrossRef]
- Candelli, M.; Franza, L.; Pignataro, G.; Ojetti, V.; Covino, M.; Piccioni, A.; Gasbarrini, A.; Franceschi, F. Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int. J. Mol. Sci. 2021, 22, 6242. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Garcia-Gil, L.J. Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity. World J. Gastrointest. Pathophysiol. 2014, 5, 213–227. [Google Scholar] [CrossRef]
- Chervy, M.; Barnich, N.; Denizot, J. Adherent-invasive E. coli: Update on the lifestyle of a troublemaker in Crohn’s disease. Int. J. Mol. Sci. 2020, 21, 3734. [Google Scholar] [CrossRef]
- Pasternak, B.A.; D’Mello, S.; Jurickova, I.I.; Han, X.; Willson, T.; Flick, L.; Petiniot, L.; Uozumi, N.; Divanovic, S.; Traurnicht, A.; et al. Lipopolysaccharide exposure is linked to activation of the acute phase response and growth failure in pediatric Crohn’s disease and murine colitis. Inflamm. Bowel Dis. 2010, 16, 856–869. [Google Scholar] [CrossRef]
- Wei, S.; Bahl, M.I.; Baunwall, S.M.D.; Hvas, C.L.; Licht, T.R. Determining gut microbial dysbiosis: A review of applied indexes for assessment of intestinal microbiota imbalances. Appl. Environ. Microbiol. 2021, 87, e00395–e21. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Zhu, S.; Wang, B.; Duan, L. Alterations of gut microbiota in patients with irritable bowel syndrome based on 16S rRNA-targeted sequencing: A systematic review. Clin. Transl. Gastroenterol. 2019, 10, e00012. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Yang, Y. Gut commensal bacteria exacerbate toxoplasmosis associated with TgSheepCHn5 (ToxoDB#2) and TgRedpandaCHn1 (ToxoDB#20) through Th1 immune response. Parasitol. Res. 2023, 122, 2795–2806. [Google Scholar] [PubMed]
- Karch, H.; Tarr, P.I.; Bielaszewska, M. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 2005, 295, 405–418. [Google Scholar] [CrossRef]
- Ashraf, K.U.; Nygaard, R.; Vickery, O.N.; Erramilli, S.K.; Herrera, C.M.; McConville, T.H.; Petrou, V.I.; Giacometti, S.I.; Dufrisne, M.B.; Nosol, K.; et al. Structural basis of lipopolysaccharide maturation by the O-antigen ligase. Nature 2022, 604, 371–376. [Google Scholar] [CrossRef]
- Whitfield, C.; Williams, D.M.; Kelly, S.D. Lipopolysaccharide O-antigens—Bacterial glycans made to measure. J. Biol. Chem. 2020, 295, 10593–10609. [Google Scholar] [CrossRef]
- Greenfield, L.K.; Whitfield, C. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 2012, 356, 12–24. [Google Scholar] [CrossRef]
- An, L.; Wirth, U.; Koch, D.; Schirren, M.; Drefs, M.; Koliogiannis, D.; Nieß, H.; Andrassy, J.; Guba, M.; Bazhin, A.V.; et al. The role of gut-derived lipopolysaccharides and the intestinal barrier in fatty liver diseases. J. Gastrointest. Surg. 2022, 26, 671–683. [Google Scholar] [CrossRef]
- Man, S.M. Inflammasomes in the gastrointestinal tract: Infection, cancer and gut microbiota homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Mi, W. Regulatory Mechanisms of lipopolysaccharide synthesis in Escherichia coli. Nat. Commun. 2022, 13, 4576. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, A.; Sendobra, A.; Maniyeri, A.; Sugalska, M.; Klein, G.; Raina, S. A new factor LapD is required for the regulation of LpxC amounts and lipopolysaccharide trafficking. Int. J. Mol. Sci. 2022, 23, 9706. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, P.G.; Lutkenhaus, J.; Young, K.; Eveland, S.S.; Anderson, M.S.; Raetz, C.R.H. Regulation of UDP-3-O-[R-3-Hydroxymyristoyl]-N-Acetylglucosamine deacetylase in Escherichia coli. J. Biol. Chem. 1996, 271, 25898–25905. [Google Scholar] [CrossRef]
- Bertani, B.; Ruiz, N. Function and biogenesis of lipopolysaccharides. EcoSal Plus. 2018, 8, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Cerávolo, I.P.; Chaves, A.C.; Bonjardim, C.A.; Sibley, D.; Romanha, A.J.; Gazzinelli, R.T. Replication of Toxoplasma gondii, but not trypanosoma cruzi, is regulated in human fibroblasts activated with gamma interferon: Requirement of a functional JAK/STAT pathway. Infect. Immun. 1999, 67, 2233–2240. [Google Scholar] [CrossRef]
- Pfefferkorn, E.R. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl. Acad. Sci. USA 1984, 81, 908–912. [Google Scholar] [CrossRef]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.-Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Dos Santos, L.M.; Commodaro, A.G.; Vasquez, A.R.R.; Kohlhoff, M.; de Paula Guerra, D.A.; Coimbra, R.S.; Martins-Filho, O.A.; Teixeira-Carvalho, A.; Rizzo, L.V.; Vieira, L.Q.; et al. Intestinal microbiota regulates tryptophan metabolism following oral infection with Toxoplasma gondii. Parasite Immunol. 2020, 42, e12720. [Google Scholar] [CrossRef] [PubMed]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Deng, C.; Sui, M.; Wei, P.; Duan, B.; Li, Z.; Zou, F. Acute Toxoplasma gondii Infection Drives Gut Microbiome Dysbiosis and Functional Disruption in Mice as Revealed by Metagenomic Sequencing. Microorganisms 2025, 13, 2056. https://doi.org/10.3390/microorganisms13092056
Wang Y, Deng C, Sui M, Wei P, Duan B, Li Z, Zou F. Acute Toxoplasma gondii Infection Drives Gut Microbiome Dysbiosis and Functional Disruption in Mice as Revealed by Metagenomic Sequencing. Microorganisms. 2025; 13(9):2056. https://doi.org/10.3390/microorganisms13092056
Chicago/Turabian StyleWang, Yidan, Caiqin Deng, Minmin Sui, Penghao Wei, Bofang Duan, Zhao Li, and Fengcai Zou. 2025. "Acute Toxoplasma gondii Infection Drives Gut Microbiome Dysbiosis and Functional Disruption in Mice as Revealed by Metagenomic Sequencing" Microorganisms 13, no. 9: 2056. https://doi.org/10.3390/microorganisms13092056
APA StyleWang, Y., Deng, C., Sui, M., Wei, P., Duan, B., Li, Z., & Zou, F. (2025). Acute Toxoplasma gondii Infection Drives Gut Microbiome Dysbiosis and Functional Disruption in Mice as Revealed by Metagenomic Sequencing. Microorganisms, 13(9), 2056. https://doi.org/10.3390/microorganisms13092056





