Study on the Plateau Adaptive Synergistic Mechanism of Rumen Microbiome-Metabolome-Resistome in Tibetan Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. Metagenomic Sequencing
2.2.1. Library Construction and Sequencing
2.2.2. Data Quality Control and Bioinformatics Analysis
2.3. Analysis of Rumen Microbial Metabolome
2.4. Metagenome-Metabolome Association Analysis
2.5. Data Analysis
3. Results
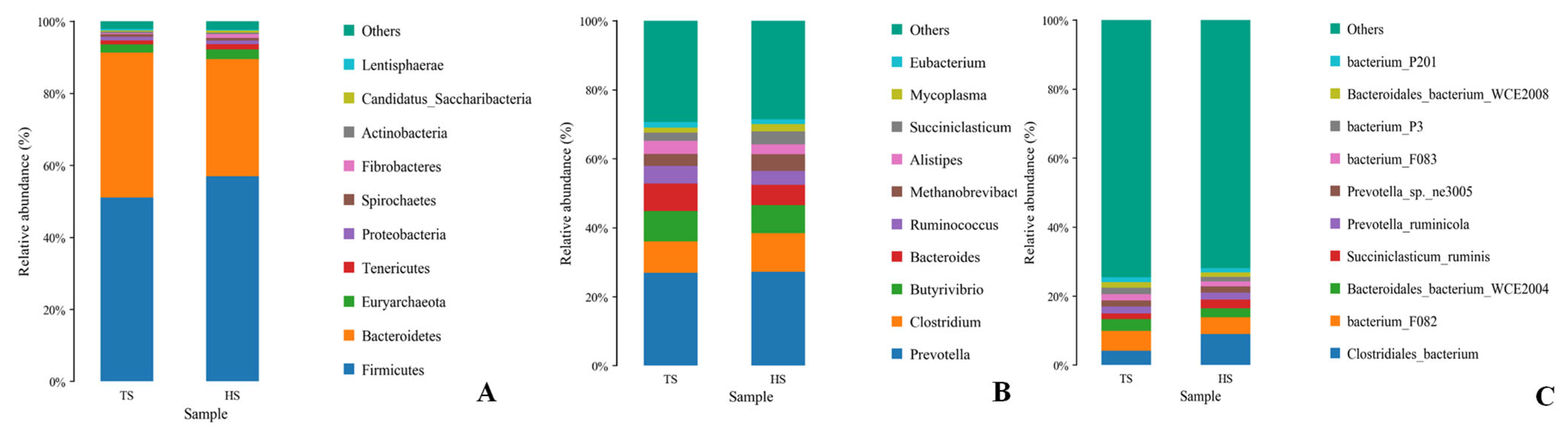
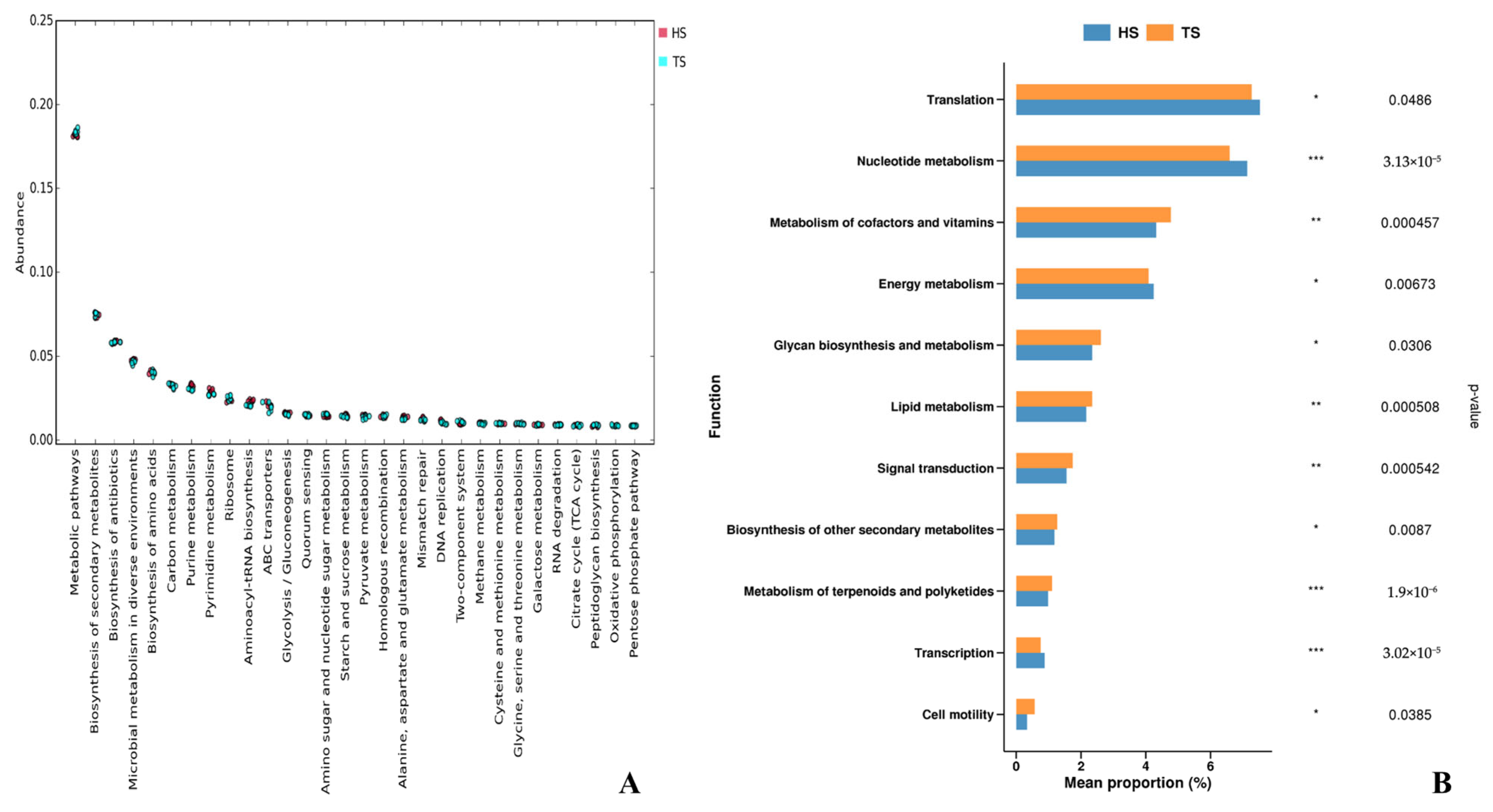
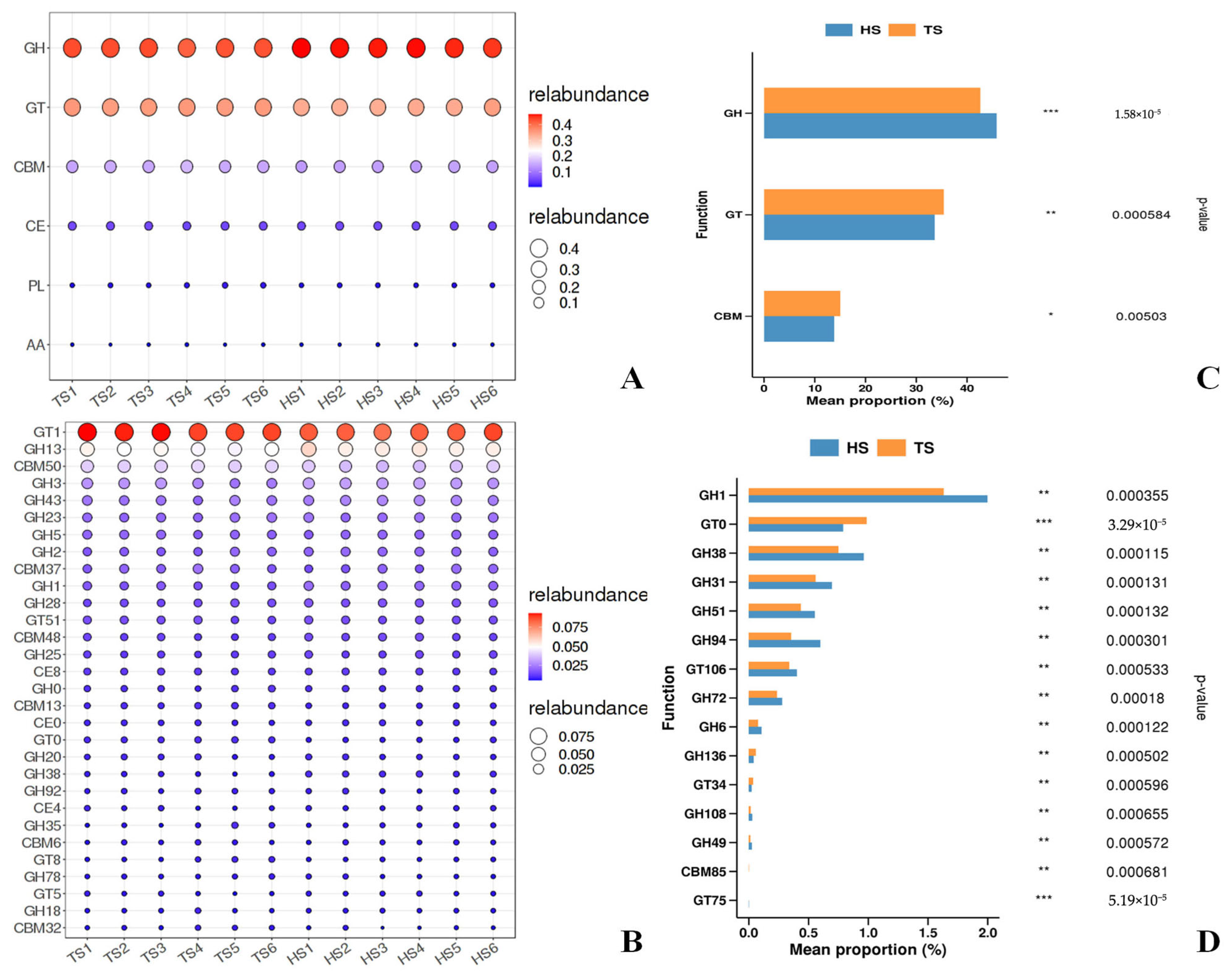
3.1. Analysis of Rumen Microbial Composition and Function in Tibetan Sheep and Hu Sheep
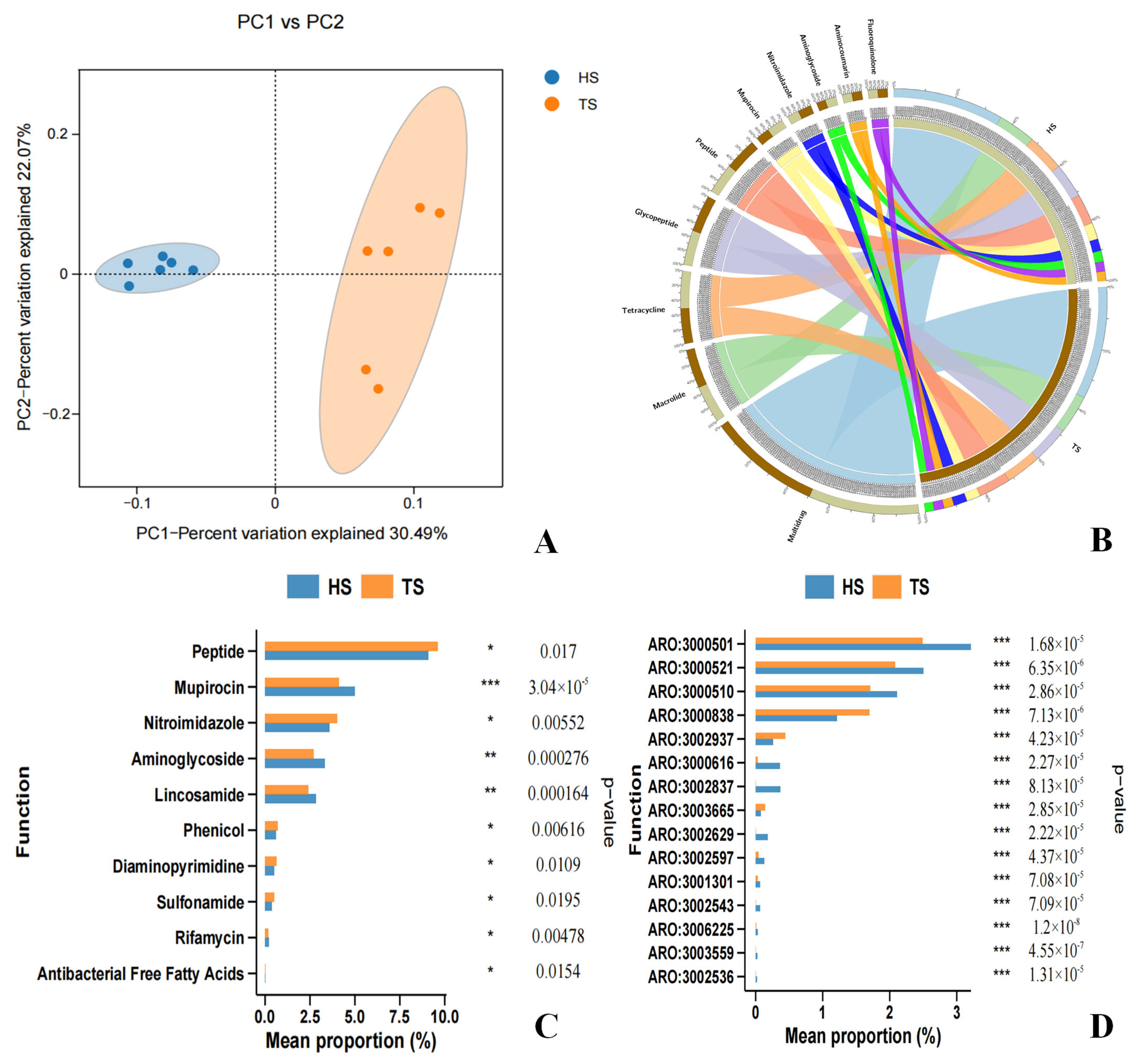
3.2. Analysis of Rumen ARGs in Tibetan Sheep and Hu Sheep
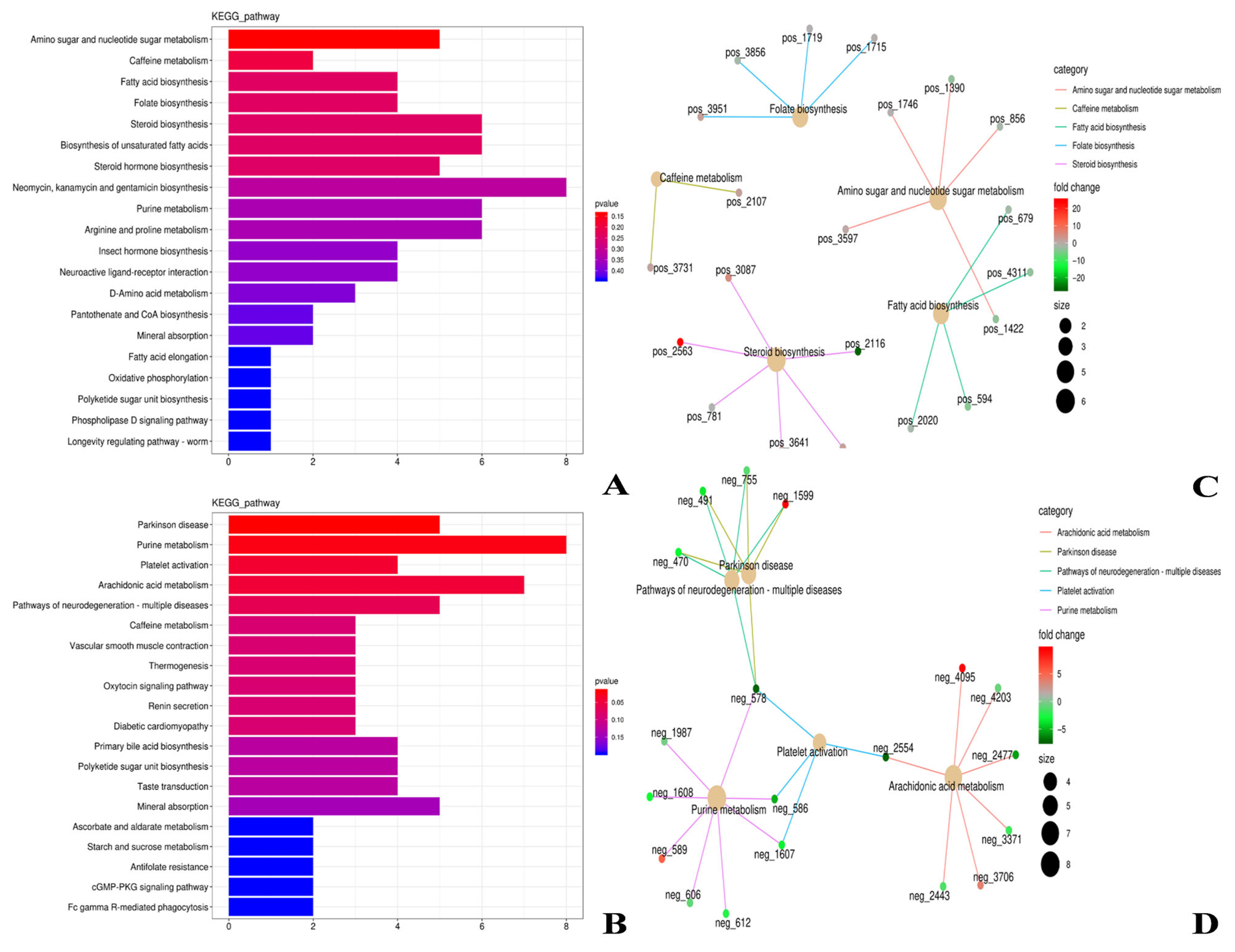
3.3. Analysis of Rumen Metabolome in Tibetan Sheep and Hu Sheep
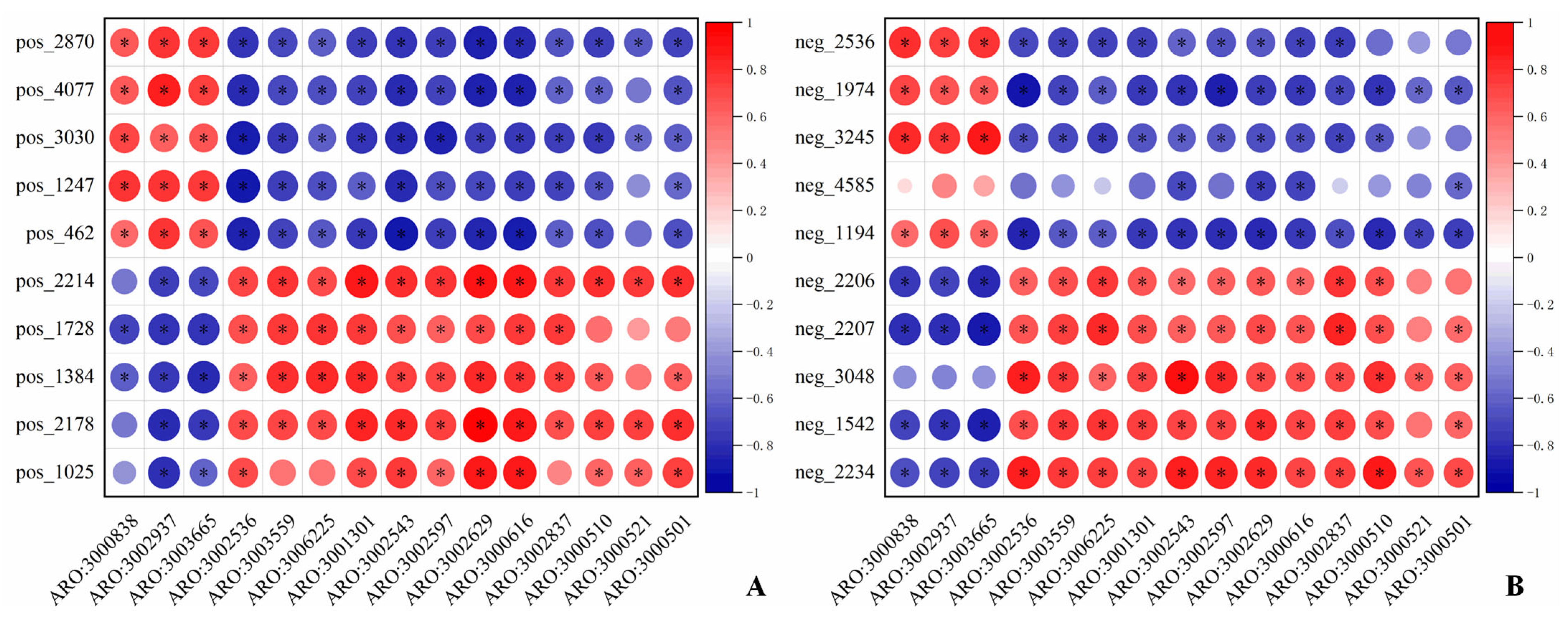
3.4. Correlation Analysis
3.4.1. Rumen Microbiota–Metabolite Interactions
3.4.2. ARGs-Differential Metabolite Analysis
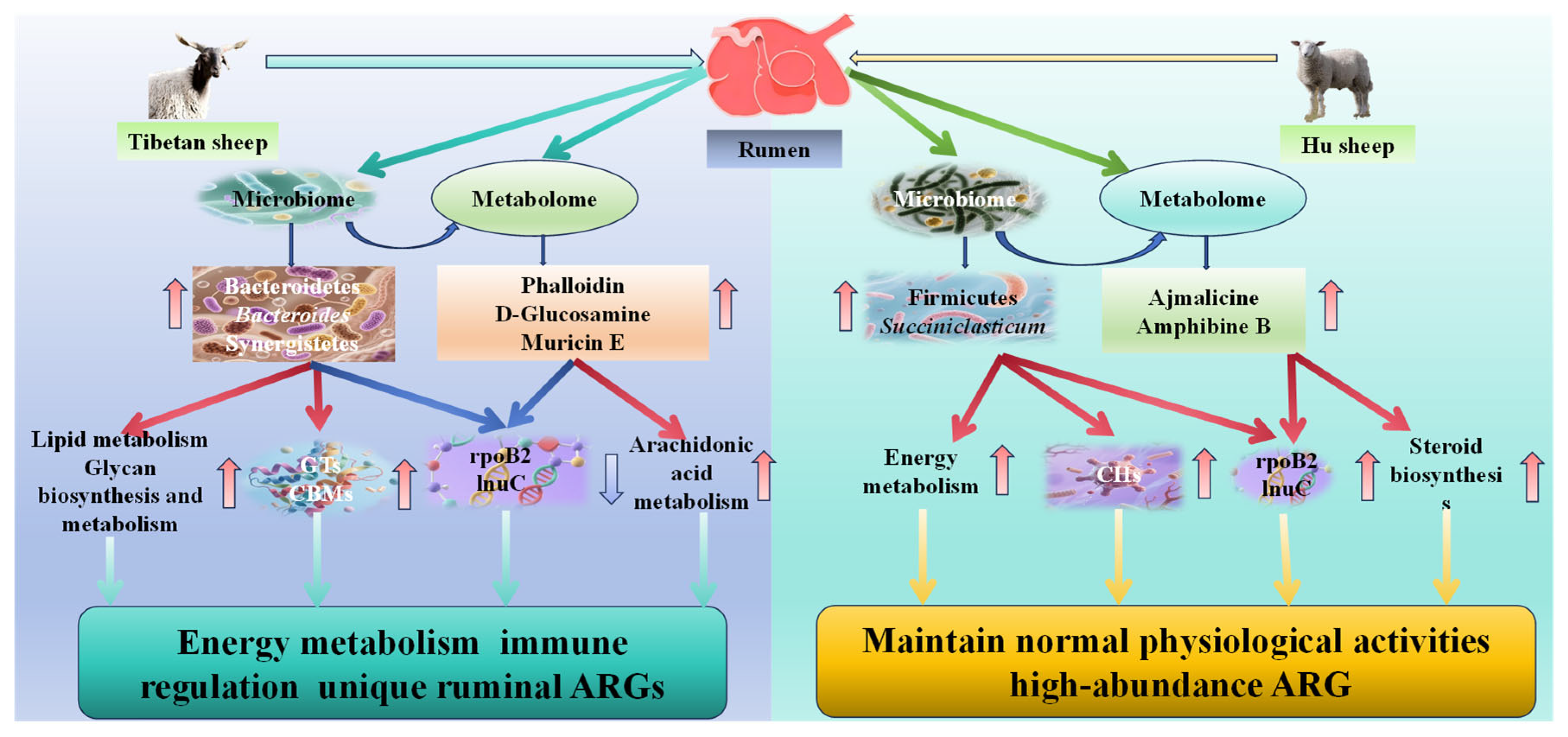
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, B.; Jia, G.; Wen, D.; Zhao, X.; Zhang, J.; Xu, Q.; Zhao, X.; Jiang, N.; Liu, Z.; Wang, Y. Rumen microbiota of indigenous and introduced ruminants and their adaptation to the qinghai-tibetan plateau. Front. Microbiol. 2022, 13, 1027138. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 2016, 26, 1873–1879. [Google Scholar] [CrossRef]
- Yang, C.; Gao, P.; Hou, F.; Yan, T.; Chang, S.; Chen, X.; Wang, Z. Relationship between chemical composition of native forage and nutrient digestibility by tibetan sheep on the qinghai-tibetan plateau. J. Anim. Sci. 2018, 96, 1140–1149. [Google Scholar] [CrossRef]
- Xue, B.; Zhao, X.Q.; Zhang, Y.S. Seasonal changes in weight and body composition of yak grazing on alpine-meadow grassland in the qinghai-tibetan plateau of china. J. Anim. Sci. 2005, 83, 1908–1913. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Chen, H.; Zhao, X.; Xu, S.; Hu, L.; Xu, T.; Jiang, L.; Zhan, W. Rumen prokaryotic communities of ruminants under different feeding paradigms on the qinghai-tibetan plateau. Syst. Appl. Microbiol. 2017, 40, 227–236. [Google Scholar] [CrossRef]
- Sun, Y.; Angerer, J.; Hou, F. Effects of grazing systems on herbage mass and liveweight gain of tibetan sheep in eastern qinghai-tibetan plateau, china. Rangel. J. 2015, 37, 181–190. [Google Scholar] [CrossRef]
- Pan, X.; Cai, Y.; Li, Z.; Chen, X.; Heller, R.; Wang, N.; Wang, Y.; Zhao, C.; Wang, Y.; Xu, H.; et al. Modes of genetic adaptations underlying functional innovations in the rumen. Sci. China Life Sci. 2021, 64, 1–21. [Google Scholar] [CrossRef]
- Lv, W.; Liu, X.; Sha, Y.; Shi, H.; Wei, H.; Luo, Y.; Wang, J.; Li, S.; Hu, J.; Guo, X.; et al. Rumen fermentation-microbiota-host gene expression interactions to reveal the adaptability of tibetan sheep in different periods. Animals 2021, 11, 3529. [Google Scholar] [CrossRef]
- Chai, J.; Zhuang, Y.; Cui, K.; Bi, Y.; Zhang, N. Metagenomics reveals the temporal dynamics of the rumen resistome and microbiome in goat kids. Microbiome 2024, 12, 14. [Google Scholar] [CrossRef]
- Auffret, M.D.; Dewhurst, R.J.; Duthie, C.; Rooke, J.A.; John Wallace, R.; Freeman, T.C.; Stewart, R.; Watson, M.; Roehe, R. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome 2017, 5, 159, Erratum in Microbiome 2019, 7, 149. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Wang, L.; Wang, K.; Yang, Y.; Ma, T.; Wang, Z.; Zhang, X.; Ni, Z.; Hou, F.; Long, R.; et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 2015, 6, 10283. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Rowland, I.R. Factors affecting metabolic activity of the intestinal microflora. Drug Metab. Rev. 1988, 19, 243–261. [Google Scholar] [CrossRef]
- Douglas, J.; Worgan, H.J.; Easton, G.L.; Poret, L.; Wolf, B.T.; Edwards, A.; Davies, E.; Ross, D.; McEwan, N.R. Microbial diversity in the digestive tract of two different breeds of sheep. J. Appl. Microbiol. 2016, 120, 1382–1389. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Zhang, H.; Wei, Q.; Zhao, C.; Yang, X.; Wu, X.; Xia, T.; Liu, G.; Zhang, L.; Gao, Y.; et al. Comparative analyses of fecal microbiota in european mouflon (ovis orientalis musimon) and blue sheep (pseudois nayaur) living at low or high altitudes. Front. Microbiol. 2019, 10, 1735. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Zhang, W.; Shen, W.; Zhang, G.; Xie, T.; Li, L.; Jinmei, J.; Liu, Y.; Kong, F.; Guo, B.; et al. Analysis of gut microbiota in chinese donkey in different regions using metagenomic sequencing. BMC Genom. 2023, 24, 524. [Google Scholar] [CrossRef]
- He, H.; Fang, C.; Liu, L.; Li, M.; Liu, W. Environmental driving of adaptation mechanism on rumen microorganisms of sheep based on metagenomics and metabolomics data analysis. Int. J. Mol. Sci. 2024, 25, 10957. [Google Scholar] [CrossRef]
- Campbell, T.P.; Sun, X.; Patel, V.H.; Sanz, C.; Morgan, D.; Dantas, G. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 2020, 14, 1584–1599. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, K.; Wu, W.; Giannoulatou, E.; Ho, J.W.K.; Li, L. Host and microbiome multi-omics integration: Applications and methodologies. Biophys. Rev. 2019, 11, 55–65. [Google Scholar] [CrossRef]
- Gao, H.; Yu, Y.; Lv, Y.; Wang, D.; Li, H.; Li, Z.; Zhang, Y.; Chen, L.; Leng, J. Metagenomic sequencing reveals the taxonomic and functional characteristics of rumen micro-organisms in gayals. Microorganisms 2023, 11, 1098. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, L.; Ke, S.; Chen, X.; Kenéz, Á.; Xu, W.; Wang, D.; Zhang, F.; Li, Y.; Cui, Z.; et al. Yak rumen microbiome elevates fiber degradation ability and alters rumen fermentation pattern to increase feed efficiency. Anim. Nutr. 2022, 11, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Peng, K.; Xue, M.; Liu, J. Metagenomics analysis revealed the distinctive ruminal microbiome and resistive profiles in dairy buffaloes. Anim. Microbiome 2021, 3, 44. [Google Scholar] [CrossRef]
- Wang, D.; Chen, L.; Tang, G.; Yu, J.; Chen, J.; Li, Z.; Cao, Y.; Lei, X.; Deng, L.; Wu, S.; et al. Multi-omics revealed the long-term effect of ruminal keystone bacteria and the microbial metabolome on lactation performance in adult dairy goats. Microbiome 2023, 11, 215. [Google Scholar] [CrossRef]
- Xue, M.; Sun, H.; Wu, X.; Liu, J.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Luo, R.; Sadakane, K.; Lam, T. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Soding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar]
- Sakaki, T.; Takeshima, T.; Tominaga, M.; Hashimoto, H.; Kawaguchi, S. Recurrence of ICA-PCoA aneurysms after neck clipping. J. Neurosurg. 1994, 80, 58–63. [Google Scholar] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sha, Y.; Liu, X.; He, Y.; Chen, X.; Yang, W.; Gao, M.; Huang, W.; Wang, J.; He, J.; et al. Unique rumen micromorphology and microbiota-metabolite interactions: Features and strategies for tibetan sheep adaptation to the plateau. Front. Microbiol. 2024, 15, 1471732. [Google Scholar] [CrossRef]
- McHardy, I.H.; Goudarzi, M.; Tong, M.; Ruegger, P.M.; Schwager, E.; Weger, J.R.; Graeber, T.G.; Sonnenburg, J.L.; Horvath, S.; Huttenhower, C.; et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 2013, 1, 17. [Google Scholar] [CrossRef]
- Furman, O.; Shenhav, L.; Sasson, G.; Kokou, F.; Honig, H.; Jacoby, S.; Hertz, T.; Cordero, O.X.; Halperin, E.; Mizrahi, I. Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 2020, 11, 1904. [Google Scholar] [CrossRef]
- Zhang, Y.K.; Zhang, X.X.; Li, F.D.; Li, C.; Li, G.Z.; Zhang, D.Y.; Song, Q.Z.; Li, X.L.; Zhao, Y.; Wang, W.M. Characterization of the rumen microbiota and its relationship with residual feed intake in sheep. Animal 2021, 15, 100161. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the role of erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial changes during pregnancy, birth, and infancy. Front. Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Oddy, V.H.; Archibald, A.L.; Vercoe, P.E.; Dalrymple, B.P. Epithelial, metabolic and innate immunity transcriptomic signatures differentiating the rumen from other sheep and mammalian gastrointestinal tract tissues. PeerJ 2016, 4, e1762. [Google Scholar] [CrossRef]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef]
- Kim, M.; Morrison, M.; Yu, Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 2011, 76, 49–63. [Google Scholar] [CrossRef]
- Cunha, I.S.; Barreto, C.C.; Costa, O.Y.A.; Bomfim, M.A.; Castro, A.P.; Kruger, R.H.; Quirino, B.F. Bacteria and archaea community structure in the rumen microbiome of goats (capra hircus) from the semiarid region of brazil. Anaerobe 2011, 17, 118–124. [Google Scholar] [CrossRef]
- Murphy, E.F.; Cotter, P.D.; Healy, S.; Marques, T.M.; O’Sullivan, O.; Fouhy, F.; Clarke, S.F.; O’Toole, P.W.; Quigley, E.M.; Stanton, C.; et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut 2010, 59, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Fernando, S.C.; Purvis, H.T.N.; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; Desilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef]
- Kawasoe, J.; Uchida, Y.; Kawamoto, H.; Miyauchi, T.; Watanabe, T.; Saga, K.; Tanaka, K.; Ueda, S.; Terajima, H.; Taura, K.; et al. Propionic acid, induced in gut by an inulin diet, suppresses inflammation and ameliorates liver ischemia and reperfusion injury in mice. Front. Immunol. 2022, 13, 862503. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, X.; Yang, S.; Zhou, J.; Zhang, T.; Qi, L.; Sun, X.; Fan, M.; Xu, S.; Cha, M.; et al. Comparative analysis of the gut microbiota composition between captive and wild forest musk deer. Front. Microbiol. 2017, 8, 1705. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, T.; Xu, S.; Ma, L.; Han, X.; Wang, X.; Zhang, X.; Hu, L.; Zhao, N.; Chen, Y.; et al. Effect of dietary concentrate to forage ratio on growth performance, rumen fermentation and bacterial diversity of tibetan sheep under barn feeding on the qinghai-tibetan plateau. PeerJ 2019, 7, e7462. [Google Scholar] [CrossRef]
- Shi, W.; Moon, C.D.; Leahy, S.C.; Kang, D.; Froula, J.; Kittelmann, S.; Fan, C.; Deutsch, S.; Gagic, D.; Seedorf, H.; et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 2014, 24, 1517–1525. [Google Scholar] [CrossRef]
- Purushe, J.; Fouts, D.E.; Morrison, M.; White, B.A.; Mackie, R.I.; Coutinho, P.M.; Henrissat, B.; Nelson, K.E. Comparative genome analysis of prevotella ruminicola and prevotella bryantii: Insights into their environmental niche. Microb. Ecol. 2010, 60, 721–729. [Google Scholar] [CrossRef]
- Teng, Z.; Zhang, N.; Zhang, L.; Zhang, L.; Liu, S.; Fu, T.; Wang, Q.; Gao, T. Integrated multi-omics reveals new ruminal microbial features associated with peanut vine efficiency in dairy cattle. Life 2024, 14, 802. [Google Scholar] [CrossRef]
- Ricke, S.C.; Martin, S.A.; Nisbet, D.J. Ecology, metabolism, and genetics of ruminal selenomonads. Crit. Rev. Microbiol. 1996, 22, 27–56. [Google Scholar] [CrossRef]
- Sawanon, S.; Koike, S.; Kobayashi, Y. Evidence for the possible involvement of selenomonas ruminantium in rumen fiber digestion. FEMS Microbiol. Lett. 2011, 325, 170–179. [Google Scholar] [CrossRef]
- Li, W.; Ma, T.; Zhang, N.; Deng, K.; Diao, Q. Dietary fat supplement affected energy and nitrogen metabolism efficiency and shifted rumen fermentation toward glucogenic propionate production via enrichment of succiniclasticum in male twin lambs. J. Integr. Agric. 2025, 24, 1285–1295. [Google Scholar] [CrossRef]
- Alvanou, M.V.; Loukovitis, D.; Melfou, K.; Giantsis, I.A. Utility of dairy microbiome as a tool for authentication and traceability. Open Life Sci. 2024, 19, 20220983. [Google Scholar] [CrossRef]
- Jenkins, T.C. Lipid metabolism in the rumen. J. Dairy Sci. 1993, 76, 3851–3863. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Hu, B.; Zhang, X.; Ge, Q.; Yan, Y.; Akresi, J.; Piyush, V.; Huang, L.; Yin, Y. DbCAN-seq update: CAZyme gene clusters and substrates in microbiomes. Nucleic Acids Res. 2023, 51, D557–D563. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Brulc, J.M.; Antonopoulos, D.A.; Miller, M.E.B.; Wilson, M.K.; Yannarell, A.C.; Dinsdale, E.A.; Edwards, R.E.; Frank, E.D.; Emerson, J.B.; Wacklin, P.; et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. USA 2009, 106, 1948–1953. [Google Scholar] [CrossRef]
- Zhu, F.; Zhang, H.; Wu, H. Glycosyltransferase-mediated sweet modification in oral streptococci. J. Dent. Res. 2015, 94, 659–665. [Google Scholar] [CrossRef]
- Jones, D.R.; Thomas, D.; Alger, N.; Ghavidel, A.; Inglis, G.D.; Abbott, D.W. SACCHARIS: An automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol. Biofuels 2018, 11, 27. [Google Scholar] [CrossRef]
- Fishbein, S.R.S.; Mahmud, B.; Dantas, G. Antibiotic perturbations to the gut microbiome. Nat. Rev. Microbiol. 2023, 21, 772–788. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Fu, Y.; Chen, Y.; Wang, Y.; Ye, D.; Wang, C.; Hu, X.; Zhou, L.; Du, J.; et al. Association of florfenicol residues with the abundance of oxazolidinone resistance genes in livestock manures. J. Hazard. Mater. 2020, 399, 123059. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Wang, Y.; Wang, S.; Wang, Z.; Du, X.; Jiang, H.; Xia, X.; Shen, Z.; Ding, S.; Wu, C.; et al. Prevalence and abundance of florfenicol and linezolid resistance genes in soils adjacent to swine feedlots. Sci. Rep. 2016, 6, 32192. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shao, B.; Shen, J.; Wang, S.; Wu, Y. Occurrence of chloramphenicol-resistance genes as environmental pollutants from swine feedlots. Environ. Sci. Technol. 2013, 47, 2892–2897. [Google Scholar] [CrossRef]
- Hitch, T.C.A.; Thomas, B.J.; Friedersdorff, J.C.A.; Ougham, H.; Creevey, C.J. Deep sequence analysis reveals the ovine rumen as a reservoir of antibiotic resistance genes. Environ. Pollut. 2018, 235, 571–575. [Google Scholar] [CrossRef]
- Li, X.; Stokholm, J.; Brejnrod, A.; Vestergaard, G.A.; Russel, J.; Trivedi, U.; Thorsen, J.; Gupta, S.; Hjelmsø, M.H.; Shah, S.A.; et al. The infant gut resistome associates with e. Coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host Microbe 2021, 29, 975–987. [Google Scholar] [CrossRef]
- Cui, G.; Bhat, S.A.; Li, W.; Wei, Y.; Kui, H.; Fu, X.; Gui, H.; Wei, C.; Li, F. Gut digestion of earthworms significantly attenuates cell-free and -associated antibiotic resistance genes in excess activated sludge by affecting bacterial profiles. Sci. Total Environ. 2019, 691, 644–653. [Google Scholar] [CrossRef]
- Gnanaprakasam, J.N.R.; López-Bañuelos, L.; Vega, L. Anacardic 6-pentadecyl salicylic acid induces apoptosis in breast cancer tumor cells, immunostimulation in the host and decreases blood toxic effects of taxol in an animal model. Toxicol. Appl. Pharmacol. 2021, 410, 115359. [Google Scholar] [CrossRef] [PubMed]
- Zorofchian Moghadamtousi, S.; Rouhollahi, E.; Karimian, H.; Fadaeinasab, M.; Firoozinia, M.; Ameen Abdulla, M.; Abdul Kadir, H. The chemopotential effect of annona muricata leaves against azoxymethane-induced colonic aberrant crypt foci in rats and the apoptotic effect of acetogenin annomuricin e in HT-29 cells: A bioassay-guided approach. PLoS ONE 2015, 10, e0122288. [Google Scholar] [CrossRef]
- Rahman, K.; Coleman, R. Selective biliary lipid secretion at low bile-salt-output rates in the isolated perfused rat liver. Effects of phalloidin. Biochem. J. 1986, 237, 301–304. [Google Scholar] [CrossRef]
- 2e,4z,6z,8z-Decatetraenedioic Acid. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/9543652 (accessed on 27 June 2025).
- Spadari, C.D.C.; Borba-Santos, L.P.; Rozental, S.; Ishida, K. Miltefosine repositioning: A review of potential alternative antifungal therapy. J. Mycol. Med. 2023, 33, 101436. [Google Scholar] [CrossRef]
- Dodsworth, T.L.; Lovejoy, D.A. Role of teneurin c-terminal associated peptides (TCAP) on intercellular adhesion and communication. Front. Neurosci. 2022, 16, 868541. [Google Scholar] [CrossRef]
- Lebidois, J.; Soifer, S.J.; Clyman, R.I.; Heymann, M.A. Piriprost: A putative leukotriene synthesis inhibitor increases pulmonary blood flow in fetal lambs. Pediatr. Res. 1987, 22, 350–354. [Google Scholar] [CrossRef]
- Dalirfardouei, R.; Karimi, G.; Jamialahmadi, K. Molecular mechanisms and biomedical applications of glucosamine as a potential multifunctional therapeutic agent. Life Sci. 2016, 152, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Günenc, A.N.; Graf, B.; Stark, H.; Chari, A. Fatty acid synthase: Structure, function, and regulation. Sub-Cell. Biochem. 2022, 99, 1–33. [Google Scholar]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V.J. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Taves, M.D.; Donahue, K.M.; Bian, J.; Cam, M.C.; Ashwell, J.D. Aire drives steroid hormone biosynthesis by medullary thymic epithelial cells. Sci. Immunol. 2023, 8, eabo7975. [Google Scholar] [CrossRef]
- Gordon, M.P.; Intrieri, O.M.; Brown, G.B. On the metabolism of purine. J. Biol. Chem. 1957, 229, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Wang, Q.; Qiao, Y.; Xu, Z.; Zhang, L.; Xiao, H.; Lin, Z.; Wu, M.; Xia, W.; Yang, H.; et al. Arachidonic acid in aging: New roles for old players. J. Adv. Res. 2025, 70, 79–101. [Google Scholar] [CrossRef]
- Melo, C.F.O.R.; Bachur, L.F.; Delafiori, J.; Dabaja, M.Z.; de Oliveira, D.N.; Guerreiro, T.M.; Tararam, C.A.; Busso-Lopes, A.F.; Moretti, M.L.; Catharino, R.R. Does leukotriene f4 play a major role in the infection mechanism of candida sp.? Microb. Pathog. 2020, 149, 104394. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent development of probiotic bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef]
- Farooqui, A.A.; Horrocks, L.A.; Farooqui, T. Glycerophospholipids in brain: Their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem. Phys. Lipids 2000, 106, 1–29. [Google Scholar] [CrossRef]
- Schink, S.J.; Christodoulou, D.; Mukherjee, A.; Athaide, E.; Brunner, V.; Fuhrer, T.; Bradshaw, G.A.; Sauer, U.; Basan, M. Glycolysis/gluconeogenesis specialization in microbes is driven by biochemical constraints of flux sensing. Mol. Syst. Biol. 2022, 18, e10704. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Y.; Liu, D.; Zhao, N.; Cheng, H.; Ren, H.; Guo, T.; Niu, H.; Zhuang, W.; Wu, J.; et al. Involvement of glycolysis/gluconeogenesis and signaling regulatory pathways in saccharomyces cerevisiae biofilms during fermentation. Front. Microbiol. 2015, 6, 139. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Chen, Q.; Sha, Y.; He, Y.; Liu, X.; Chen, X.; Shao, P.; Huang, W.; He, Y.; Li, M.; et al. Study on the Plateau Adaptive Synergistic Mechanism of Rumen Microbiome-Metabolome-Resistome in Tibetan Sheep. Microorganisms 2025, 13, 2049. https://doi.org/10.3390/microorganisms13092049
Gao X, Chen Q, Sha Y, He Y, Liu X, Chen X, Shao P, Huang W, He Y, Li M, et al. Study on the Plateau Adaptive Synergistic Mechanism of Rumen Microbiome-Metabolome-Resistome in Tibetan Sheep. Microorganisms. 2025; 13(9):2049. https://doi.org/10.3390/microorganisms13092049
Chicago/Turabian StyleGao, Xu, Qianling Chen, Yuzhu Sha, Yanyu He, Xiu Liu, Xiaowei Chen, Pengyang Shao, Wei Huang, Yapeng He, Mingna Li, and et al. 2025. "Study on the Plateau Adaptive Synergistic Mechanism of Rumen Microbiome-Metabolome-Resistome in Tibetan Sheep" Microorganisms 13, no. 9: 2049. https://doi.org/10.3390/microorganisms13092049
APA StyleGao, X., Chen, Q., Sha, Y., He, Y., Liu, X., Chen, X., Shao, P., Huang, W., He, Y., Li, M., Hao, Z., Shi, B., & Xu, J. (2025). Study on the Plateau Adaptive Synergistic Mechanism of Rumen Microbiome-Metabolome-Resistome in Tibetan Sheep. Microorganisms, 13(9), 2049. https://doi.org/10.3390/microorganisms13092049





