Functions of the Three Common Fungal Extracellular Membrane (CFEM) Domain-Containing Genes of Arthrobotrys flagrans in the Process of Nematode Trapping
Abstract
1. Introduction
2. Materials and Methods
2.1. Organisms and Media
2.2. Formation of Traps in A. flagrans After Induction by C. elegans
2.3. Transcriptome Analysis of A. flagrans Interacted with C. elegans
2.4. Analysis of Physicochemical Properties and Domains of AfCFEM1-3
2.5. Deletion and Overexpression of AfCFEM1-3
2.6. Determination of the Effect of AfCFEM1-3 on Trap Formation and Pathogenicity
2.7. Cryo-Scanning Electron Microscope and Transmission Electron Microscope Observation
2.8. Determination of the Relative Transcriptional Level of AfCFEM1-8 in ΔAfCFEM1-3 Mutants
2.9. Proteomic Analysis of ΔAfCFEM1 and ΔAfCFEM2 Mutants
2.10. Statistical Analysis
3. Results
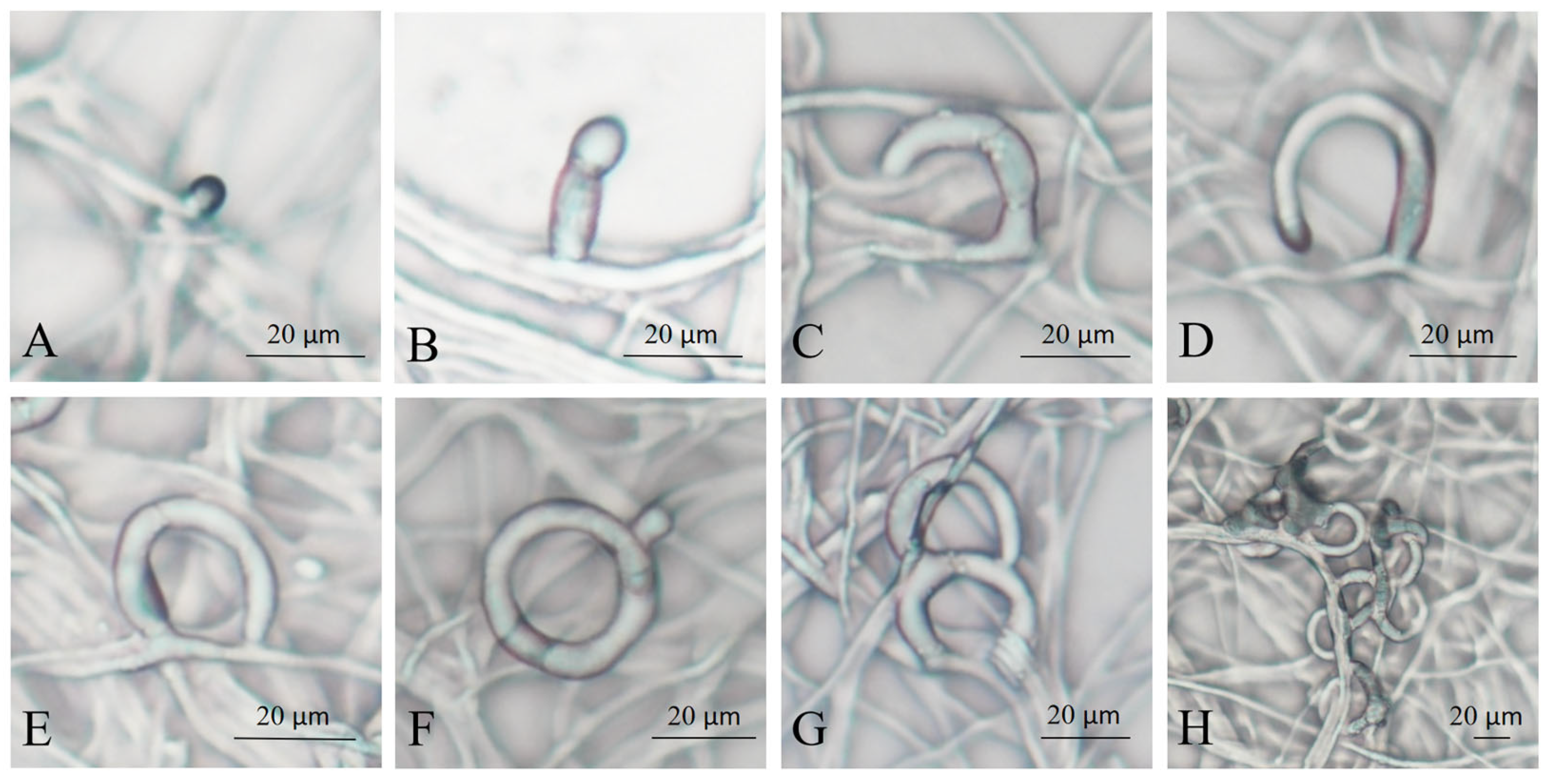
3.1. The Process of Trap Formation of A. flagrans in Response to C. elegans Induction
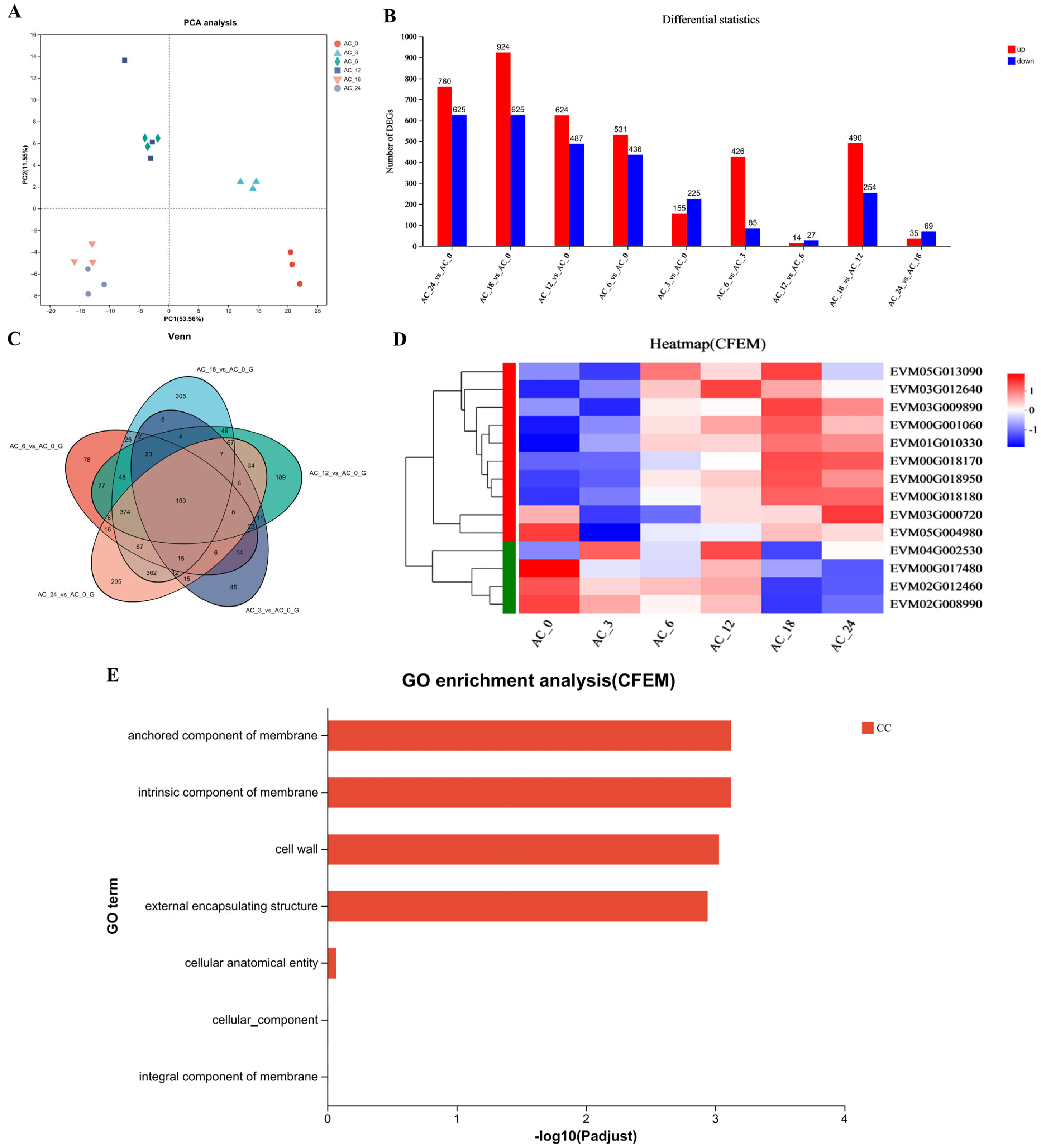
3.2. C. elegans Induction Altered Gene Expression in A. flagrans
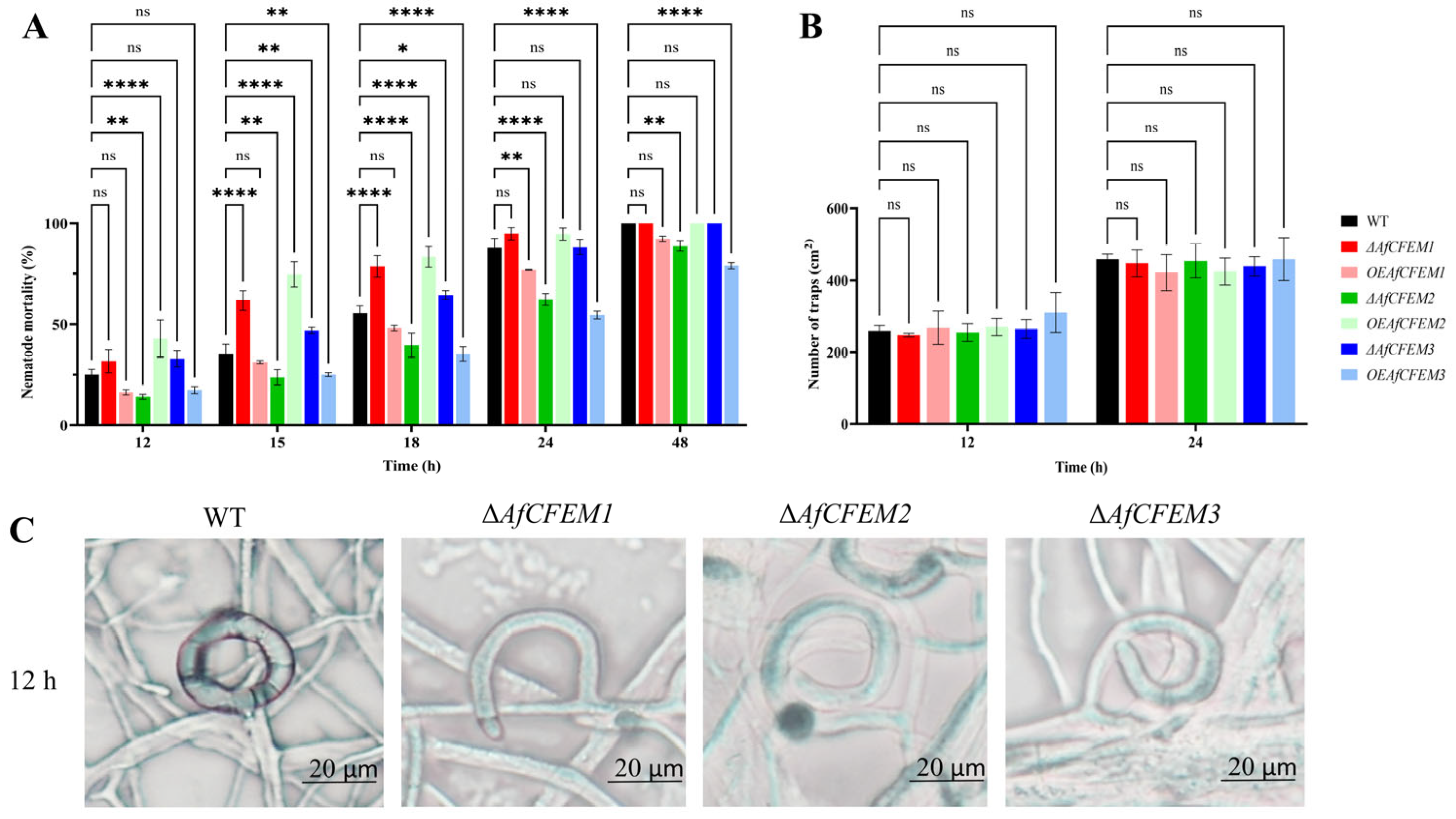
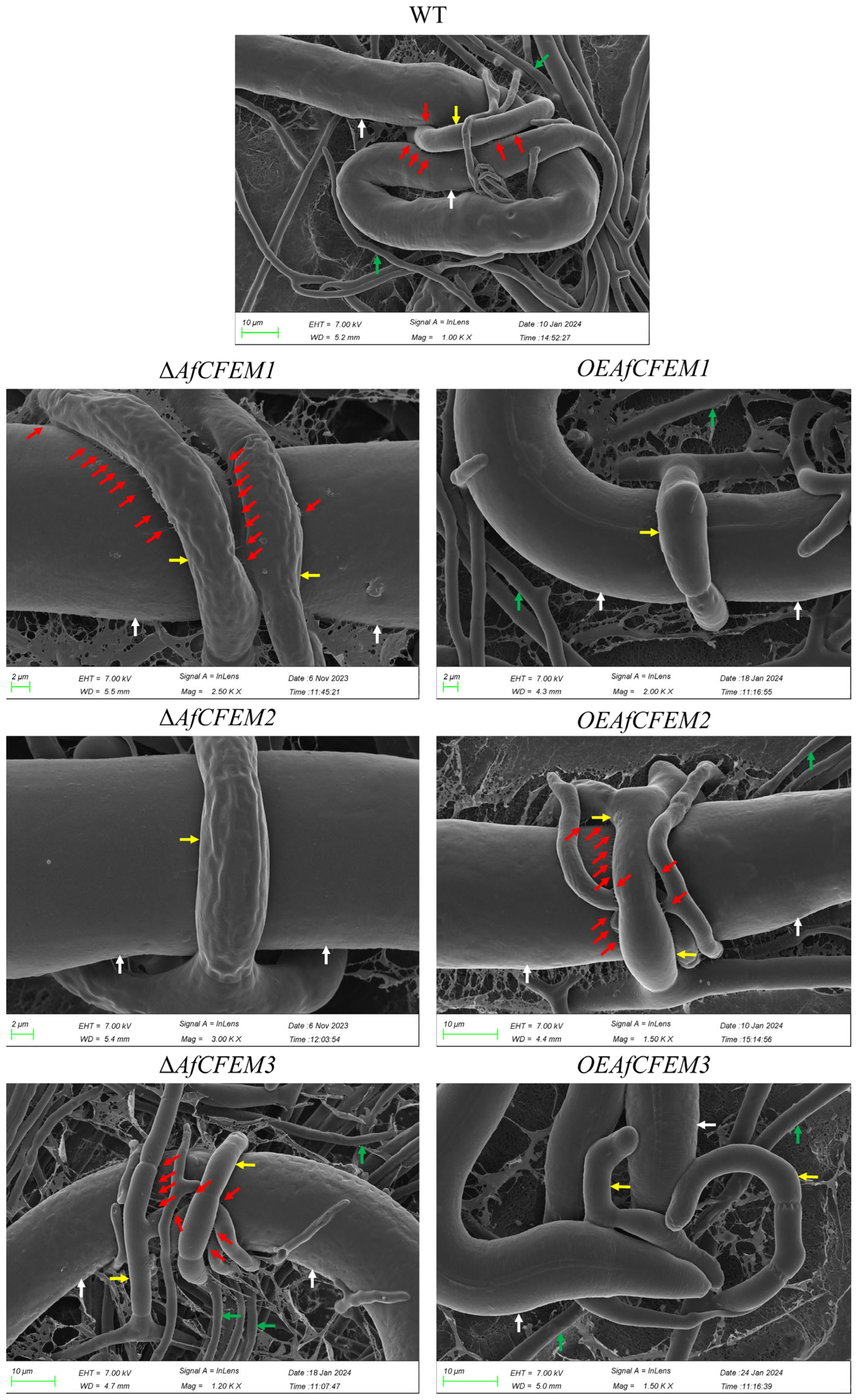
3.3. AfCFEM1-3 Play a Vital Role in the Trap’s Morphology and Pathogenicity of A. flagrans
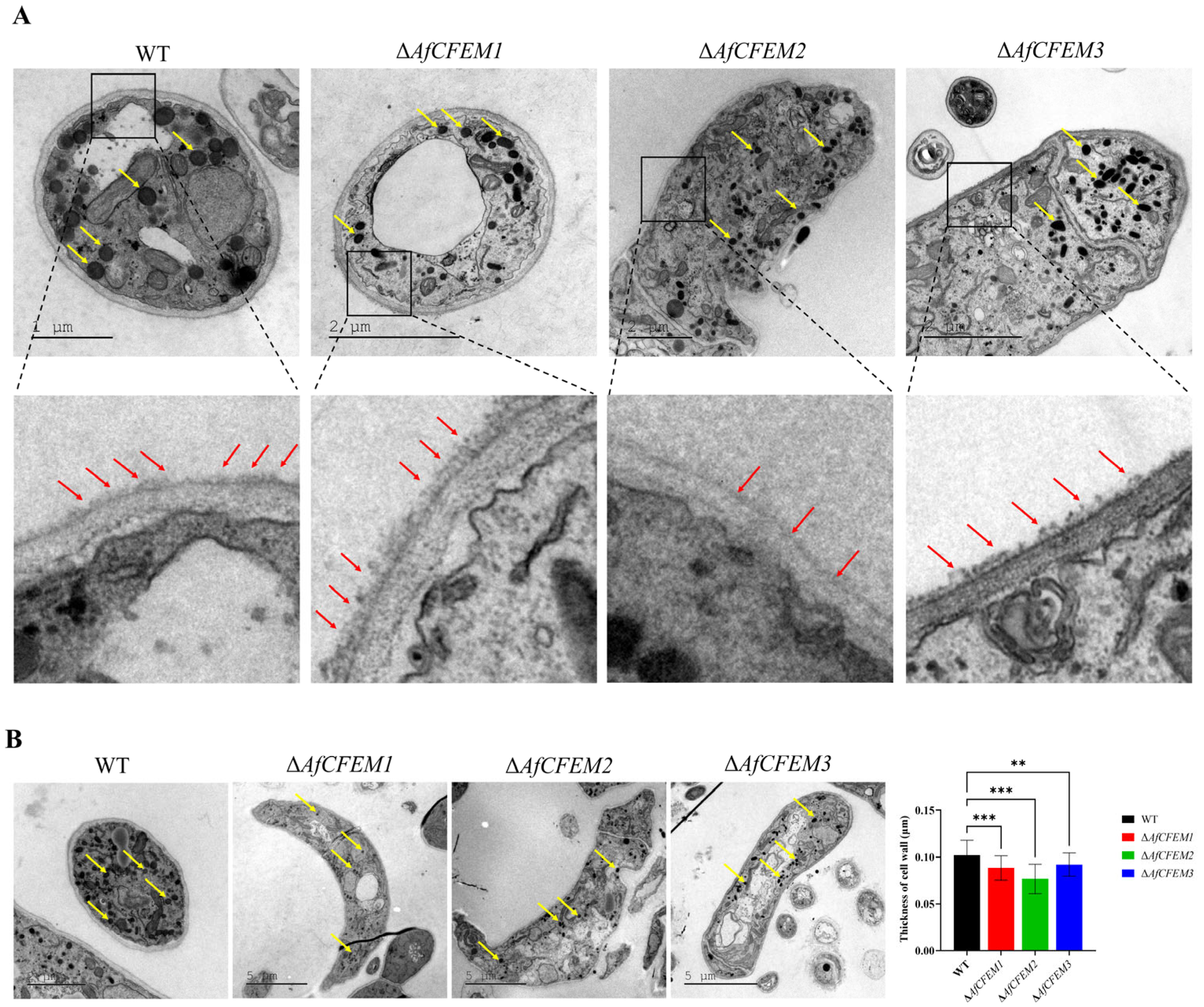
3.4. AfCFEM1-3 Associated with the Cell Wall Formation and Adhesive Proteins in Traps of A. flagrans
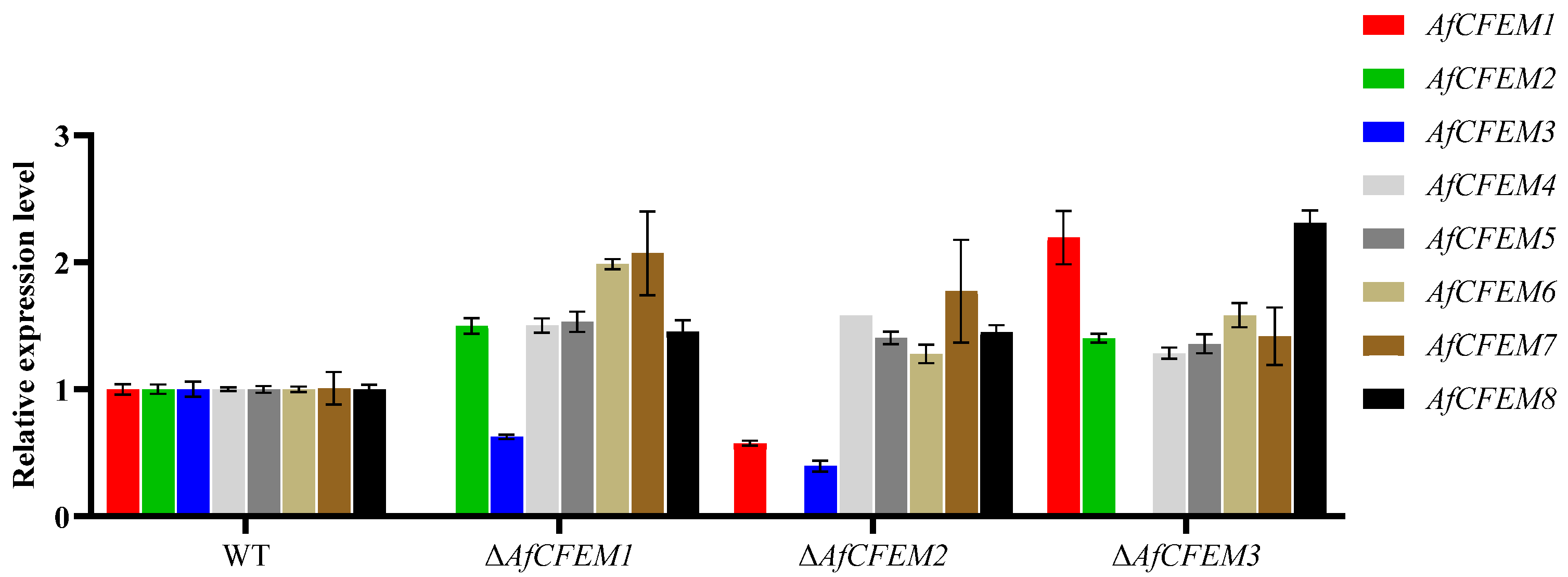
3.5. The Compensating Effects Exist Among AfCFEM1-8 of A. flagrans
3.6. AfCFEM1 and AfCFEM2 Affected the Protein Expression in A. flagrans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, G.; Khan, A.; Khan, A.A.; Ali, A.; Mohhamad, H.I. Biological control: A novel strategy for the control of the plant parasitic nematodes. Antonie Van Leeuwenhoek 2021, 114, 885–912. [Google Scholar] [CrossRef]
- Ibrahim, H.; Nchiozem-Ngnitedem, V.A.; Dandurand, L.M.; Popova, I. Naturally-occurring nematicides of plant origin: Two decades of novel chemistries. Pest Manag. Sci. 2025, 81, 540–571. [Google Scholar] [CrossRef]
- Fitzpatrick, J.L. Global food security: The impact of veterinary parasites and parasitologists. Vet. Parasitol. 2013, 195, 233–248. [Google Scholar] [CrossRef]
- Ma, G.; Wang, T.; Korhonen, P.K.; Hofmann, A.; Sternberg, P.W.; Young, N.D.; Gasser, R.B. Chapter Four—Elucidating the molecular and developmental biology of parasitic nematodes: Moving to a multiomics paradigm. In Advances in Parasitology; Rollinson, D., Stothard, R., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 108, pp. 175–229. [Google Scholar]
- Yang, Y.; Yang, E.; An, Z.; Liu, X. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. USA 2007, 104, 8379–8384. [Google Scholar] [CrossRef] [PubMed]
- Terrill, T.H.; Larsen, M.; Samples, O.; Husted, S.; Miller, J.E.; Kaplan, R.M.; Gelaye, S. Capability of the nematode-trapping fungus Duddingtonia flagrans to reduce infective larvae of gastrointestinal nematodes in goat feces in the southeastern United States: Dose titration and dose time interval studies. Vet. Parasitol. 2004, 120, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.S.; Larsen, M.; Wolstrup, J.; Grønvold, J.; Nansen, P.; Bjørn, H. Growth rate and trapping efficacy of nematode-trapping fungi under constant and fluctuating temperatures. Parasitol. Res. 1999, 85, 661–668. [Google Scholar] [CrossRef]
- Facchini Rodrigues, J.V.; Braga, F.R.; Campos, A.K.; de Carvalho, L.M.; Araujo, J.M.; Aguiar, A.R.; Ferraz, C.M.; da Silveira, W.F.; Valadão, M.C.; de Oliveira, T. Duddingtonia flagrans formulated in rice bran in the control of Oesophagostomum spp. intestinal parasite of swine. Exp. Parasitol. 2018, 184, 11–15. [Google Scholar] [CrossRef]
- Monteiro, T.S.A.; Balbino, H.M.; Mello, I.N.K.; Coutinho, R.R.; de Araújo, J.V.; Freitas, L.G. Duddingtonia flagrans preying a plant parasitic nematode. Braz. J. Biol. 2020, 80, 197–198. [Google Scholar] [CrossRef]
- Balbino, H.M.; Monteiro, T.S.A.; Coutinho, R.R.; Pacheco, P.V.M.; Freitas, L.G.d. Association of Duddingtonia flagrans with microorganisms for management of Meloidogyne javanica and acquisition of nutrients in soybean. Biol. Control 2021, 159, 104626. [Google Scholar] [CrossRef]
- Monteiro, T.S.A.; Valadares, S.V.; de Mello, I.N.K.; Moreira, B.C.; Kasuya, M.C.M.; de Araújo, J.V.; de Freitas, L.G. Nematophagus fungi increasing phosphorus uptake and promoting plant growth. Biol. Control 2018, 123, 71–75. [Google Scholar] [CrossRef]
- Mei, X.; Wang, X.; Li, G. Pathogenicity and volatile nematicidal metabolites from Duddingtonia flagrans against Meloidogyne incognita. Microorganisms 2021, 9, 2268. [Google Scholar] [CrossRef]
- Wernet, V.; Fischer, R. Establishment of Arthrobotrys flagrans as biocontrol agent against the root pathogenic nematode Xiphinema index. Environ. Microbiol. 2023, 25, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Dijksterhuis, J.; Veenhuis, M.; Harder, W.; Nordbring-Hertz, B. Nematophagous fungi: Physiological aspects and structure-function relationships. Adv. Microb. Physiol. 1994, 36, 111–143. [Google Scholar]
- Nordbring-Hertz, B.; Stålhammar-Carlemalm, M. Capture of nematodes by Arthrobotrys oligospora, an electron microscope study. Can. J. Bot. 1978, 56, 1297–1307. [Google Scholar] [CrossRef]
- den Belder, E.; Jansen, E.; Donkers, J. Adhesive hyphae of Arthrobotrys oligospora: An ultrastructural study. Eur. J. Plant Pathol. 1996, 102, 471–478. [Google Scholar] [CrossRef]
- Ji, X.; Yu, Z.; Yang, J.; Xu, J.; Zhang, Y.; Liu, S.; Zou, C.; Li, J.; Liang, L.; Zhang, K.Q. Expansion of adhesion genes drives pathogenic adaptation of nematode-trapping fungi. iScience 2020, 23, 101057. [Google Scholar] [CrossRef]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef]
- Liang, L.; Shen, R.; Mo, Y.; Yang, J.; Ji, X.; Zhang, K.-Q. A proposed adhesin AoMad1 helps nematode-trapping fungus Arthrobotrys oligospora recognizing host signals for life-style switching. Fungal Genet. Biol. 2015, 81, 172–181. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, J.; Chen, X.; Huang, Y.; Wu, Y.; Zhu, J.; Li, W. Effect of CFEM proteins on pathogenicity, patulin accumulation and host immunity of postharvest apple pathogens Penicillium expansum. Int. J. Food Microbiol. 2025, 435, 111180. [Google Scholar] [CrossRef]
- Kou, Y.; Tan, Y.H.; Ramanujam, R.; Naqvi, N.I. Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 2017, 214, 330–342. [Google Scholar] [CrossRef]
- Kornitzer, D.; Roy, U. Pathways of heme utilization in fungi. Biochim. Biophys. Acta-Mol. Cell Res. 2020, 1867, 118817. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.D.; Thon, M.R.; Pan, H.; Dean, R.A. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 2005, 6, R24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.N.; Wu, Q.Y.; Zhang, G.Z.; Zhu, Y.Y.; Murphy, R.W.; Liu, Z.; Zou, C.G. Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Sci. Rep. 2015, 5, 13032. [Google Scholar] [CrossRef]
- Mrsa, V.; Ecker, M.; Strahl-Bolsinger, S.; Nimtz, M.; Lehle, L.; Tanner, W. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 1999, 181, 3076–3086. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, W.; Lai, Y.; Xiang, M.; Wang, X.; Zhang, X.; Liu, X. Drechslerella stenobrocha genome illustrates the mechanism of constricting rings and the origin of nematode predation in fungi. BMC Genom. 2014, 15, 114. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Ran, Y.; Zhang, K.Q.; Li, G.H. AfLaeA, a global regulator of mycelial growth, chlamydospore production, pathogenicity, secondary metabolism, and energy metabolism in the nematode-trapping fungus Arthrobotrys flagrans. Microbiol. Spectr. 2023, 11, e0018623. [Google Scholar] [CrossRef]
- Mallick, S.; Mishra, N.; Barik, B.K.; Negi, V.D. Salmonella typhimurium fepB negatively regulates C. elegans behavioral plasticity. J. Infect. 2022, 84, 518–530. [Google Scholar] [CrossRef]
- Fischer, R.; Requena, N. Small-secreted proteins as virulence factors in nematode-trapping fungi. Trends Microbiol. 2022, 30, 615–617. [Google Scholar] [CrossRef]
- Zhang, H.X.; Tan, J.L.; Wei, L.X.; Wang, Y.L.; Zhang, C.P.; Wu, D.K.; Zhu, C.Y.; Zhang, Y.; Zhang, K.Q.; Niu, X.M. Morphology Regulatory Metabolites from Arthrobotrys oligospora. J. Nat. Prod. 2012, 75, 1419–1423. [Google Scholar] [CrossRef]
- Hsueh, Y.P.; Mahanti, P.; Schroeder, F.C.; Sternberg, P.W. Nematode-trapping fungi eavesdrop on nematode pheromones. Curr. Biol. 2013, 23, 83–86. [Google Scholar] [CrossRef]
- Yu, X.; Hu, X.; Pop, M.; Wernet, N.; Kirschhöfer, F.; Brenner-Weiß, G.; Keller, J.; Bunzel, M. Fatal attraction of Caenorhabditis elegans to predatory fungi through 6-methyl-salicylic acid. Nat. Commun. 2021, 12, 5462. [Google Scholar] [CrossRef]
- Xie, H.; Aminuzzaman, F.M.; Xu, L.; Lai, Y.; Li, F.; Liu, X. Trap induction and trapping in eight nematode-trapping fungi (Orbiliaceae) as affected by juvenile stage of Caenorhabditis elegans. Mycopathologia 2010, 169, 467–473. [Google Scholar] [CrossRef]
- Su, H.; Zhao, Y.; Zhou, J.; Feng, H.; Jiang, D.; Zhang, K.Q.; Yang, J. Trapping devices of nematode-trapping fungi: Formation, evolution, and genomic perspectives. Biol. Rev. Camb. Philos. Soc. 2017, 92, 357–368. [Google Scholar] [CrossRef]
- Kulkarni, R.D.; Kelkar, H.S.; Dean, R.A. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 2003, 28, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.; Ramage, G.; Blanes, R.; Murgui, A.; Casanova, M.; Martínez, J.P. Some biological features of Candida albicans mutants for genes coding fungal proteins containing the CFEM domain. FEMS Yeast Res. 2011, 11, 273–284. [Google Scholar] [CrossRef]
- Lin, H.C.; de Ulzurrun, G.V.; Chen, S.A.; Yang, C.T.; Tay, R.J.; Iizuka, T.; Huang, T.Y.; Kuo, C.Y.; Gonçalves, A.P.; Lin, S.Y.; et al. Key processes required for the different stages of fungal carnivory by a nematode-trapping fungus. PLoS Biol. 2023, 21, e3002400. [Google Scholar] [CrossRef]
- Cao, Y.; Zhu, X.; Jiao, R.; Xia, Y. The Magas1 gene is involved in pathogenesis by affecting penetration in Metarhizium acridum. J. Microbiol. Biotechnol. 2012, 22, 889–893. [Google Scholar] [CrossRef]
- Dupres, V.; Alsteens, D.; Wilk, S.; Hansen, B.; Heinisch, J.J.; Dufrêne, Y.F. The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat. Chem. Biol. 2009, 5, 857–862. [Google Scholar] [CrossRef]
- Linder, T.; Gustafsson, C.M. Molecular phylogenetics of ascomycotal adhesins--a novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 2008, 45, 485–497. [Google Scholar] [CrossRef]
- Rodicio, R.; Heinisch, J.J. Together we are strong--cell wall integrity sensors in yeasts. Yeast 2010, 27, 531–540. [Google Scholar] [CrossRef]
- Maddi, A.; Dettman, A.; Fu, C.; Seiler, S.; Free, S.J. WSC-1 and HAM-7 are MAK-1 MAP kinase pathway sensors required for cell wall integrity and hyphal fusion in Neurospora crassa. PLoS ONE 2012, 7, e42374. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.M.; Meerupati, T.; Levander, F.; Friman, E.; Ahrén, D.; Tunlid, A. Proteome of the nematode-trapping cells of the fungus Monacrosporium haptotylum. Appl. Environ. Microbiol. 2013, 79, 4993–5004. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.; Pedrós, B.; Murgui, A.; Casanova, M.; López-Ribot, J.L.; Martínez, J.P. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res. 2006, 6, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Vidanes, G.M.; Maguire, S.L.; Guida, A.; Synnott, J.M.; Andes, D.R.; Butler, G. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS ONE 2011, 6, e28151. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, W.; Chen, Y.; Xiang, M.; Liu, X. DdaSTE12 is involved in trap formation, ring inflation, conidiation, and vegetative growth in the nematode-trapping fungus Drechslerella dactyloides. Appl. Microbiol. Biotechnol. 2021, 105, 7379–7393. [Google Scholar] [CrossRef]
- Moreno-García, J.; Coi, A.L.; Zara, G.; García-Martínez, T.; Mauricio, J.C.; Budroni, M. Study of the role of the covalently linked cell wall protein (Ccw14p) and yeast glycoprotein (Ygp1p) within biofilm formation in a flor yeast strain. FEMS Yeast Res. 2018, 18, foy005. [Google Scholar] [CrossRef]
- Feng, L.; Dong, M.; Huang, Z.; Wang, Q.; An, B.; He, C.; Wang, Q.; Luo, H. CgCFEM1 is required for the full virulence of Colletotrichum gloeosporioides. Int. J. Mol. Sci. 2024, 25, 2937. [Google Scholar] [CrossRef]
- Kanda, S.; Aimi, T.; Kano, S.; Ishihara, S.; Kitamoto, Y.; Morinaga, T. Ambient pH signaling regulates expression of the serine protease gene (spr1) in pine wilt nematode-trapping fungus, Monacrosporium megalosporum. Microbiol. Res. 2008, 163, 63–72. [Google Scholar] [CrossRef]
- Tunlid, A.; Rosén, S.; Ek, B.; Rask, L. Purification and characterization of an extracellular serine protease from the nematode-trapping fungus Arthrobotrys oligospora. Microbiology 1994, 140 Pt 7, 1687–1695. [Google Scholar] [CrossRef]
- Szabó, M.; Urbán, P.; Virányi, F.; Kredics, L.; Fekete, C. Comparative gene expression profiles of Trichoderma harzianum proteases during in vitro nematode egg-parasitism. Biol. Control 2013, 67, 337–343. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.; Zhao, Y.; Huang, Y.; Zhang, K.Q.; Yang, J. Malate synthase gene AoMls in the nematode-trapping fungus Arthrobotrys oligospora contributes to conidiation, trap formation, and pathogenicity. Appl. Microbiol. Biotechnol. 2014, 98, 2555–2563. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, Y.; Bai, N.; Yang, L.; Xie, M.; Yang, J.; Zhu, M.; Zhang, K.Q.; Yang, J. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2022, 65, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, M.; Liu, Y.; Yang, L.; Yang, J. Aoatg11 and Aoatg33 are indispensable for mitophagy, and contribute to conidiation, the stress response, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Res. 2023, 266, 127252. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Zhu, Y.; Bai, N.; Xie, M.; Zhang, K.-Q.; Yang, J. Aolatg1 and Aolatg13 regulate autophagy and play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Cell. Infect. Microbiol. 2022, 11, 824407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, G.; Chen, B.; Peng, Y. The virulence contribution of the CFEM family genes of Beauveria bassiana is closely influenced by the external iron environment. Microbiol. Spectr. 2025, 13, e0309624. [Google Scholar] [CrossRef] [PubMed]
- Youssar, L.; Wernet, V.; Hensel, N.; Yu, X.; Hildebrand, H.G.; Schreckenberger, B.; Kriegler, M.; Hetzer, B.; Frankino, P.; Dillin, A.; et al. Intercellular communication is required for trap formation in the nematode-trapping fungus Duddingtonia flagrans. PLoS Genet. 2019, 15, e1008029. [Google Scholar] [CrossRef]
- Nordbring-Hertz, B.; Friman, E.; Veenhuis, M. Hyphal fusion during initial stages of trap formation in Arthrobotrys oligospora. Antonie Van Leeuwenhoek 1989, 55, 237–244. [Google Scholar] [CrossRef]
- Daskalov, A.; Heller, J.; Herzog, S.; Fleißner, A.; Glass, N.L. Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. Microbiol. Spectr. 2017, 5, FUNK-0015-2016. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, T.; Deng, X.; Zhang, Y.; Li, G. Functions of the Three Common Fungal Extracellular Membrane (CFEM) Domain-Containing Genes of Arthrobotrys flagrans in the Process of Nematode Trapping. Microorganisms 2025, 13, 2001. https://doi.org/10.3390/microorganisms13092001
Shi T, Deng X, Zhang Y, Li G. Functions of the Three Common Fungal Extracellular Membrane (CFEM) Domain-Containing Genes of Arthrobotrys flagrans in the Process of Nematode Trapping. Microorganisms. 2025; 13(9):2001. https://doi.org/10.3390/microorganisms13092001
Chicago/Turabian StyleShi, Tingting, Xiaotong Deng, Yu Zhang, and Guohong Li. 2025. "Functions of the Three Common Fungal Extracellular Membrane (CFEM) Domain-Containing Genes of Arthrobotrys flagrans in the Process of Nematode Trapping" Microorganisms 13, no. 9: 2001. https://doi.org/10.3390/microorganisms13092001
APA StyleShi, T., Deng, X., Zhang, Y., & Li, G. (2025). Functions of the Three Common Fungal Extracellular Membrane (CFEM) Domain-Containing Genes of Arthrobotrys flagrans in the Process of Nematode Trapping. Microorganisms, 13(9), 2001. https://doi.org/10.3390/microorganisms13092001





