Functional Genomic Characteristics of Marine Sponge-Associated Microbulbifer spongiae MI-GT
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Genome Sequencing, and Assembly
2.2. Genome Annotation and Analysis
2.3. Comparative Genomic Analysis
3. Results
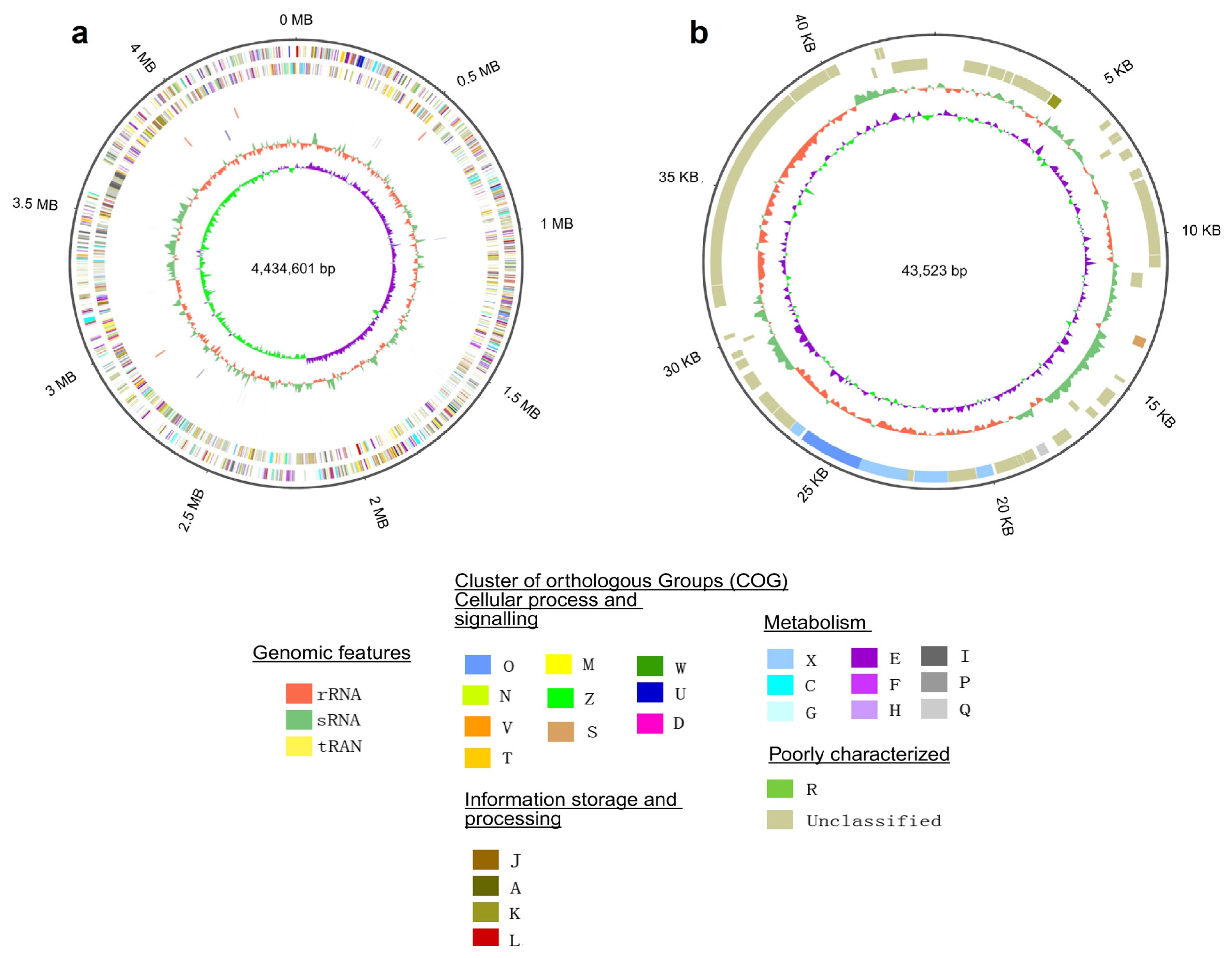
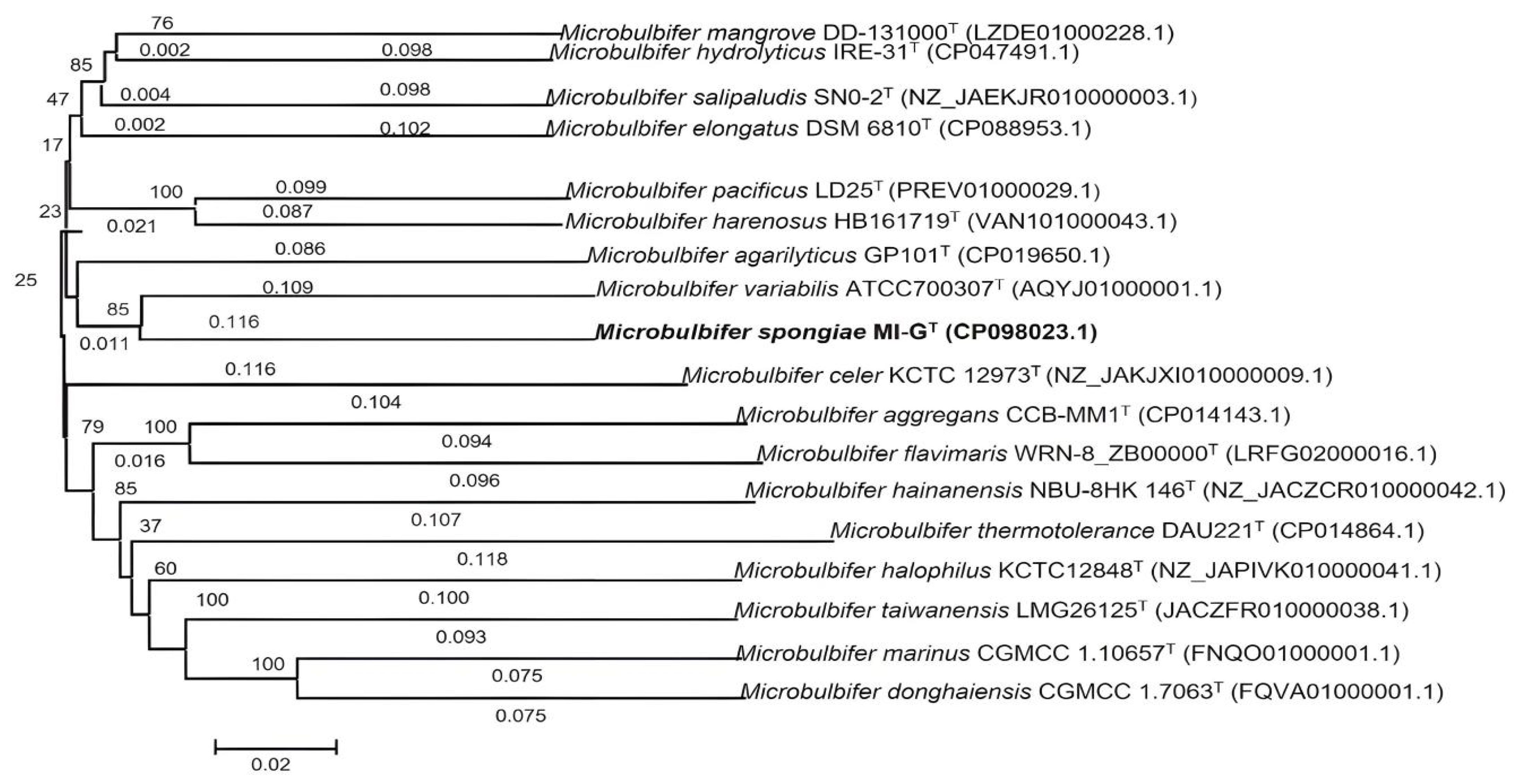
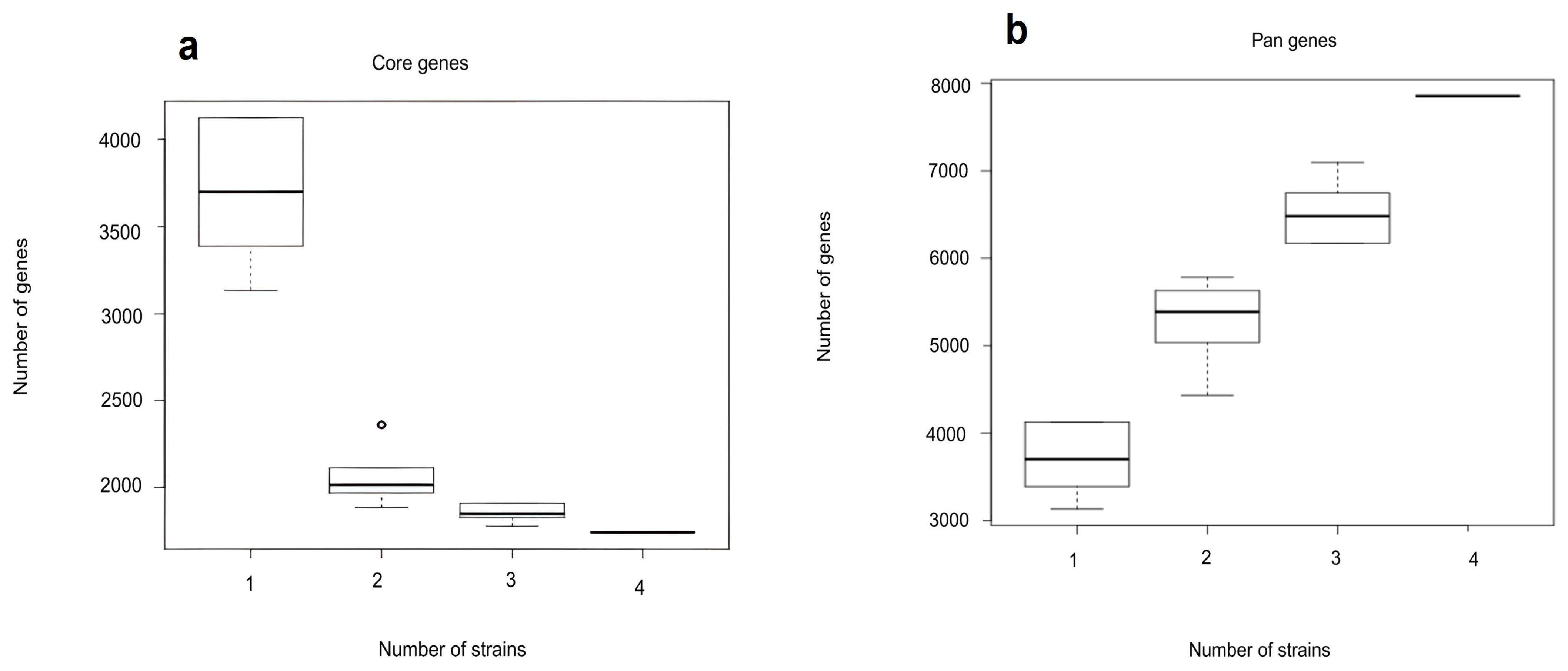
3.1. Genome Features of Microbulbifer Spongiae MI-GT and Three Microbulbifer Reference Strains
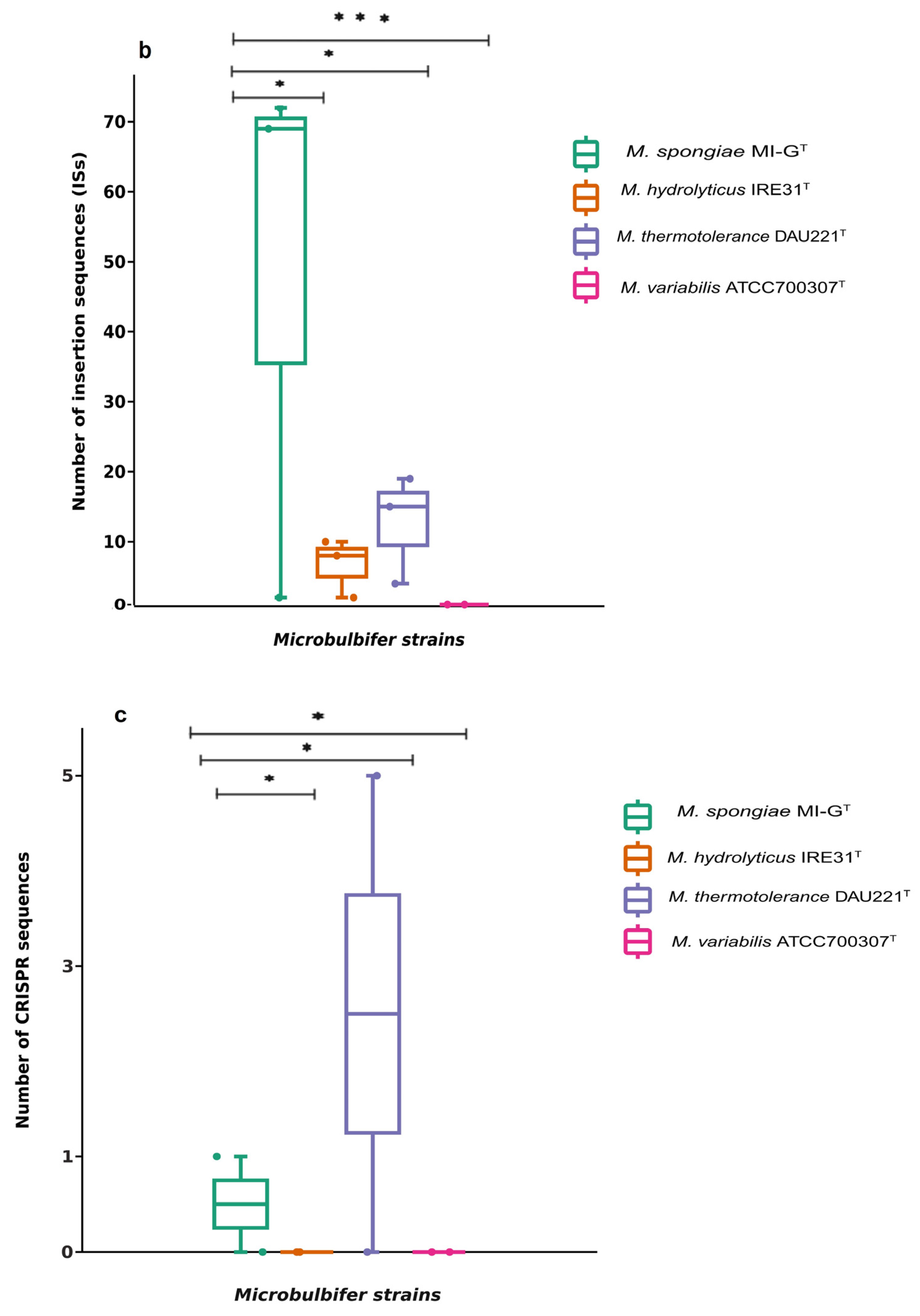
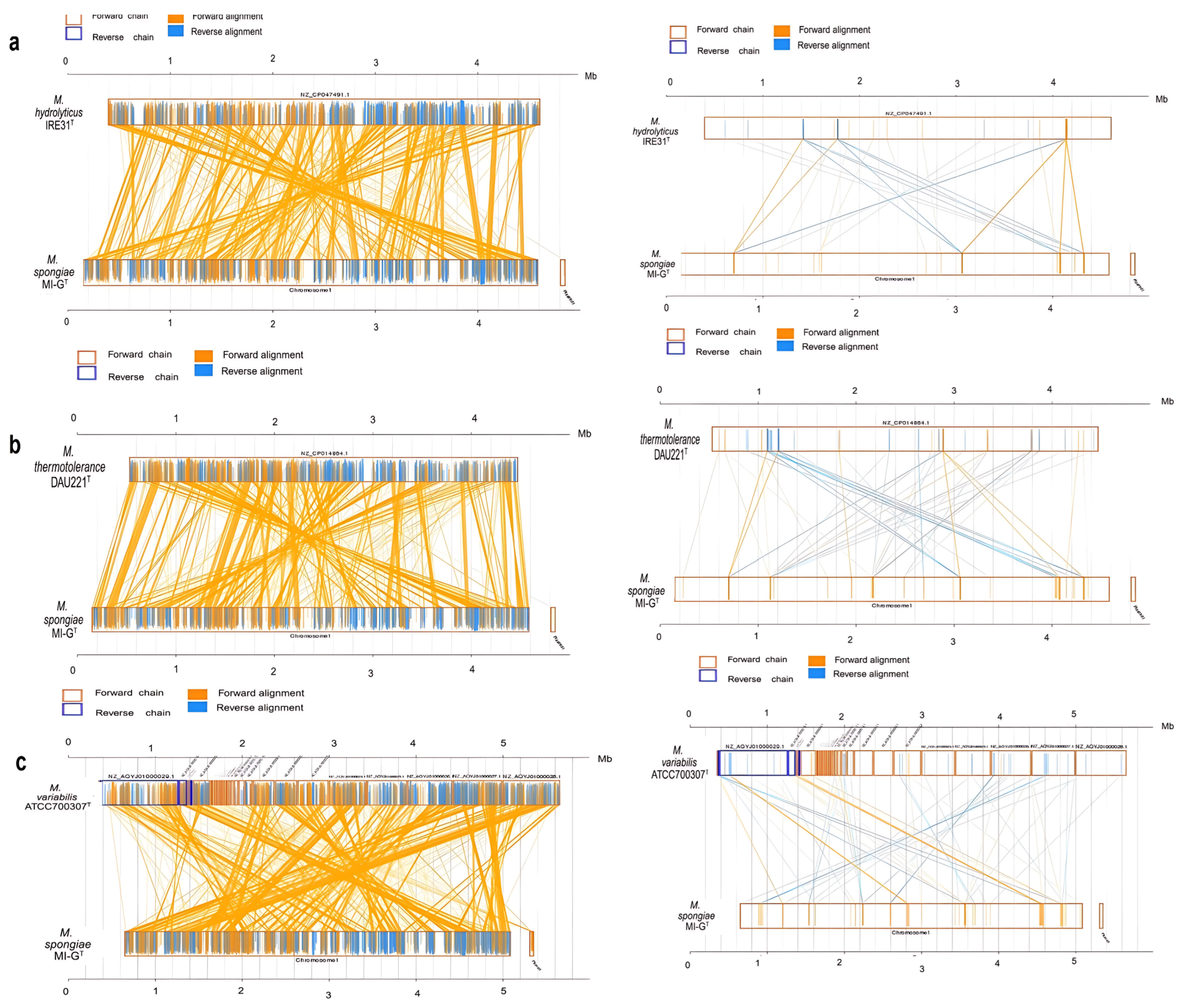
3.2. Mobile Gene Elements in Genomes of Four Microbulbifer Strains
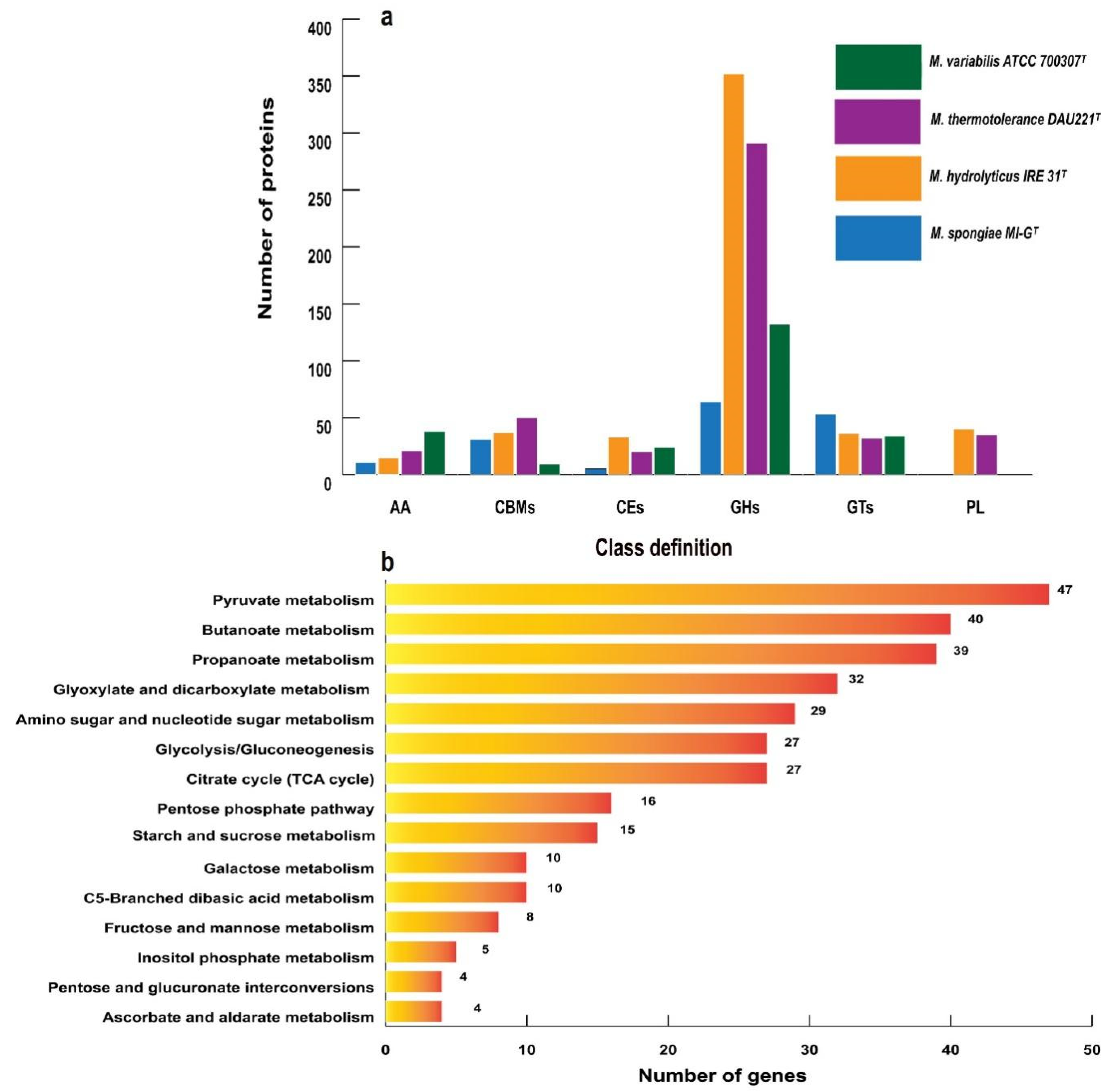
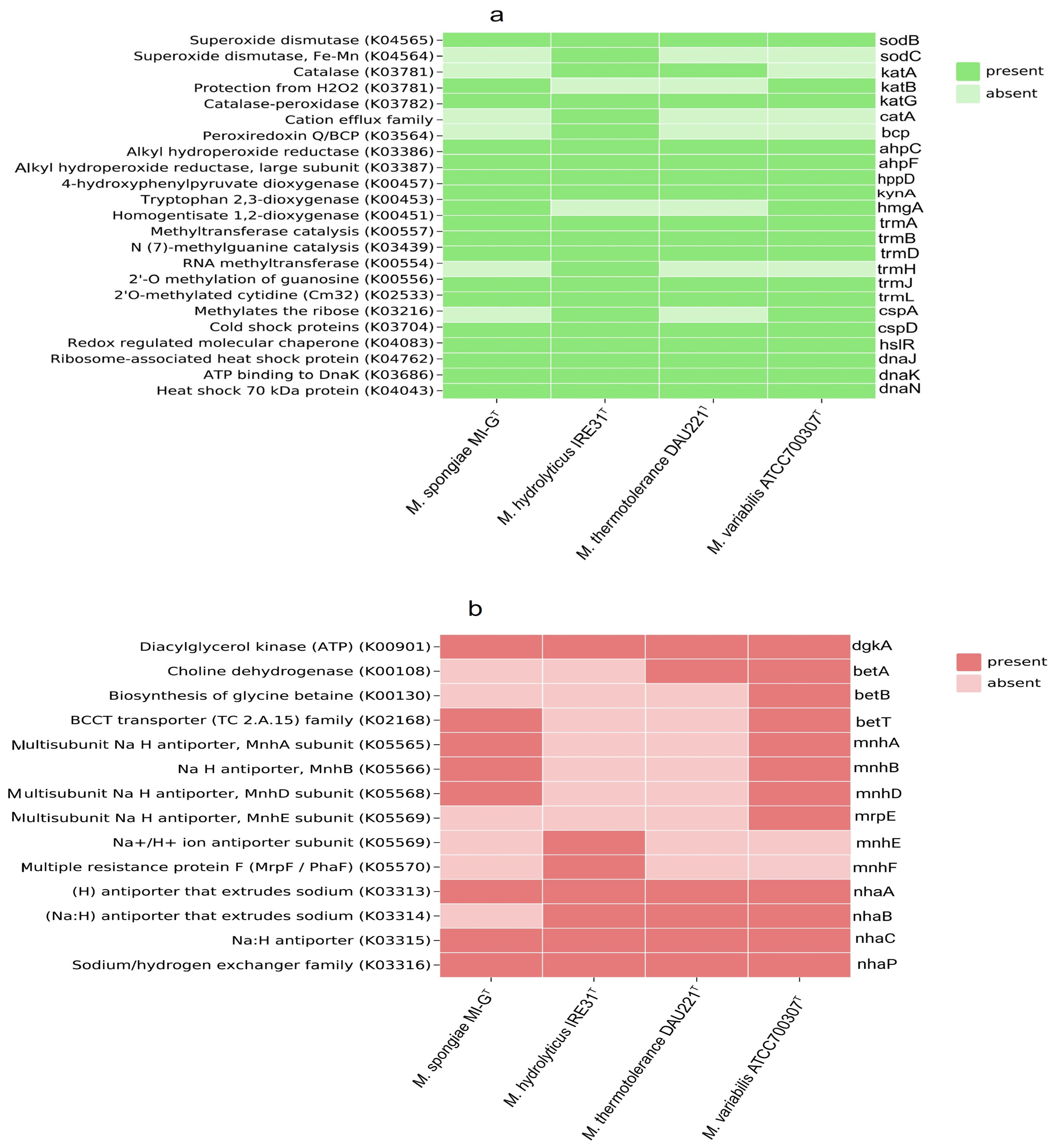
3.3. Carbohydrate Metabolism and Stress Response
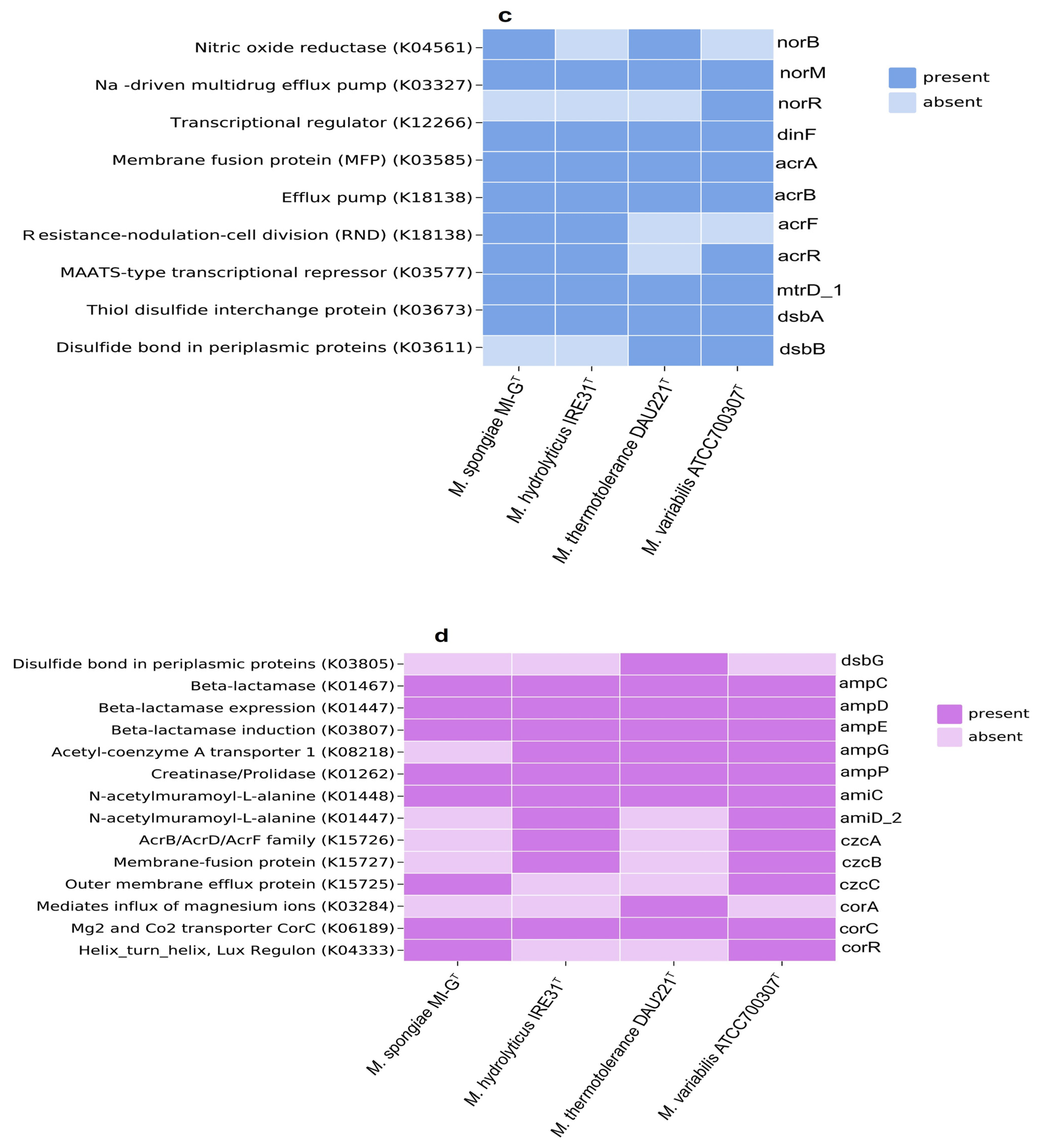
3.4. Different Genomic Characteristics of M. spongiae MI-GT from Other Microbulbifer Strains
3.5. Sponge-Bacteria Metabolic Interaction Indicated by Genome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef]
- Bell, J.J.; Strano, F.; Broadribb, M.; Wood, G.; Harris, B.; Resende, A.C.; Novak, E.; Micaroni, V. Sponge functional roles in a changing world. Adv. Mar. Biol. 2023, 95, 27–89. [Google Scholar] [PubMed]
- Beazley, L.I.; Kenchington, E.L.; Murillo, F.J.; Sacau, M.d.M. Deep-sea sponge grounds enhance diversity and abundance of epibenthic megafauna in the Northwest Atlantic. ICES J. Mar. Sci. 2013, 70, 1471–1490. [Google Scholar] [CrossRef]
- Webster, N.S.; Thomas, T. The Sponge Hologenome. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Bordenstein, S.R.; Theis, K.R. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef]
- Easson, C.; Thacker, R. Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 2014, 5, 532. [Google Scholar] [CrossRef]
- Rix, L.; Ribes, M.; Coma, R.; Jahn, M.T.; de Goeij, J.M.; van Oevelen, D.; Escrig, S.; Meibom, A.; Hentschel, U. Heterotrophy in the earliest gut: A single-cell view of heterotrophic carbon and nitrogen assimilation in sponge-microbe symbioses. ISME J. 2020, 14, 2554–2567. [Google Scholar] [CrossRef] [PubMed]
- Moeller, F.U.; Webster, N.S.; Herbold, C.W.; Behnam, F.; Domman, D.; Albertsen, M.; Mooshammer, M.; Markert, S.; Turaev, D.; Becher, D.; et al. Characterization of a thaumarchaeal symbiont that drives incomplete nitrification in the tropical sponge Ianthella basta. Environ. Microbiol. 2019, 21, 3831–3854. [Google Scholar] [CrossRef]
- Engelberts, J.P.; Robbins, S.J.; de Goeij, J.M.; Aranda, M.; Bell, S.C.; Webster, N.S. Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 2020, 14, 1100–1110. [Google Scholar] [CrossRef]
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, M.W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 2012, 10, 641–654. [Google Scholar] [CrossRef]
- Eastman, A.W.; Heinrichs, D.E.; Yuan, Z.-C. Comparative and genetic analysis of the four sequenced Paenibacillus polymyxa genomes reveals a diverse metabolism and conservation of genes relevant to plant-growth promotion and competitiveness. BMC Genom. 2014, 15, 851. [Google Scholar] [CrossRef] [PubMed]
- Steiner, L.X.; Wiese, J.; Rahn, T.; Borchert, E.; Slaby, B.M.; Hentschel, U. Maribacter halichondriae sp. nov. isolated from the marine sponge Halichondria panicea, displays features of a sponge-associated life style. Antonie Van Leeuwenhoek 2024, 117, 56. [Google Scholar] [CrossRef]
- Yang, B.; Yue, Y.; Chen, Y.; Ding, M.; Li, B.; Wang, L.; Wang, Q.; Stanton, C.; Ross, R.P.; Zhao, J.; et al. Lactobacillus plantarum CCFM1143 Alleviates Chronic Diarrhea via Inflammation Regulation and Gut Microbiota Modulation: A Double-Blind, Randomized, Placebo-Controlled Study. Front. Immunol. 2021, 12, 746585. [Google Scholar] [CrossRef]
- Slaby, B.M.; Hackl, T.; Horn, H.; Bayer, K.; Hentschel, U. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 2017, 11, 2465–2478. [Google Scholar] [CrossRef]
- Qin, X.; Wang, H.; Miao, C.; Yang, X.; Zhang, Y.; Feng, J.; Forsythe, S.J.; Man, C.; Jiang, Y. Comparative genomics reveals environmental adaptation differences between Cronobacter species. Food Res. Int. 2021, 147, 110541. [Google Scholar] [CrossRef]
- Vale, F.F.; Lehours, P.; Yamaoka, Y. Editorial: The Role of Mobile Genetic Elements in Bacterial Evolution and Their Adaptability. Front. Microbiol. 2022, 13, 849667. [Google Scholar] [CrossRef]
- Jahn, M.T.; Lachnit, T.; Markert, S.M.; Stigloher, C.; Pita, L.; Ribes, M.; Dutilh, B.E.; Hentschel, U. Lifestyle of sponge symbiont phages by host prediction and correlative microscopy. ISME J. 2021, 15, 2001–2011. [Google Scholar] [CrossRef]
- Rodriguez Jimenez, A.; Guiglielmoni, N.; Goetghebuer, L.; Dechamps, E.; George, I.F.; Flot, J.F. Comparative genome analysis of Vagococcus fluvialis reveals abundance of mobile genetic elements in sponge-isolated strains. BMC Genom. 2022, 23, 618. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhang, L.; Hu, H.; Liu, X.; Zhang, C.; Xin, Y.; Liu, B.; Chen, Z.; Xu, K.; Liu, Y. Complete genome sequencing and comparative genomic analyses of a new spotted-fever Rickettsia heilongjiangensis strain B8. Emerg. Microbes Infect. 2023, 12, 2153085. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Wang, D.; Dong, X.; Liu, D.; Liu, Y.; Li, P.; Wu, G.; Luo, Y.; Zhang, R.; Liu, S.; et al. Microbulbifer flavimaris sp. nov. a halophilic Gammaproteobacteria isolated from marine sediment of the Yellow Sea, China. Int. J. Syst. Evol. Microbiol. 2019, 69, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Hu, Y.; Zhang, G.; Li, M. Comparative genomic analysis reveals metabolic diversity of different Paenibacillus groups. Appl. Microbiol. Biotechnol. 2020, 104, 10133–10143. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhu, S.; Guo, C.; Xie, F.; Jung, D.; Li, S.; Zhang, W.; He, S. Microbulbifer hainanensis sp. nov. a moderately halopilic bacterium isolated from mangrove sediment. Antonie Van Leeuwenhoek 2021, 114, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, P.S.; Hyun, D.W.; Kim, H.S.; Shin, N.R.; Jung, M.J.; Yun, J.H.; Kim, M.S.; Whon, T.W.; Bae, J.W. Microbulbifer echini sp. nov. isolated from the gastrointestinal tract of a purple sea urchin, Heliocidaris crassispina. Int. J. Syst. Evol. Microbiol. 2017, 67, 998–1004. [Google Scholar] [CrossRef]
- Zhang, D.S.; Huo, Y.Y.; Xu, X.W.; Wu, Y.H.; Wang, C.S.; Xu, X.F.; Wu, M. Microbulbifer marinus sp. nov. and Microbulbifer yueqingensis sp. nov. isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 3, 505–510. [Google Scholar] [CrossRef]
- Wang, C.; Yu, Q.Y.; Ji, N.N.; Zheng, Y.; Taylor, J.W.; Guo, L.D.; Gao, C. Bacterial genome size and gene functional diversity negatively correlate with taxonomic diversity along a pH gradient. Nat. Commun. 2023, 14, 7437. [Google Scholar] [CrossRef]
- Thomas, T.; Rusch, D.; DeMaere, M.Z.; Yung, P.Y.; Lewis, M.; Halpern, A.; Heidelberg, K.B.; Egan, S.; Steinberg, P.D.; Kjelleberg, S. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010, 4, 1557–1567. [Google Scholar] [CrossRef]
- Ishaq, N.; Zhang, M.; Gao, L.; Ilan, M.; Li, Z. Microbulbifer spongiae sp. nov. isolated from marine sponge Diacarnus erythraeanus. Int. J. Syst. Evol. Microbiol. 2024, 74. [Google Scholar] [CrossRef]
- Stiffler, A.K.; Hesketh-Best, P.J.; Varona, N.S.; Zagame, A.; Wallace, B.A.; Lapointe, B.E.; Silveira, C.B. Genomic and induction evidence for bacteriophage contributions to sargassum-bacteria symbioses. Microbiome 2024, 12, 143. [Google Scholar] [CrossRef]
- Gao, L.; Song, Q.; Sang, J.; Xiao, Y.; Li, Z. Cytobacillus spongiae sp. nov. isolated from sponge Diacarnus spinipoculum. Int. J. Syst. Evol. Microbiol. 2023, 73, 005903. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Lomsadze, A.; Gemayel, K.; Tang, S.; Borodovsky, M. Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res. 2018, 28, 1079–1089. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, X.; Liu, M.; Liu, W.; Xu, J.; Li, Y. Comparative evaluation of 16S rRNA primer pairs in identifying nitrifying guilds in soils under long-term organic fertilization and water management. Front. Microbiol. 2024, 15, 1424795. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Ontiveros-Palacios, N.; Cooke, E.; Nawrocki, E.P.; Triebel, S.; Marz, M.; Rivas, E.; Griffiths-Jones, S.; Petrov, A.I.; Bateman, A.; Sweeney, B. Rfam 15: RNA families database in 2025. Nucleic Acids Res. 2024, 53, D258–D267. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–w35. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Vargas, W.A.; Martín, J.M.S.; Rech, G.E.; Rivera, L.P.; Benito, E.P.; Díaz-Mínguez, J.M.; Thon, M.R.; Sukno, S.A. Plant Defense Mechanisms Are Activated during Biotrophic and Necrotrophic Development of Colletotricum graminicola in Maize. Plant Physiol. 2012, 158, 1342–1358. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Thanki, A.S.; Soranzo, N.; Haerty, W.; Davey, R.P. GeneSeqToFamily: A Galaxy workflow to find gene families based on the Ensembl Compara GeneTrees pipeline. Gigascience 2018, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Huang, F.; Liu, X.; Guo, J.; Cui, N.; Liang, C.; Lian, Y.; Deng, J.; Wu, H.; Yin, H.; et al. Phylogenetic analysis based on single-copy orthologous proteins in highly variable chloroplast genomes of Corydalis. Sci. Rep. 2022, 12, 14241. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Cui, Z.; Xu, J.; Hao, D.; Shi, J.; Wang, D.; Xiao, H.; Duan, X.; Chen, R.; Li, W. NG-Circos: Next-generation Circos for data vis-ualization and interpretation. NAR Genome Bioinform. 2020, 2, lqaa069. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, M.; Liu, M.; Thomas, T. Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 2014, 23, 1635–1645. [Google Scholar] [CrossRef]
- Sugden, S.; Holert, J.; Cardenas, E.; Mohn, W.W.; Stein, L.Y. Microbiome of the freshwater sponge Ephydatia muelleri shares compositional and functional similarities with those of marine sponges. ISME J. 2022, 16, 2503–2512. [Google Scholar] [CrossRef]
- Reynolds, D.; Thomas, T. Evolution and function of eukaryotic-like proteins from sponge symbionts. Mol. Ecol. 2016, 25, 5242–5253. [Google Scholar] [CrossRef] [PubMed]
- Horn, H.; Slaby, B.M.; Jahn, M.T.; Bayer, K.; Moitinho-Silva, L.; Förster, F.; Abdelmohsen, U.R.; Hentschel, U. An Enrichment of CRISPR and Other Defense-Related Features in Marine Sponge-Associated Microbial Metagenomes. Front. Microbiol. 2016, 7, 1751. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.A.; Robbins, S.J.; Tan, S.; Rix, L.; Miller, D.J.; Webster, N.S.; Zhang, G.; Bourne, D.G. Comparative genomics identifies key adaptive traits of sponge-associated microbial symbionts. Environ. Microbiol. 2024, 26, e16690. [Google Scholar] [CrossRef]
- Goodhead, I.; Darby, A.C. Taking the pseudo out of pseudogenes. Curr. Opin. Microbiol. 2015, 23, 102–109. [Google Scholar] [CrossRef]
- Romanenko, L.; Kurilenko, V.; Otstavnykh, N.; Velansky, P.; Isaeva, M.; Mikhailov, V. Microbulbifer okhotskensis sp. nov. isolated from a deep bottom sediment of the Okhotsk Sea. Arch. Microbiol. 2022, 204, 548. [Google Scholar] [CrossRef]
- Frank, A.C. Molecular host mimicry and manipulation in bacterial symbionts. FEMS Microbiol. Lett. 2019, 366, fnz038. [Google Scholar] [CrossRef]
- Pan, X.; Lührmann, A.; Satoh, A.; Laskowski-Arce, M.A.; Roy, C.R. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 2008, 320, 1651–1654. [Google Scholar] [CrossRef]
- O’Brien, P.A.; Tan, S.; Frade, P.R.; Robbins, S.J.; Engelberts, J.P.; Bell, S.C.; Vanwonterghem, I.; Miller, D.J.; Webster, N.S.; Zhang, G.; et al. Validation of key sponge symbiont pathways using genome-centric metatranscriptomics. Environ. Microbiol. 2023, 25, 3207–3224. [Google Scholar] [CrossRef]
- Kamke, J.; Sczyrba, A.; Ivanova, N.; Schwientek, P.; Rinke, C.; Mavromatis, K.; Woyke, T.; Hentschel, U. Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J. 2013, 7, 2287–2300. [Google Scholar] [CrossRef]
- Moh, T.H.; Furusawa, G.; Amirul, A.A. Microbulbifer aggregans sp. nov. isolated from estuarine sediment from a mangrove forest. Int. J. Syst. Evol. Microbiol. 2017, 67, 4089–4094. [Google Scholar] [CrossRef] [PubMed]
- Berlemont, R.; Martiny, A.C. Glycoside Hydrolases across Environmental Microbial Communities. PLoS Comput. Biol. 2016, 12, e1005300. [Google Scholar] [CrossRef]
- Nguyen, S.T.C.; Freund, H.L.; Kasanjian, J.; Berlemont, R. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl. Microbiol. Biotechnol. 2018, 102, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Hallam, S.J.; Konstantinidis, K.T.; Putnam, N.; Schleper, C.; Watanabe, Y.; Sugahara, J.; Preston, C.; de la Torre, J.; Richardson, P.M.; DeLong, E.F. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. USA 2006, 103, 18296–18301. [Google Scholar] [CrossRef]
- Khizar, Y.; Farooq, U.; Attia, K.A.; Rehman, O.U.; Abushady, A.M.; Fiaz, S.; Zeb, U.; Iqbal, R.; Uzair, M. Genome-wide identification and characterization of stress-responsive genes in Chlorella vulgaris. BMC Genom. Data 2025, 26, 20. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Raghu, G.; Sharma, Y.V.; Biju, A.R.; Rajasekharan, M.V.; Shivaji, S. Increase in oxidative stress at low temperature in an antarctic bacterium. Curr. Microbiol. 2011, 62, 544–546. [Google Scholar] [CrossRef]
- Moreno-Pino, M.; Manrique-de-la-Cuba, M.F.; López-Rodríguez, M.; Parada-Pozo, G.; Rodríguez-Marconi, S.; Ribeiro, C.G.; Flores-Herrera, P.; Guajardo, M.; Trefault, N. Unveiling microbial guilds and symbiotic relationships in Antarctic sponge microbiomes. Sci. Rep. 2024, 14, 6371. [Google Scholar] [CrossRef]
- Chung, I.Y.; Kim, B.O.; Jang, H.J.; Cho, Y.H. Dual promoters of the major catalase (KatA) govern distinct survival strategies of Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 31185. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Choi, Y.S.; Cho, Y.H. Unusual properties of catalase A (KatA) of Pseudomonas aeruginosa PA14 are associated with its biofilm peroxide resistance. J. Bacteriol. 2008, 190, 2663–2670. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Nian, Y.; Lin, H.; Li, J.; Lin, X.; Li, T.; Wang, R.; Wang, L.; Beattie, G.A.; Zhang, J.; et al. Structure and mechanism of the osmoregulated choline transporter BetT. Sci. Adv. 2024, 10, eado6229. [Google Scholar] [CrossRef]
- Shen, L.; Zang, X.; Sun, K.; Chen, H.; Che, X.; Sun, Y.; Wang, G.; Zhang, S.; Chen, G. Complete genome sequencing of Bacillus sp. TK-2, analysis of its cold evolution adaptability. Sci. Rep. 2021, 11, 4836. [Google Scholar] [CrossRef]
- Lin, J.; Xu, L.; Yang, J.; Wang, Z.; Shen, X. Beyond dueling: Roles of the type VI secretion system in microbiome modulation, pathogenesis and stress resistance. Stress. Biol. 2021, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Gogarten, J.P.; Doolittle, W.F.; Lawrence, J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002, 19, 2226–2238. [Google Scholar] [CrossRef]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.J.; Rocha, E.P. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011, 7, e1001284. [Google Scholar] [CrossRef]
- Bobay, L.M.; Ochman, H. The Evolution of Bacterial Genome Architecture. Front. Genet. 2017, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.; Polónia, A.R.M.; Cleary, D.F.R.; Huang, Y.M.; de Voogd, N.J.; Keller-Costa, T.; Costa, R.; Gomes, N.C.M. Assessing the genomic composition, putative ecological relevance and biotechnological potential of plasmids from sponge bacterial symbionts. Microbiol. Res. 2022, 265, 127183. [Google Scholar] [CrossRef]
- Stasiak, G.; Mazur, A.; Wielbo, J.; Marczak, M.; Zebracki, K.; Koper, P.; Skorupska, A. Functional relationships between plasmids and their significance for metabolism and symbiotic performance of Rhizobium leguminosarum bv. trifolii. J. Appl. Genet. 2014, 55, 515–527. [Google Scholar] [CrossRef]
- Yang, L.-L.; Jiang, Z.; Li, Y.; Wang, E.-T.; Zhi, X.-Y. Plasmids Related to the Symbiotic Nitrogen Fixation Are Not Only Cooperated Functionally but Also May Have Evolved over a Time Span in Family Rhizobiaceae. Genome Biol. Evol. 2020, 12, 2002–2014. [Google Scholar] [CrossRef]
- Gil, R.; Sabater-Muñoz, B.; Perez-Brocal, V.; Silva, F.J.; Latorre, A. Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes. A puzzling evolutionary story. Gene 2006, 370, 17–25. [Google Scholar] [CrossRef]
- Touchon, M.; Rocha, E.P.C. Causes of Insertion Sequences Abundance in Prokaryotic Genomes. Mol. Biol. Evol. 2007, 24, 969–981. [Google Scholar] [CrossRef]
- Munshi, I.D.; Mathuria, A.; Sharma, H.; Acharya, M.; Chaudhary, A.; Jain, K.; Ragini; Dahiya, S.; Arora, R.; Singh, V.; et al. Emerging concept of genomic islands in bacterial adaptation and pathogenicity. Res. Microbiol. 2025, 104303. [Google Scholar] [CrossRef]
- Valle, J.; Vergara-Irigaray, M.; Merino, N.; Penadés, J.R.; Lasa, I. sigmaB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J. Bacteriol. 2007, 189, 2886–2896. [Google Scholar] [CrossRef]
- Leavis, H.L.; Willems, R.J.; van Wamel, W.J.; Schuren, F.H.; Caspers, M.P.; Bonten, M.J. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 2007, 3, e7. [Google Scholar] [CrossRef]
- Siegl, A.; Kamke, J.; Hochmuth, T.; Piel, J.; Richter, M.; Liang, C.; Dandekar, T.; Hentschel, U. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011, 5, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Hong, Y.; Xu, R.; Guo, W.; Zhang, M.; Lu, Z.; Han, Q.; Mo, Z.; Dan, X.; Li, Y. Genomic evidence of genetic diversity and functional evolution in Flavobacterium columnare. Front. Microbiol. 2023, 14, 1240471. [Google Scholar] [CrossRef]
- Zhang, Q.; Rho, M.; Tang, H.; Doak, T.G.; Ye, Y. CRISPR-Cas systems target a diverse collection of invasive mobile genetic elements in human microbiomes. Genome Biol. 2013, 14, R40. [Google Scholar] [CrossRef]
- Deem, K.D.; Brisson, J.A. Problems with Paralogs: The Promise and Challenges of Gene Duplicates in Evo-Devo Research. Integr. Comp. Biol. 2024, 64, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; French, S.; El Zahed, S.S.; Ong, W.K.; Karp, P.D.; Brown, E.D. Gene Dispensability in Escherichia coli Grown in Thirty Different Carbon Environments. mBio 2020, 11, e02259-20. [Google Scholar] [CrossRef]
- Karimi, E.; Ramos, M.; Gonçalves, J.M.S.; Xavier, J.R.; Reis, M.P.; Costa, R. Comparative Metagenomics Reveals the Distinctive Adaptive Features of the Spongia officinalis Endosymbiotic Consortium. Front. Microbiol. 2017, 8, 2499. [Google Scholar] [CrossRef]
- Konstantinou, D.; Popin, R.V.; Fewer, D.P.; Sivonen, K.; Gkelis, S. Genome Reduction and Secondary Metabolism of the Marine Sponge-Associated Cyanobacterium Leptothoe. Mar. Drugs 2021, 19, 298. [Google Scholar] [CrossRef] [PubMed]
- Karimi, E.; Keller-Costa, T.; Slaby, B.M.; Cox, C.J.; da Rocha, U.N.; Hentschel, U.; Costa, R. Genomic blueprints of sponge-prokaryote symbiosis are shared by low abundant and cultivatable Alphaproteobacteria. Sci. Rep. 2019, 9, 1999. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, L.; Guo, Z.; Tang, T.; Zhu, B. Alginate degrading enzymes: An updated comprehensive review of the structure, catalytic mechanism, modification method and applications of alginate lyases. Crit. Rev. Biotechnol. 2021, 41, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Reynolds, D.; Liu, M.; Stark, M.; Kjelleberg, S.; Webster, N.S.; Thomas, T. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. USA 2012, 109, E1878–E1887. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, S.; Jóhannsson, R.; Marteinsson, V. Genome analysis of sponge symbiont ‘Candidatus Halichondribacter symbioticus’ shows genomic adaptation to a host-dependent lifestyle. Environ. Microbiol. 2020, 22, 483–498. [Google Scholar] [CrossRef]

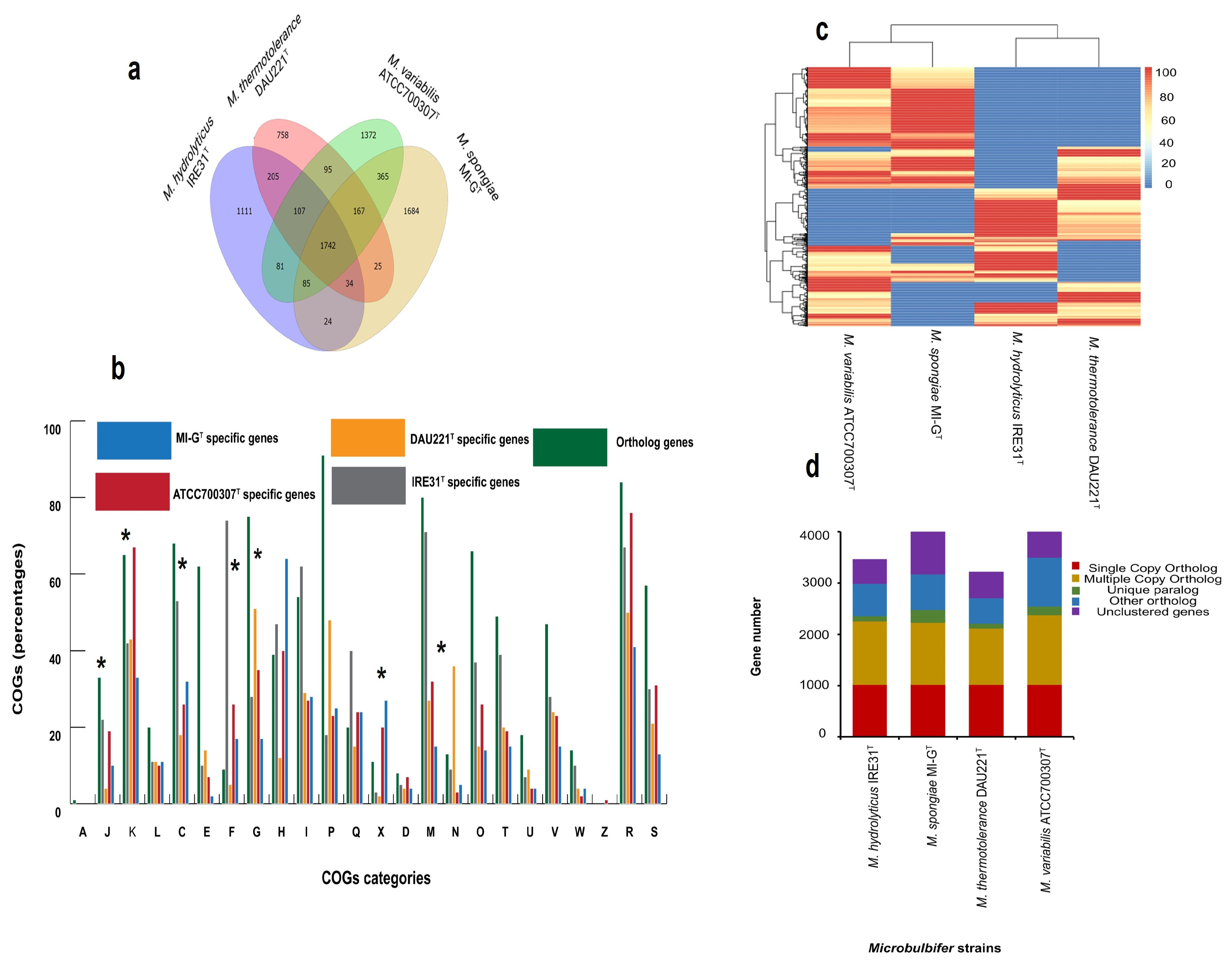

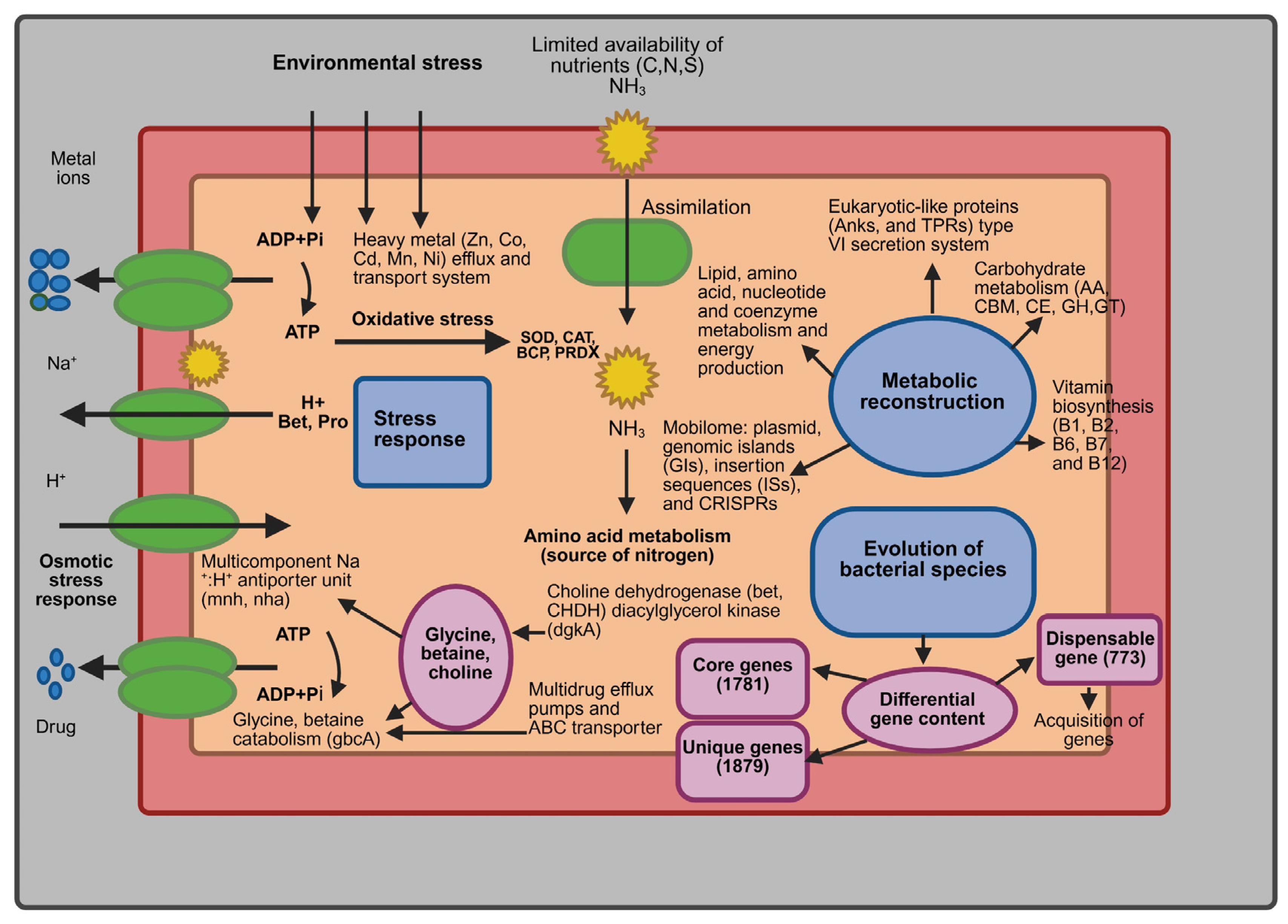
| Category | MI-GT | IRE31T | DAU221T | ATCC700307T |
|---|---|---|---|---|
| Genome size (nt) | 4,478,124 (bp) | 4,209,307 (bp) | 3,938,396 (bp) | 4,855,835 (bp) |
| Plasmid | 1 | N/A | N/A | N/A |
| GC content (%) | 54.51 | 57.6 | 56.6 | 49.5 |
| Protein-coding genes | 4433 | 3549 | 3273 | 4253 |
| Genes with functions | 3683 | 3440 | 3145 | 4142 |
| Contig Number | 2 | 1 | 1 | 1 |
| Completeness (%) | 100 | 100 | 99.44 | 99.44 |
| Contamination (%) | 0.56 | 0.56 | 0 | 0.56 |
| ncRNA | 4 | 4 | 4 | 4 |
| tRNA | 50 | 66 | 48 | 59 |
| 5s_rRNA | 4 | 4 | 3 | 5 |
| 16s_rRNA | 4 | 4 | 3 | 5 |
| 23s_rRNA | 4 | 4 | 3 | 5 |
| Pseudogenes | 83 | 27 | 67 | 33 |
| Genome coverage | 267.0x | 400.0x | 197.8x | 100x |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishaq, N.; Song, Q.; Ilan, M.; Li, Z. Functional Genomic Characteristics of Marine Sponge-Associated Microbulbifer spongiae MI-GT. Microorganisms 2025, 13, 1940. https://doi.org/10.3390/microorganisms13081940
Ishaq N, Song Q, Ilan M, Li Z. Functional Genomic Characteristics of Marine Sponge-Associated Microbulbifer spongiae MI-GT. Microorganisms. 2025; 13(8):1940. https://doi.org/10.3390/microorganisms13081940
Chicago/Turabian StyleIshaq, Nabila, Qianqian Song, Micha Ilan, and Zhiyong Li. 2025. "Functional Genomic Characteristics of Marine Sponge-Associated Microbulbifer spongiae MI-GT" Microorganisms 13, no. 8: 1940. https://doi.org/10.3390/microorganisms13081940
APA StyleIshaq, N., Song, Q., Ilan, M., & Li, Z. (2025). Functional Genomic Characteristics of Marine Sponge-Associated Microbulbifer spongiae MI-GT. Microorganisms, 13(8), 1940. https://doi.org/10.3390/microorganisms13081940







