Automated Platform for the Analysis of Multi-Plate Growth and Reporter Data
Abstract
1. Introduction
2. Materials and Methods
3. The Architecture of GROOT Software
3.1. General
3.2. Data-Ingest Layer
- Automated block discovery: Each selected Excel sheet is parsed once. A vectorized search for characteristic matrix header starts with coordinates; this permits arbitrarily located blocks and mixed orientations in a single workbook.
- Orientation and integrity checks: The helper DetermineVectorType classifies matrices as vertical- or horizontal-time, after which empty margins, NaN seams, trailing zero segments, and Excel’s > 24 h rollover are excised. Misaligned time vectors (>180 s) are rejected early, raising non-blocking warnings via the logging service.
- Canonicalization: Time stamps are coerced to seconds, re-exported in user-chosen units, and welded to numeric payloads whose columns represent wells. Text labels are id-prefixed (MX01–MXnn) to remain unique across sheets.
- The resulting objects: Time, data, and label form the model layer.
3.3. Model Layer
- Hierarchical grouping: A dual-tree structure stores native groups (sheet→well) and selected groups (Z01–Z99), or automatic groups by the plate letters (A1-A12→MXnA). The mover widget clones checked well into custom parents without mutating source nodes; this is implemented as lightweight handle copies, that is, memory scales with selections, not data size.
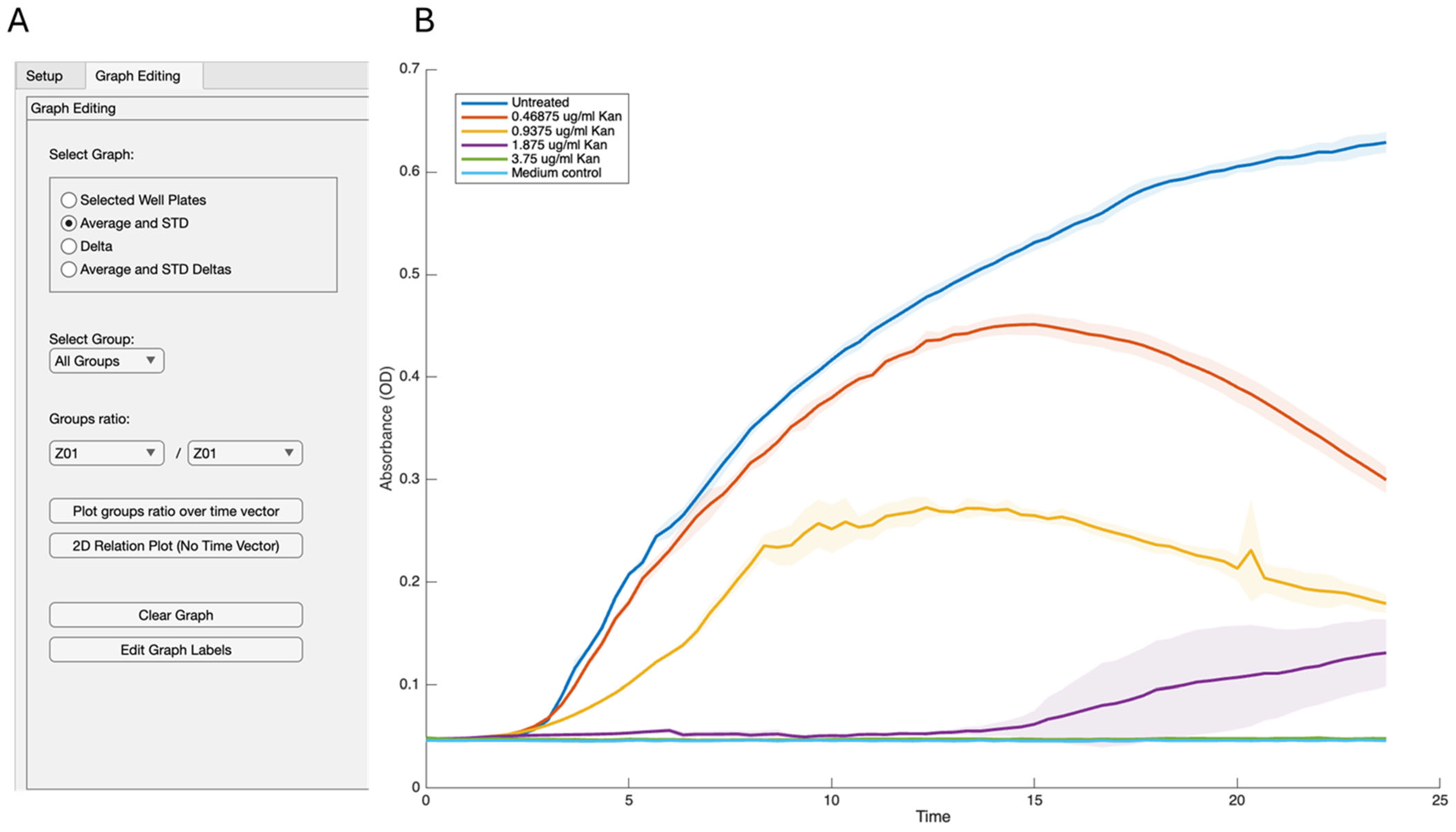
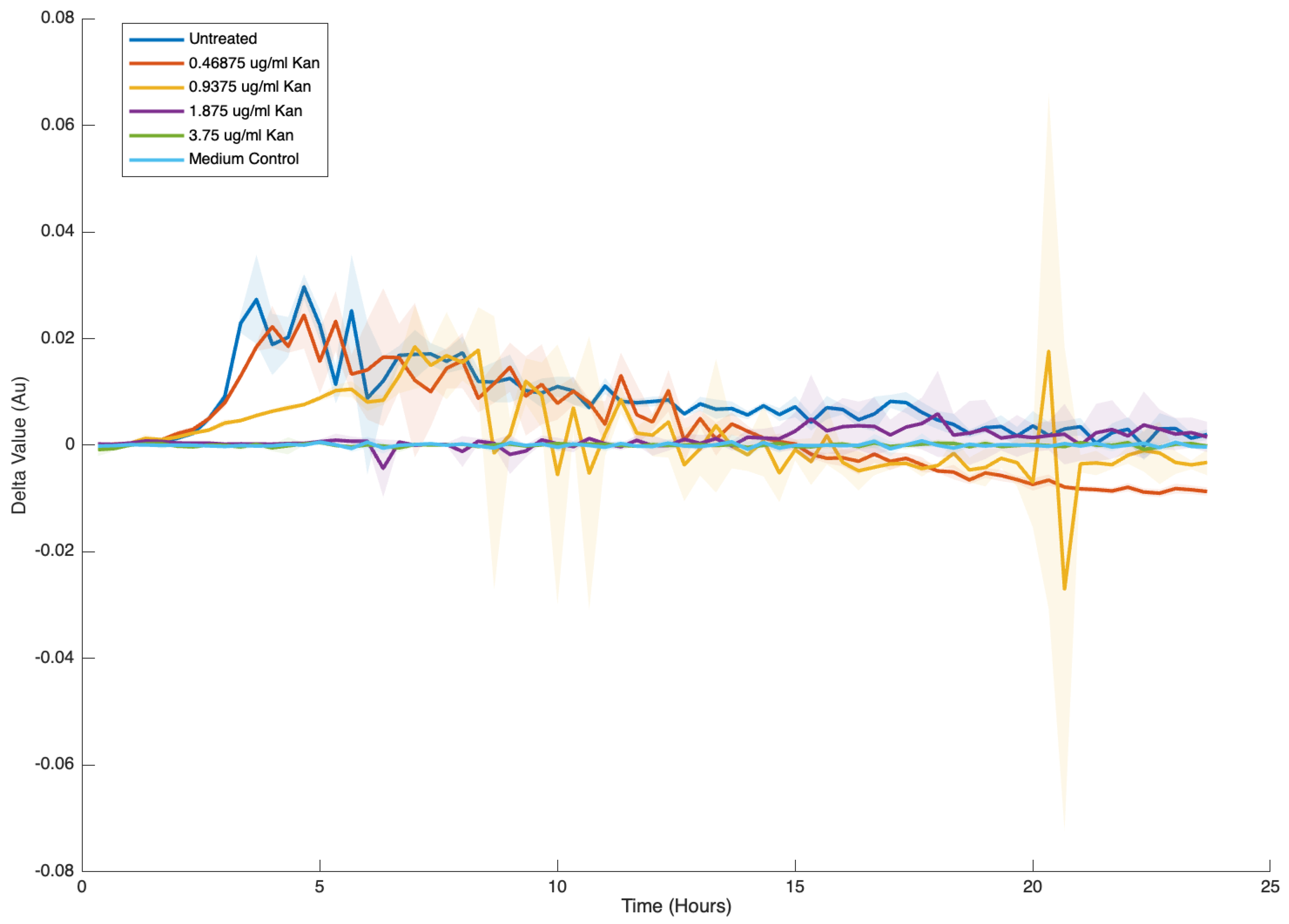
- Derived statistics: Vectorized routines expose on-demand views: mean ± SD envelopes, first differences, and propagated ratio errors. All statistics preserve NaN positions, ensuring downstream plots reflect missingness transparently.
3.4. Calculations Layer
3.5. Presentation Layer
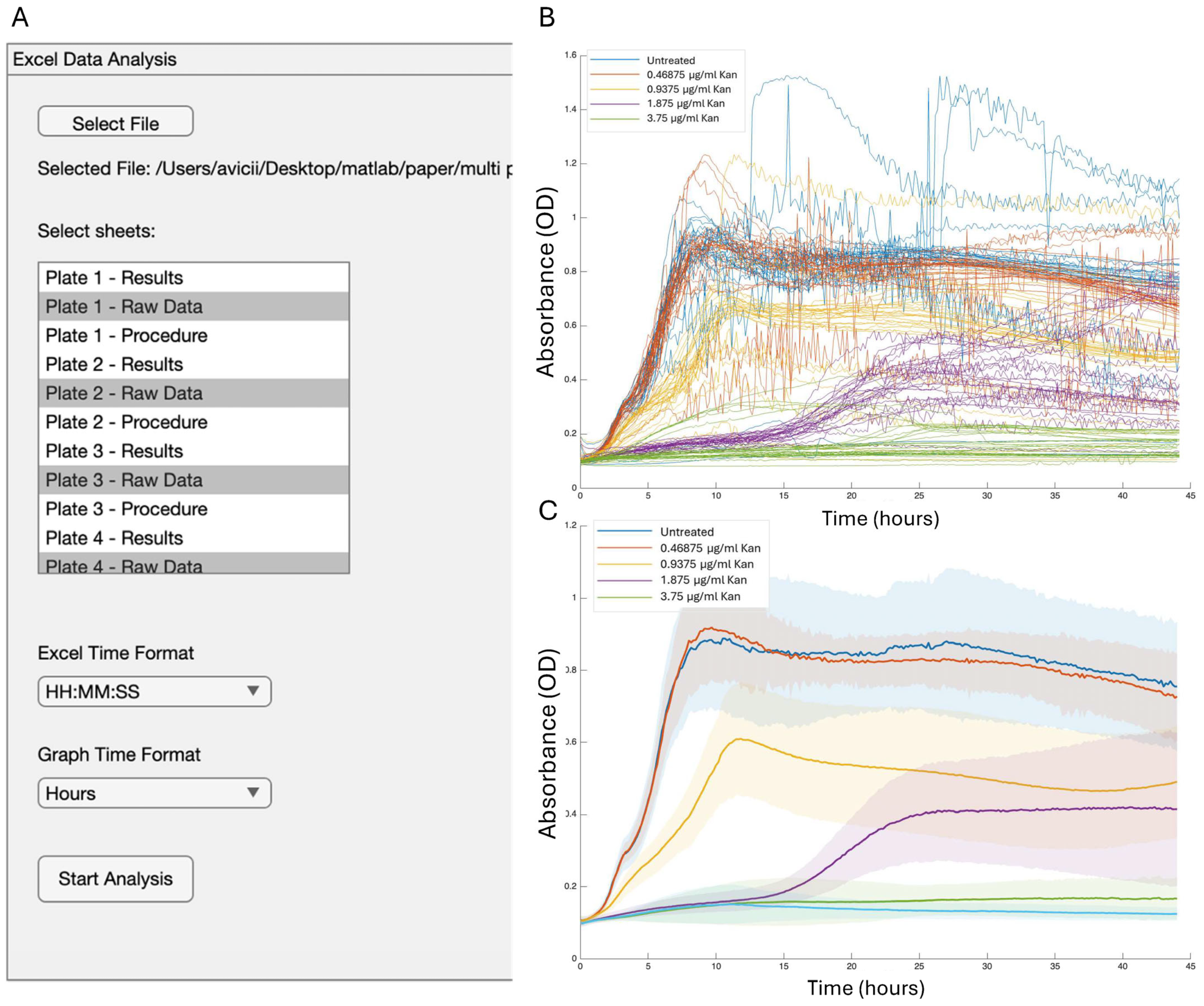
- Setup tab: Orchestrates file selection, sheet list, time-unit dropdowns, and ingest progress. A blinking uilamp is toggled by a timer that is instantiated and deterministically destroyed inside analyzeData, preventing orphan threads.
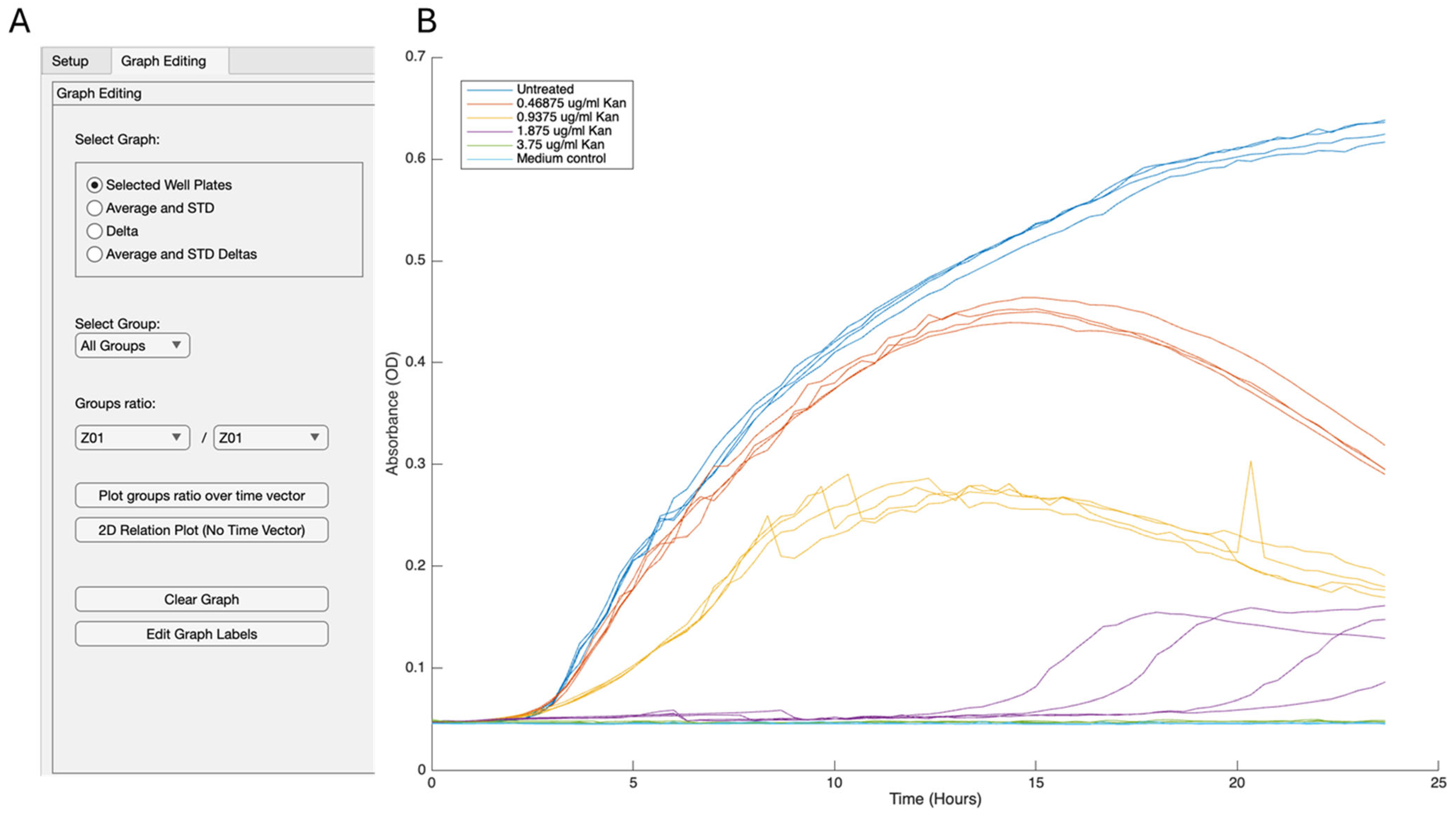
- Graph-editing tab (selected manually): Radio-button groups choose the visual mode. Dropdowns directly connect to live group lists via observers, Edit Graph Labels button enables customization of the axis and main title labels, and legend box can be modified manually. Plot routines share a single UI axes object, cleared only when the user requests a reset, allowing multilayer storytelling. Custom data tips and HSV/greyscale palettes enhance traceability.
- Audit trail: All status messages funnel through custom_fprintf—which has timestamps and color-coded errors—and stream simultaneously to a pop-up log window and in-panel label.
3.6. Statistics
4. Results
4.1. GROOT User Guide
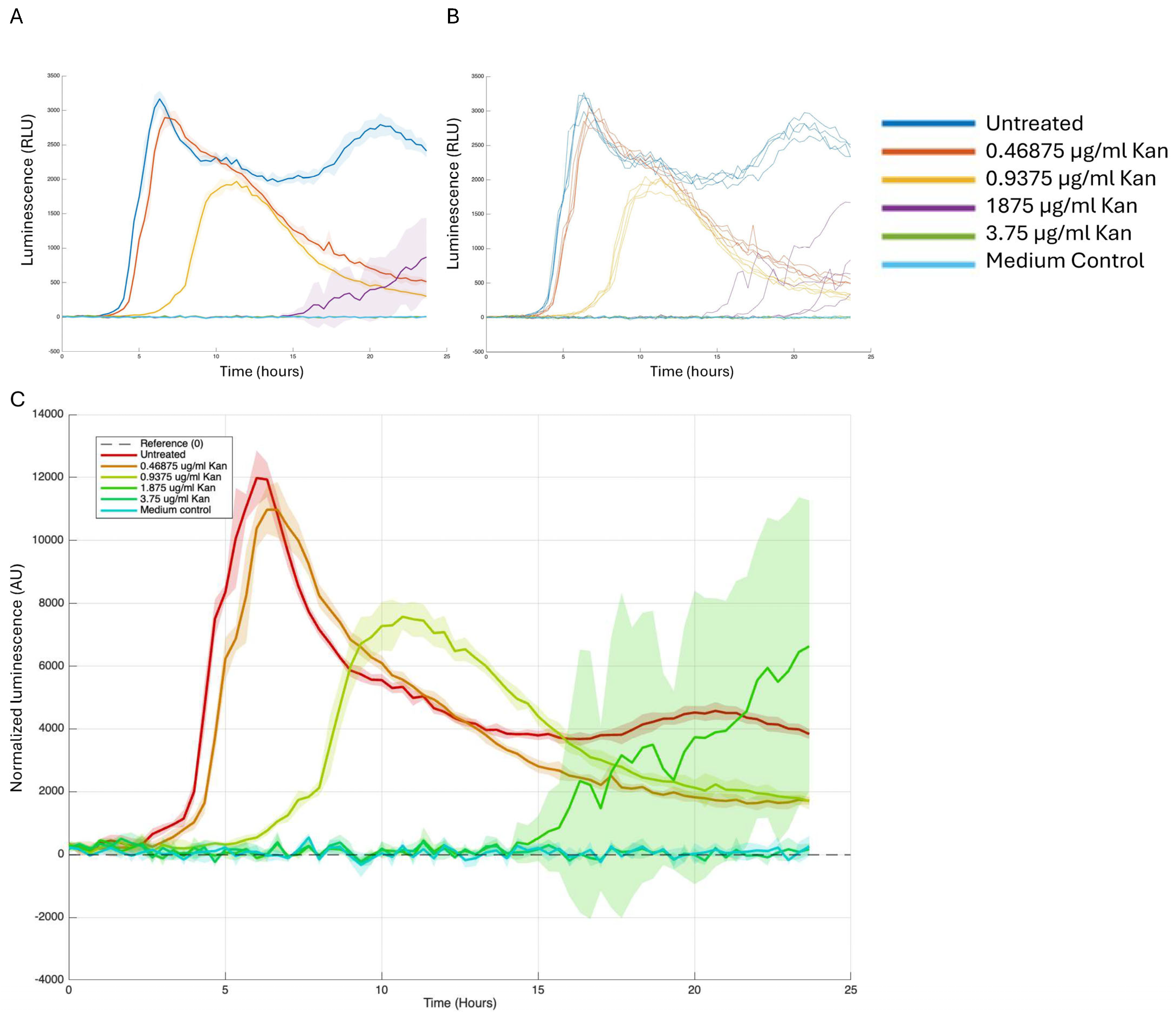
4.2. Growth and Reporter Data Analysis
4.3. Statistical Analysis Features of GROOT
Data Export and Housekeeping
4.4. Growth-Expression Dynamics
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mira, P.; Barlow, M.; Meza, J.C.; Hall, B.G. Statistical Package for Growth Rates Made Easy. Mol. Biol. Evol. 2017, 34, 3303–3309. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, W.R.; Jones, S.E.; Muscarella, M.E.; Behringer, M.G.; Lehmkuhl, B.K.; Lennon, J.T. Microbial population dynamics and evolutionary outcomes under extreme energy limitation. Proc. Natl. Acad. Sci. USA 2021, 118, e2101691118. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Nguyen, T.V.P.; Homaee, E.; Golding, I. Using bacterial population dynamics to count phages and their lysogens. Nat. Commun. 2024, 15, 7814. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.; McMeekin, T.A. Predictive microbiology. Int. J. Food Microbiol. 1994, 23, 241–264. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth rates made easy. Mol. Biol. Evol. 2014, 31, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Mira, P.M.; Crona, K.; Greene, D.; Meza, J.C.; Sturmfels, B.; Barlow, M. Rational design of antibiotic treatment plans: A treatment strategy for managing evolution and reversing resistance. PLoS ONE 2015, 10, e0122283. [Google Scholar] [CrossRef]
- Jung, P.P.; Christian, N.; Kay, D.P.; Skupin, A.; Linster, C.L. Protocols and programs for high-throughput growth and aging phenotyping in yeast. PLoS ONE 2015, 10, e0119807. [Google Scholar] [CrossRef] [PubMed]
- Maan, H.; Itkin, M.; Malitsky, S.; Friedman, J.; Kolodkin-Gal, I. Resolving the conflict between antibiotic production and rapid growth by recognition of peptidoglycan of susceptible competitors. Nat. Commun. 2022, 13, 431. [Google Scholar] [CrossRef] [PubMed]
- Bucher, T.; Keren-Paz, A.; Hausser, J.; Olender, T.; Cytryn, E.; Kolodkin-Gal, I. An active beta-lactamase is a part of an orchestrated cell wall stress resistance network of Bacillus subtilis and related rhizosphere species. Environ. Microbiol. 2019, 21, 1068–1085. [Google Scholar] [CrossRef] [PubMed]
- Gilhar, O.; Ben-Navi, L.R.; Olender, T.; Aharoni, A.; Friedman, J.; Kolodkin-Gal, I. Multigenerational inheritance drives symbiotic interactions of the bacterium Bacillus subtilis with its plant host. Microbiol. Res. 2024, 286, 127814. [Google Scholar] [CrossRef] [PubMed]
- Image-based high-content reporter assays: Limitations and advantages. Drug Discov. Today Technol. 2010, 7, e1–e94. [CrossRef]
- Waidmann, M.S.; Bleichrodt, F.S.; Laslo, T.; Riedel, C.U. Bacterial luciferase reporters: The Swiss army knife of molecular biology. Bioeng. Bugs 2011, 2, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Arnaouteli, S.; Bamford, N.C.; Stanley-Wall, N.R.; Kovacs, A.T. Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 2021, 19, 600–614. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Chu, F.; Kolter, R.; Losick, R. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008, 67, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.; Kearns, D.B.; Branda, S.S.; Kolter, R.; Losick, R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 2006, 59, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- McLoon, A.L.; Kolodkin-Gal, I.; Rubinstein, S.M.; Kolter, R.; Losick, R. Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J. Bacteriol. 2011, 193, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Povolotsky, T.L.; Keren-Paz, A.; Kolodkin-Gal, I. Metabolic Microenvironments Drive Microbial Differentiation and Antibiotic Resistance. Trends Genet. 2021, 37, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer-Shaanan, Y.; Sibony-Nevo, O.; Bloom-Ackermann, Z.; Suissa, R.; Steinberg, N.; Kartvelishvily, E.; Brumfeld, V.; Kolodkin-Gal, I. Spatio-temporal assembly of functional mineral scaffolds within microbial biofilms. NPJ Biofilms Microbiomes 2016, 2, 15031. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, L.M.; Spagnoli, R. Biological transformation of kanamycin A to amikacin (BBK-8). J. Antibiot. 1983, 36, 328–330. [Google Scholar] [CrossRef] [PubMed][Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nahami, A.; Kain, D.; Cohen, Y.; Kolodkin-Gal, Y.; Assouline, Y.; Yona, A.H.; Kolodkin-Gal, I.; Dorfan, Y. Automated Platform for the Analysis of Multi-Plate Growth and Reporter Data. Microorganisms 2025, 13, 1889. https://doi.org/10.3390/microorganisms13081889
Nahami A, Kain D, Cohen Y, Kolodkin-Gal Y, Assouline Y, Yona AH, Kolodkin-Gal I, Dorfan Y. Automated Platform for the Analysis of Multi-Plate Growth and Reporter Data. Microorganisms. 2025; 13(8):1889. https://doi.org/10.3390/microorganisms13081889
Chicago/Turabian StyleNahami, Avichay, Dor Kain, Yonatan Cohen, Yuval Kolodkin-Gal, Yohanan Assouline, Avihu H. Yona, Ilana Kolodkin-Gal, and Yuval Dorfan. 2025. "Automated Platform for the Analysis of Multi-Plate Growth and Reporter Data" Microorganisms 13, no. 8: 1889. https://doi.org/10.3390/microorganisms13081889
APA StyleNahami, A., Kain, D., Cohen, Y., Kolodkin-Gal, Y., Assouline, Y., Yona, A. H., Kolodkin-Gal, I., & Dorfan, Y. (2025). Automated Platform for the Analysis of Multi-Plate Growth and Reporter Data. Microorganisms, 13(8), 1889. https://doi.org/10.3390/microorganisms13081889







