Alteromonas nitratireducens sp. nov., a Novel Nitrate-Reducing Bacterium Isolated from Marine Sediments, and the Evolution of Nitrate-Reducing Genes in the Genus Alteromonas
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation, Cultivation, and Preservation
2.2. Phylogenetic Reconstruction and Genomic Analysis
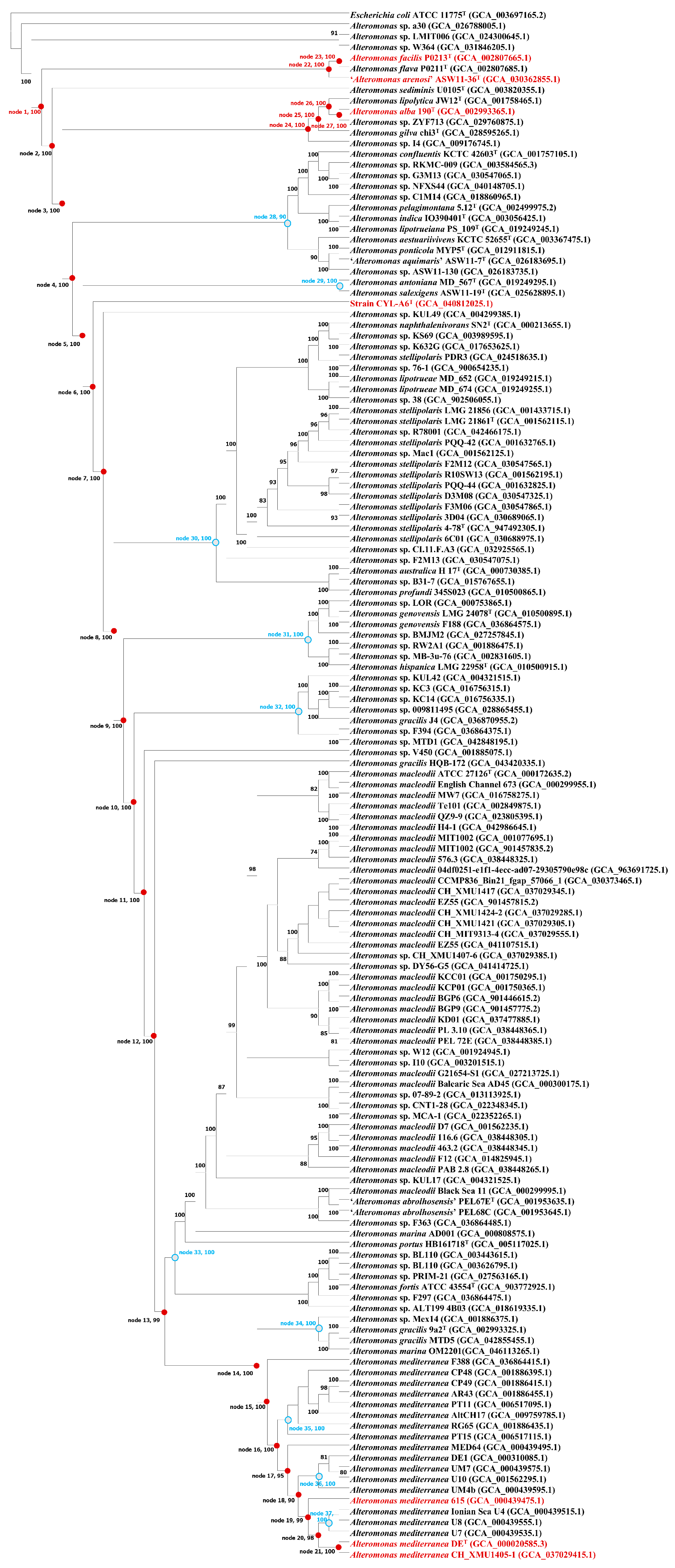
2.2.1. Phylogenetic Reconstruction Based on 16S rRNA Gene Sequences
2.2.2. Genomic Sequencing, Assembly, and Annotation
2.2.3. Comparative Genomic Analysis and Phylogenomic Reconstruction
2.3. Determinations of Phenotypic Characteristics
2.3.1. Determination of Biochemical Characteristics
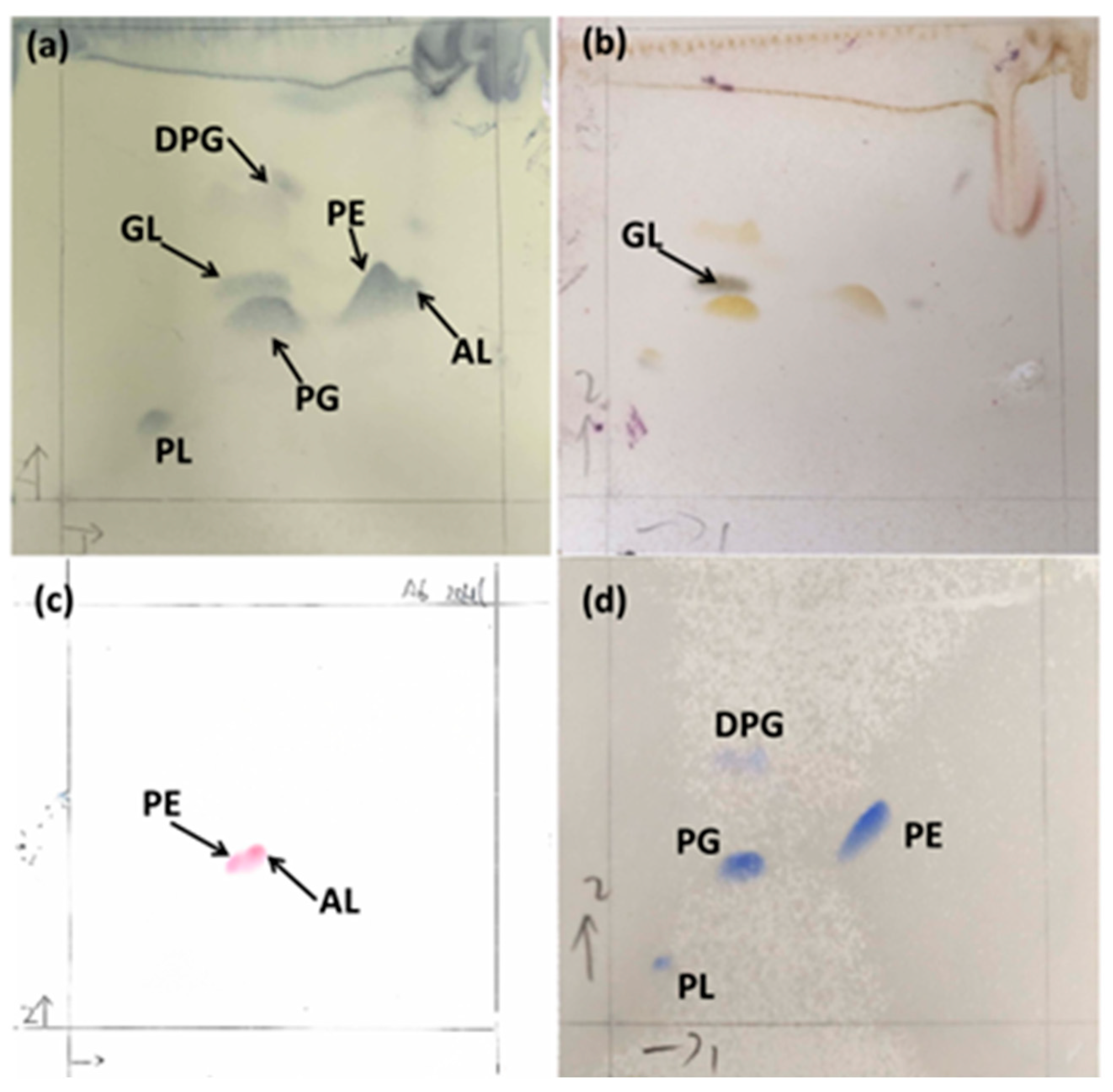
2.3.2. Determination of Chemotaxonomic Characteristics
2.4. Nitrate Reduction Test and Its Evolutionary Trajectory Analysis
3. Results
3.1. Taxonomic Identification of Cultivated Strains Based on the 16S rRNA Gene Sequences
3.2. Polyphasic Taxonomy of Strain CYL-A6T
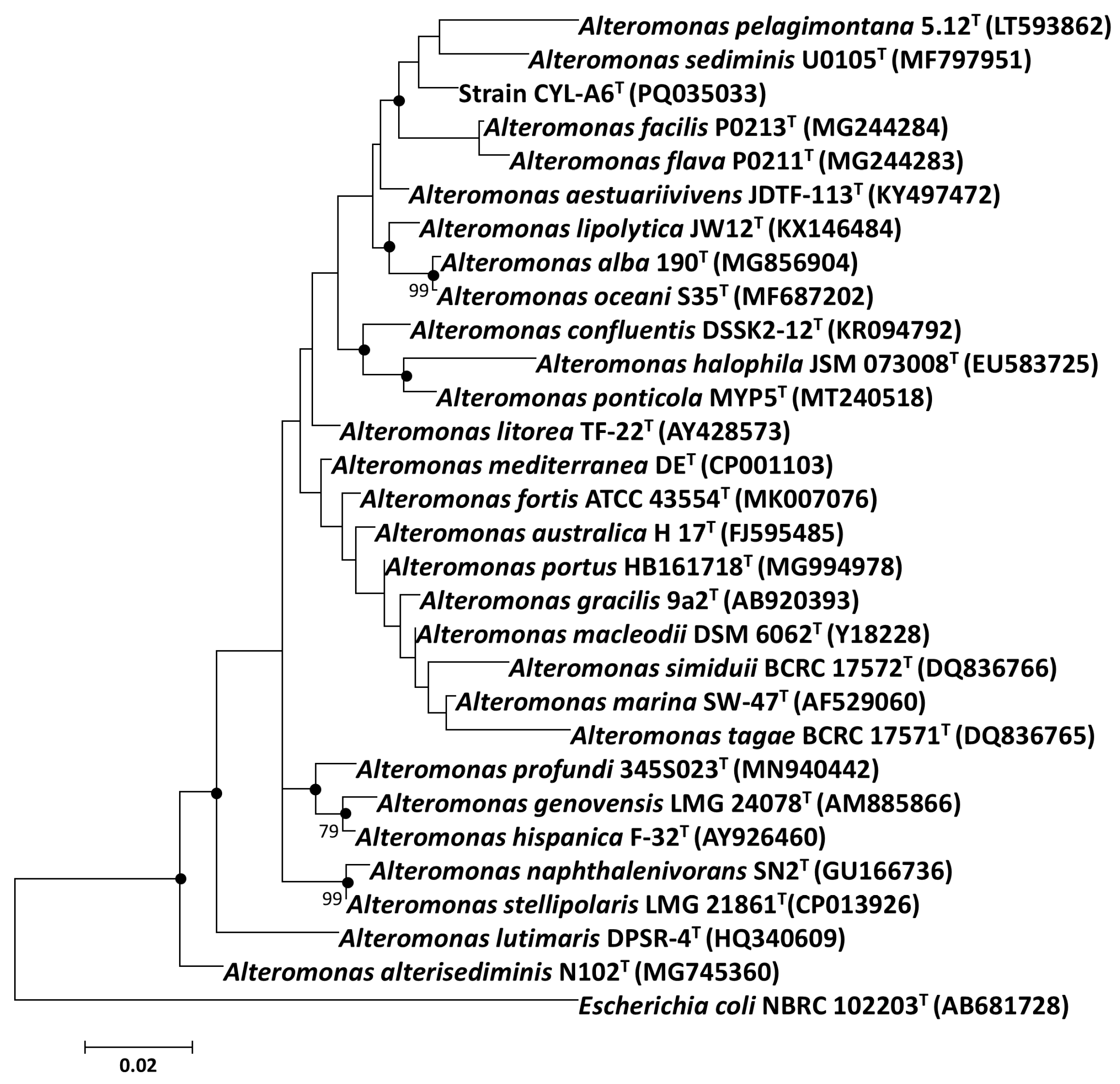
3.2.1. 16S rRNA Gene Sequence Identity and Phylogenetic Relationship
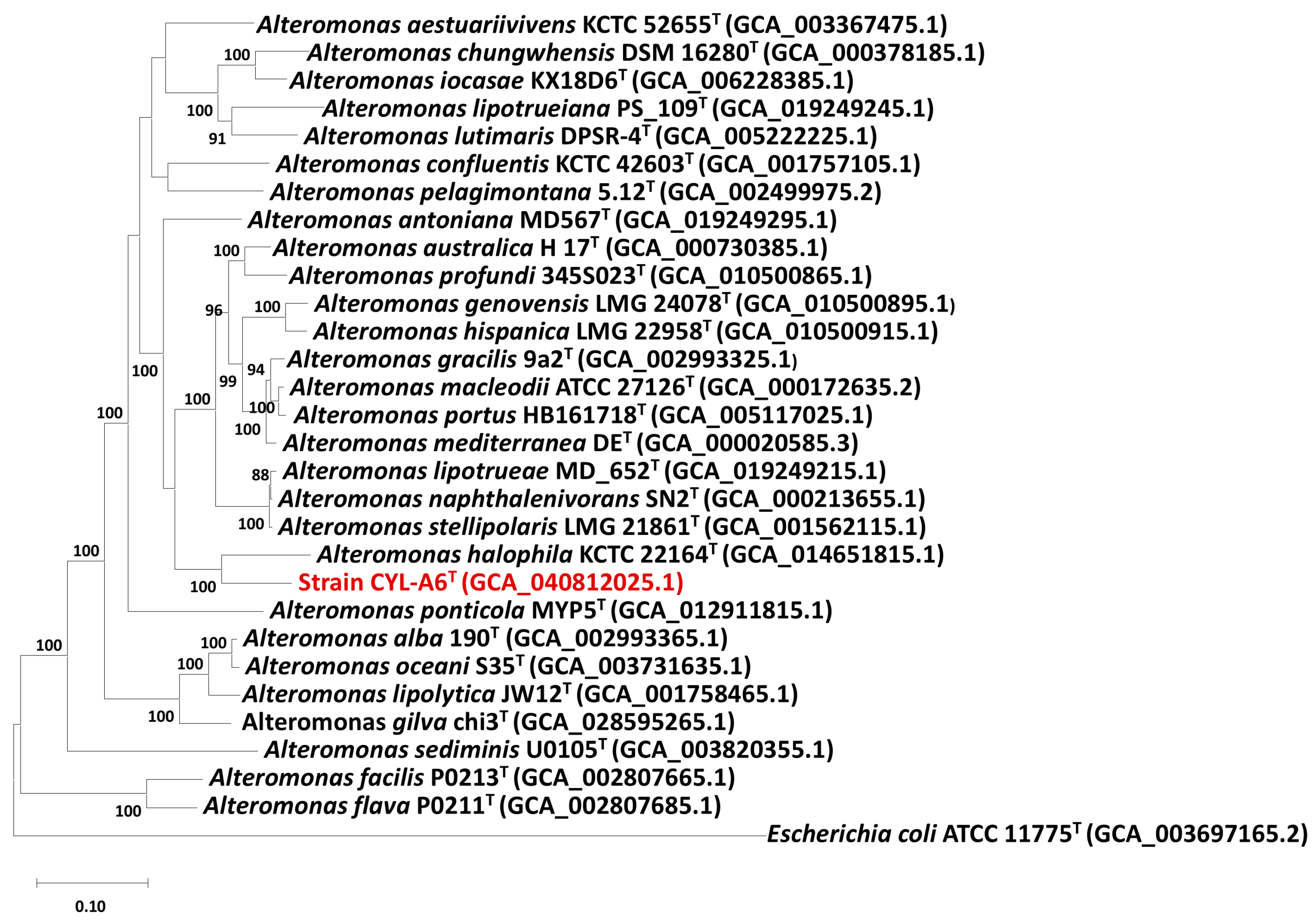
3.2.2. Genomic Comparisons and Phylogenomic Relationship
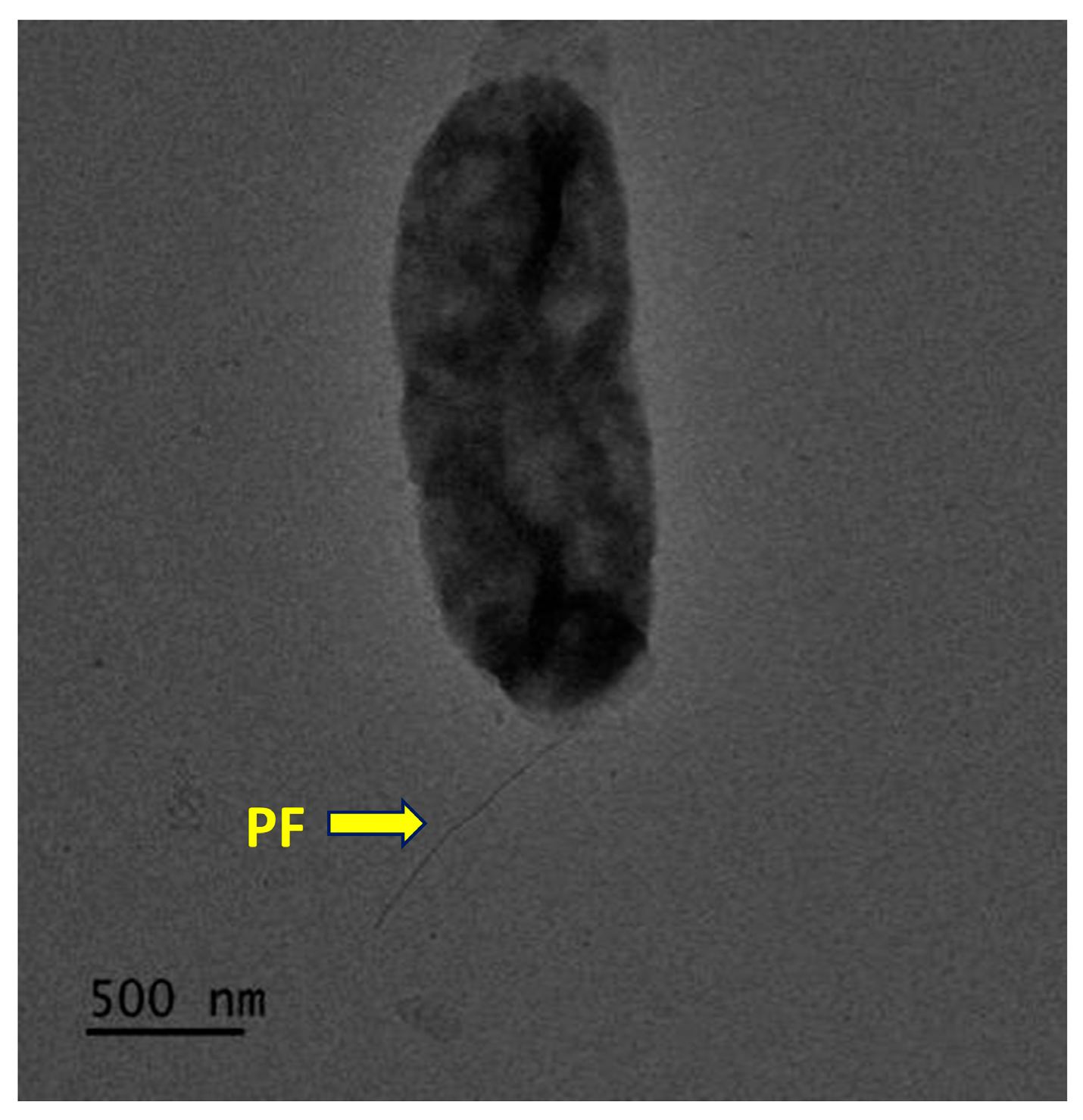
3.3. Phenotypic Characteristics of Strain CYL-A6T
3.4. Evolutionary Trajectory of Nitrate-Reducing Genes in the Genus Alteromonas
4. Discussion
4.1. Proposal of Alteromonas nitratireducens sp. nov.
4.2. Description of Alteromonas nitratireducens sp. nov.
4.3. Loss of Nitrate-Reducing Genes and Gain of Other Metabolism Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANI | Average Nucleotide Identity |
| COG | Clusters of Orthologous Groups of proteins |
| GO | Gene Ontology |
| isDDH | in silico DNA–DNA Hybridization |
| KCTC | Korean Collection of Type Cultures |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MA | Marine Agar 2216 |
| MCCC | Marine Culture Collection of China |
| Q-8 | Ubiquinone-8 |
References
- Mattoo, R.; Suman, B.M. Microbial roles in the terrestrial and aquatic nitrogen cycle—Implications in climate change. FEMS Microbiol. Lett. 2023, 370, fnad061. [Google Scholar] [CrossRef]
- Mermer, A.; Keles, T.; Sirin, Y. Recent studies of nitrogen containing heterocyclic compounds as novel antiviral agents: A review. Bioorg. Chem. 2021, 114, 105076. [Google Scholar] [CrossRef] [PubMed]
- Albright, M.B.N.; Timalsina, B.; Martiny, J.B.H.; Dunbar, J. Comparative Genomics of Nitrogen Cycling Pathways in Bacteria and Archaea. Microb. Ecol. 2019, 77, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, D.A.; Capone, D.G. The marine nitrogen cycle: New developments and global change. Nat. Rev. Microbiol. 2022, 20, 401–414, Erratum in Nat. Rev. Microbiol. 2022, 20, 444. [Google Scholar] [CrossRef]
- Bourceau, O.M.; Ferdelman, T.; Lavik, G.; Mussmann, M.; Kuypers, M.M.M.; Marchant, H.K. Simultaneous sulfate and nitrate reduction in coastal sediments. ISME Commun. 2023, 3, 17. [Google Scholar] [CrossRef]
- Jiang, X.; Jiao, N. Nitrate assimilation by marine heterotrophic bacteria. Sci. China Earth Sci. 2016, 59, 477–483. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, S.; Zhang, D.; Sha, Z. The diversity and metabolism of culturable nitrate-reducing bacteria from the photic zone of the western north Pacific Ocean. Microb. Ecol. 2023, 86, 2781–2789. [Google Scholar] [CrossRef]
- Kaviraj, M.; Kumar, U.; Snigdha, A.; Chatterjee, S. Nitrate reduction to ammonium: A phylogenetic, physiological, and genetic aspects in prokaryotes and eukaryotes. Arch. Microbiol. 2024, 206, 297. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.; Stolz, J.F.; Basu, P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014, 43, 676–706. [Google Scholar] [CrossRef]
- Ren, J.; Tang, J.; Min, H.; Tang, D.; Jiang, R.; Liu, Y.; Huang, X. Nitrogen removal characteristics of novel bacterium Klebsiella sp. TSH15 by assimilatory/dissimilatory nitrate reduction and ammonia assimilation. Bioresour. Technol. 2024, 394, 130184. [Google Scholar] [CrossRef]
- You, X.; Wang, S.; Chen, J. Magnetic biochar accelerates microbial succession and enhances assimilatory nitrate reduction during pig manure composting. Environ. Int. 2024, 184, 108469. [Google Scholar] [CrossRef]
- Yuan, W.; Yang, D.; Zhang, X.; Jiang, C.; Wang, D.; Zuo, J.; Xu, S.; Zhuang, X. Enhanced nitrogen removal of the anaerobic ammonia oxidation process by coupling with an efficient nitrate reducing bacterium (Bacillus velezensis M3-1). J. Environ. Sci. 2024, 146, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Gago, J.F.; Viver, T.; Urdiain, M.; Pastor, S.; Kämpfer, P.; Ferreira, E.; Rossello-Mora, R. Description of three new Alteromonas species Alteromonas antoniana sp. nov., Alteromonas lipotrueae sp. nov. and Alteromonas lipotrueiana sp. nov. isolated from marine environments, and proposal for reclassification of the genus Salinimonas as Alteromonas. Syst. Appl. Microbiol. 2021, 44, 126226. [Google Scholar] [CrossRef]
- Park, S.; Kim, I.; Chhetri, G.; So, Y.; Jung, Y.; Woo, H.; Seo, T. Alteromonas gilva sp. nov. and Erythrobacter fulvus sp. nov., isolated from a tidal mudflat. Int. J. Syst. Evol. Microbiol. 2023, 73, 006032. [Google Scholar] [CrossRef]
- Shen, X.; Zhu, S.; Dong, B.; Chen, Y.; Xue, Z.; Ren, N.; Chen, T.; Chen, X.; Yang, J.; Chen, J. Alteromonas profundi sp. nov., isolated from the Indian Ocean. Int. J. Syst. Evol. Microbiol. 2020, 70, 4531–4536. [Google Scholar] [CrossRef]
- Sinha, R.K.; Krishnan, K.P.; Singh, A.; Thomas, F.A.; Jain, A.; Kurian, P.J. Alteromonas pelagimontana sp. nov., a marine exopolysaccharide-producing bacterium isolated from the Southwest Indian Ridge. Int. J. Syst. Evol. Microbiol. 2017, 67, 4032–4038. [Google Scholar] [CrossRef]
- Sun, C.; Xamxidin, M.; Wu, Y.-H.; Cheng, H.; Wang, C.-S.; Xu, X.-W. Alteromonas alba sp. nov., a marine bacterium isolated from seawater of the West Pacific Ocean. Int. J. Syst. Evol. Microbiol. 2019, 69, 278–284, Erratum in Int. J. Syst. Evol. Microbiol. 2020, 70, 2956. [Google Scholar] [CrossRef]
- Cai, G.; Yu, X.; Wang, H.; Zheng, T.; Azam, F. Nutrient-dependent interactions between a marine copiotroph Alteromonas and a diatom Thalassiosira pseudonana. mBio 2023, 14, e00940-23. [Google Scholar] [CrossRef]
- He, W.; Xue, H.-P.; Liu, C.; Zhang, A.H.; Huang, J.-K.; Zhang, D.-F. Biomineralization of struvite induced by indigenous marine bacteria of the genus Alteromonas. Front. Mar. Sci. 2023, 10, 1085345. [Google Scholar] [CrossRef]
- Henríquez-Castillo, C.; Plominsky, A.M.; Ramírez-Flandes, S.; Bertagnolli, A.D.; Stewart, F.J.; Ulloa, O. Metaomics unveils the contribution of Alteromonas bacteria to carbon cycling in marine oxygen minimum zones. Front. Mar. Sci. 2022, 9, 993667. [Google Scholar] [CrossRef]
- Lu, Z.; Entwistle, E.; Kuhl, M.D.; Durrant, A.R.; Barreto Filho, M.M.; Goswami, A.; Morris, J.J. Coevolution of marine phytoplankton and Alteromonas bacteria in response to pCO2 and coculture. ISME J. 2025, 19, wrae259. [Google Scholar] [CrossRef]
- Manck, L.E.; Park, J.; Tully, B.J.; Poire, A.M.; Bundy, R.M.; Dupont, C.L.; Barbeau, K.A. Petrobactin, a siderophore produced by Alteromonas, mediates community iron acquisition in the global ocean. ISME J. 2022, 16, 358–369. [Google Scholar] [CrossRef]
- Sher, D.; George, E.E.; Wietz, M.; Gifford, S.; Zoccarato, L.; Weissberg, O.; Koedooder, C.; Valiya Kalladi, W.B.; Barreto Filho, M.M.; Mireles, R.; et al. Collaborative metabolic curation of an emerging model marine bacterium, Alteromonas macleodii ATCC 27126. PLoS ONE 2025, 20, e0321141. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; McMeekin, T.A. Alteromonas. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–7. [Google Scholar] [CrossRef]
- Ivanova, E.P.; López-Pérez, M.; Zabalos, M.; Nguyen, S.H.; Webb, H.K.; Ryan, J.; Lagutin, K.; Vyssotski, M.; Crawford, R.J.; Rodriguez-Valera, F. Ecophysiological diversity of a novel member of the genus Alteromonas, and description of Alteromonas mediterranea sp. nov. Antonie Van Leeuwenhoek 2015, 107, 119–132. [Google Scholar] [CrossRef]
- Luo, B.; Su, J.-Y.; Zhang, Y.-F.; Xiao, Y.-H.; Peng, Y.-L.; Sun, M.-L.; Li, Y. Alteromonas arenosi sp. nov., a novel bioflocculant-producing bacterium, isolated from intertidal sand. Antonie Van Leeuwenhoek 2024, 117, 28. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.; Han, J.-R.; Chen, G.-J.; Du, Z.-J. Alteromonas flava sp. nov. and Alteromonas facilis sp. nov., two novel copper tolerating bacteria isolated from a sea cucumber culture pond in China. Syst. Appl. Microbiol. 2019, 42, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-J.; Xu, L.; Wang, N.; Sun, H.-M.; Chen, X.-L.; Zhang, Y.-Z.; Shi, M.; Zhang, X.-Y. Putridiphycobacter roseus gen. nov., sp. nov., isolated from Antarctic rotten seaweed. Int. J. Syst. Evol. Microbiol. 2020, 70, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.-S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 006421. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Shen, W.; Sipos, B.; Zhao, L. SeqKit2: A Swiss army knife for sequence and alignment processing. iMeta 2024, 3, e191. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Royo-Llonch, M.; Sánchez, P.; Ruiz-González, C.; Salazar, G.; Pedrós-Alió, C.; Sebastián, M.; Labadie, K.; Paoli, L.; Ibarbalz, F.M.; Zinger, L.; et al. Compendium of 530 metagenome-assembled bacterial and archaeal genomes from the polar Arctic Ocean. Nat. Microbiol. 2021, 6, 1561–1574. [Google Scholar] [CrossRef]
- Lee, I.; Kim, O.Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory friendly classification with the genome taxonomy database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hua, J.; Ying, J.-J.; Dong, H.; Li, H.; Xamxidin, M.; Han, B.-N.; Sun, C.; Xu, L. Erythrobacter aurantius sp. nov., isolated from intertidal seawater in Taizhou. Int. J. Syst. Evol. Microbiol. 2022, 72, 005616. [Google Scholar] [CrossRef]
- Sun, X.-Y.; Dong, H.; Zhang, Y.; Gao, J.-W.; Zhou, P.; Sun, C.; Xu, L. Isolation and cultivation of carotenoid-producing strains from tidal flat sediment and proposal of Croceibacterium aestuarii sp. nov., a novel carotenoid-producing species in the family Erythrobacteraceae. J. Mar. Sci. Eng. 2024, 12, 99. [Google Scholar] [CrossRef]
- Xu, L.; Huo, Y.-Y.; Li, Z.-Y.; Wang, C.-S.; Oren, A.; Xu, X.-W. Chryseobacterium profundimaris sp. nov., a new member of the family Flavobacteriaceae isolated from deep-sea sediment. Antonie Van Leeuwenhoek 2015, 107, 979–989. [Google Scholar] [CrossRef]
- Minnikin, D.E. Chemical principles in the organization of lipid components in the mycobacterial cell envelope. Res. Microbiol. 1991, 142, 423–427. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome. Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef]
- Csűös, M. Count: Evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 2010, 26, 1910–1912. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Rinke, C.; Chuvochina, M.; Mussig, A.J.; Chaumeil, P.-A.; Davín, A.A.; Waite, D.W.; Whitman, W.B.; Parks, D.H.; Hugenholtz, P. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat. Microbiol. 2021, 6, 946–959. [Google Scholar] [CrossRef]
- Riesco, R.; Trujillo, M.E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2024, 74, 006300. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-G.; Xiao, H.-D.; Tang, S.-K.; Zhang, Y.-Q.; Borrathybay, E.; Cui, X.-L.; Li, W.-J.; Liu, Y.-Q. Alteromonas halophila sp. nov., a new moderately halophilic bacterium isolated from a sea anemone. Antonie Van Leeuwenhoek 2009, 96, 259–266. [Google Scholar] [CrossRef]
- Sevillya, G.; Adato, O.; Snir, S. Detecting horizontal gene transfer: A probabilistic approach. BMC Genom. 2020, 21, 106. [Google Scholar] [CrossRef] [PubMed]
- Chon, N.L.; Schultz, N.J.; Zheng, H.; Lin, H. Anion pathways in the NarK nitrate/nitrite exchanger. J. Chem. Inf. Model. 2023, 63, 5142–5152. [Google Scholar] [CrossRef]
- Durand, S.; Guillier, M. Transcriptional and post-transcriptional control of the nitrate respiration in bacteria. Front. Mol. Biosci. 2021, 8, 667758. [Google Scholar] [CrossRef]
- Fukuda, M.; Takeda, H.; Kato, H.E.; Doki, S.; Ito, K.; Maturana, A.D.; Ishitani, R.; Nureki, O. Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK. Nat. Commun. 2015, 6, 7097. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, I.; Aleksenko, V.A.; Orekhov, P.; Goncharov, I.M.; Nazarenko, V.V.; Semenov, O.; Remeeva, A.; Gordeliy, V. Nitrate- and nitrite-sensing histidine kinases: Function, structure, and natural diversity. Int. J. Mol. Sci. 2021, 22, 5933. [Google Scholar] [CrossRef]
- Hird, K.; Campeciño, J.O.; Hegg, E.L. From genes to function: Regulation, maturation, and evolution of cytochrome c nitrite reductase in nitrate reduction to ammonium. Appl. Environ. Microbiol. 2025, 91, e00292-25. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Weisener, C.G.; Ni, J.; He, B.; Xie, D.; Li, Z. Nitrate assimilation, dissimilatory nitrate reduction to ammonium, and denitrification coexist in Pseudomonas putida Y-9 under aerobic conditions. Bioresour. Technol. 2020, 312, 123597. [Google Scholar] [CrossRef]
- Kraft, B.; Strous, M.; Tegetmeyer, H.E. Microbial nitrate respiration—Genes, enzymes and environmental distribution. J. Biotechnol. 2011, 155, 104–117. [Google Scholar] [CrossRef]
- Kaviraj, M.; Kumar, U.; Chatterjee, S.; Parija, S.; Padbhushan, R.; Nayak, A.K.; Gupta, V.V.S.R. Dissimilatory nitrate reduction to ammonium (DNRA): A unique biogeochemical cycle to improve nitrogen (N) use efficiency and reduce N-loss in rice paddy. Rhizosphere 2024, 30, 100875. [Google Scholar] [CrossRef]
- Li, X.; Sardans, J.; Hou, L.; Gao, D.; Liu, M.; Peñuelas, J. Dissimilatory nitrate/nitrite reduction processes in river sediments across climatic gradient: Influences of biogeochemical controls and climatic temperature regime. J. Geophys. Res Biogeosci. 2019, 124, 2305–2320. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, J.; Shen, G.; Zhou, Y.; Zhang, X.; Zhou, Y.; Yu, Z.; Ma, J. Dissimilatory nitrate reduction to ammonia in the natural environment and wastewater treatment facilities: A comprehensive review. Environ. Technol. Innov. 2025, 37, 104011. [Google Scholar] [CrossRef]
- Iranzo, J.; Wolf, Y.I.; Koonin, E.V.; Sela, I. Gene gain and loss push prokaryotes beyond the homologous recombination barrier and accelerate genome sequence divergence. Nat. Commun. 2019, 10, 5376. [Google Scholar] [CrossRef]
- Berg, G.; Jørgensen, N. Purine and pyrimidine metabolism by estuarine bacteria. Aquat. Microb. Ecol. 2006, 42, 215–226. [Google Scholar] [CrossRef]
- Chen, Q.; Xu, W.; Wu, H.; Guang, C.; Zhang, W.; Mu, W. An overview of D-galactose utilization through microbial fermentation and enzyme-catalyzed conversion. Appl. Microbiol. Biotechnol. 2021, 105, 7161–7170. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zeng, X.; Zheng, J.; Zou, Y.; Qiu, S.; Dai, Y. AHL-mediated quorum sensing to regulate bacterial substance and energy metabolism: A review. Microbiol. Res. 2022, 262, 127102. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Villadsen, J. Oxidative phosphorylation revisited. Biotechnol. Bioeng. 2015, 112, 429–437. [Google Scholar] [CrossRef]
- Schink, S.J.; Christodoulou, D.; Mukherjee, A.; Athaide, E.; Brunner, V.; Fuhrer, T.; Bradshaw, G.A.; Sauer, U.; Basan, M. Glycolysis/gluconeogenesis specialization in microbes is driven by biochemical constraints of flux sensing. Mol. Syst. Biol. 2022, 18, e10704. [Google Scholar] [CrossRef]
- Thi Quynh Le, H.; Lee, E.Y. Methanotrophs: Metabolic versatility from utilization of methane to multi-carbon sources and perspectives on current and future applications. Bioresour. Technol. 2023, 384, 129296. [Google Scholar] [CrossRef]
- Xu, X.; Li, C.; Cao, W.; Yan, L.; Cao, L.; Han, Q.; Gao, M.; Chen, Y.; Shen, Z.; Jiang, J.; et al. Bacterial growth and environmental adaptation via thiamine biosynthesis and thiamine-mediated metabolic interactions. ISME J. 2024, 18, wrae157. [Google Scholar] [CrossRef]
- Yuan, W.; Du, Y.; Yu, K.; Xu, S.; Liu, M.; Wang, S.; Yang, Y.; Zhang, Y.; Sun, J. The production of pyruvate in biological technology: A critical review. Microorganisms 2022, 10, 2454. [Google Scholar] [CrossRef] [PubMed]


| Type Strain | NCBI Assembly Accession Number | No. of Contigs | Size (Mbp) | GC Content (%) |
|---|---|---|---|---|
| A. aestuariivivens KCTC 52655T | GCA_003367475.1 | 35 | 3.85 | 50.5 |
| A. alba 190T | GCA_002993365.1 | 221 | 5.16 | 48.7 |
| A. antoniana MD_567T | GCA_019249295.1 | 90 | 4.95 | 48.9 |
| A. australica H 17T | GCA_000730385.1 | 1 | 4.31 | 44.9 |
| A. chungwhensis DSM 16280T | GCA_000378185.1 | 30 | 3.98 | 47.0 |
| A. confluentis KCTC 42603T | GCA_001757105.1 | 46 | 4.86 | 48.0 |
| A. facilis P0213T | GCA_002807665.1 | 23 | 3.91 | 44.7 |
| A. flava P0211T | GCA_002807685.1 | 42 | 3.65 | 45.7 |
| A. genovensis LMG 24078T | GCA_010500895.1 | 37 | 4.07 | 44.3 |
| A. gilva chi3T | GCA_028595265.1 | 7 | 4.57 | 48.4 |
| A. gracilis 9a2T | GCA_002993325.1 | 31 | 4.32 | 44.3 |
| A. halophila KCTC 22164T | GCA_014651815.1 | 36 | 3.95 | 50.8 |
| A. hispanica LMG 22958T | GCA_010500915.1 | 31 | 4.07 | 43.8 |
| A. iocasae KX18D6T | GCA_006228385.1 | 2 | 4.16 | 47.3 |
| A. lipolytica JW12T | GCA_001758465.1 | 21 | 5.02 | 48.4 |
| A. lipotrueae MD_652T | GCA_019249215.1 | 44 | 4.75 | 43.7 |
| A. lipotrueiana PS_109T | GCA_019249245.1 | 47 | 3.76 | 45.8 |
| A. lutimaris DPSR-4T | GCA_005222225.1 | 1 | 4.12 | 49.8 |
| A. macleodii ATCC 27126T | GCA_000172635.2 | 1 | 4.65 | 44.7 |
| A. mediterranea DET | GCA_000020585.3 | 1 | 4.48 | 44.9 |
| A. naphthalenivorans SN2T | GCA_000213655.1 | 1 | 4.97 | 43.5 |
| A. oceani S35T | GCA_003731635.1 | 90 | 4.99 | 48.7 |
| A. pelagimontana 5.12T | GCA_002499975.2 | 1 | 4.31 | 46.1 |
| A. ponticola MYP5T | GCA_012911815.1 | 22 | 3.60 | 46.1 |
| A. portus HB161718T | GCA_005117025.1 | 32 | 4.54 | 44.1 |
| A. profundi 345S023T | GCA_010500865.1 | 99 | 4.39 | 44.4 |
| A. sediminis U0105T | GCA_003820355.1 | 14 | 3.96 | 45.3 |
| A. stellipolaris LMG 21861T | GCA_001562115.1 | 2 | 4.90 | 43.5 |
| Type Strain | ANI (%) | isDDH (%) |
|---|---|---|
| A. aestuariivivens KCTC 52655T | 72.0 | 19.7 |
| A. alba 190T | 71.6 | 20.7 |
| A. antoniana MD_567T | 72.7 | 19.3 |
| A. australica H 17T | 71.0 | 19.4 |
| A. chungwhensis DSM 16280T | 70.5 | 18.9 |
| A. confluentis KCTC 42603T | 71.2 | 20.4 |
| A. facilis P0213T | 69.8 | 23.8 |
| A. flava P0211T | 69.0 | 20.5 |
| A. genovensis LMG 24078T | 70.8 | 19.9 |
| A. gilva chi3T | 70.5 | 20.1 |
| A. gracilis 9a2T | 71.2 | 20.6 |
| A. halophila KCTC 22164T | 73.7 | 19.0 |
| A. hispanica LMG 22958T | 70.7 | 19.8 |
| A. iocasae KX18D6T | 70.9 | 19.6 |
| A. lipolytica JW12T | 71.0 | 20.2 |
| A. lipotrueae MD_652T | 70.5 | 19.0 |
| A. lipotrueiana PS_109T | 70.6 | 18.8 |
| A. lutimaris DPSR-4T | 71.6 | 19.5 |
| A. macleodii ATCC 27126T | 71.4 | 20.8 |
| A. mediterranea DET | 71.8 | 21.7 |
| A. naphthalenivorans SN2T | 71.0 | 19.5 |
| A. oceani S35T | 70.9 | 19.9 |
| A. pelagimontana 5.12T | 70.9 | 19.2 |
| A. ponticola MYP5T | 70.7 | 19.0 |
| A. portus HB161718T | 71.5 | 20.3 |
| A. profundi 345S023T | 70.5 | 19.5 |
| A. sediminis U0105T | 69.0 | 20.7 |
| A. stellipolaris LMG 21861T | 70.7 | 19.3 |
| Characteristic | Strain CYL-A6T | A. halophila KCTC 22164T |
|---|---|---|
| Temperature for growth (°C): | ||
| Range | 20–45 | 10–40 * |
| Optimum | 37 | 25–30 * |
| NaCl for growth (w/v, %): | ||
| Range | 1.0–12.0 | 1.0–20.0 * |
| Optimum | 3.5 | 5.0–10.0 * |
| pH for growth: | ||
| Range | 5.5–9.0 | 6.0–10.0 * |
| Optimum | 7.0 | 7.5 * |
| Hydrolysis of | ||
| Tween 40 | − | + |
| API ZYM: | ||
| α- and β-Galactosidase | − | + |
| β-Glucosidase | − | + |
| API 20NE: | ||
| Hydrolysis of urea | + | − |
| β-Galactosidase | − | + |
| Nitrate reduction | + | − |
| Utilization of: | ||
| D-Mannitol | − | + |
| Acid production from: | ||
| D-Glucose and D-mannose | + | − |
| DNA G+C content (%) | 51.8 | 50.8 |
| Fatty Acids | Strain CYL-A6T | A. halophila KCTC 22164T |
|---|---|---|
| Straight-chain | ||
| C12:0 | 2.0 | 3.5 |
| C14:0 | 3.9 | 3.6 |
| C16:0 | 22.6 | 23.7 |
| C17:0 | 5.8 | 2.2 |
| C18:0 | 2.1 | 1.2 |
| Branched-chain | ||
| iso-C14:0 | 0.6 | 0.2 |
| iso-C16:0 | 1.3 | 0.6 |
| iso-C18:0 | 0.6 | 0.2 |
| Unsaturated | ||
| C15:1ω8c | 1.6 | 0.5 |
| C17:1ω8c | 6.6 | 2.9 |
| Hydroxy | ||
| C10:0 3-OH | 2.1 | 2.2 |
| C11:0 3-OH | 1.6 | 0.5 |
| C12:0 3-OH | 0.9 | 0.7 |
| C12:1 3-OH | 1.4 | 2.6 |
| Summed feature 2 | 3.7 | 3.2 |
| Summed feature 3 | 20.3 | 28.0 |
| Summed feature 7 | 1.6 | 1.3 |
| Summed feature 8 | 12.4 | 17.6 |
| OCs | KO Number | Annotation | COG Category |
|---|---|---|---|
| OG0000020 | K16264 | czcD; cobalt-zinc-cadmium efflux system protein | P |
| OG0000050 | K07814 | cyclic di-GMP phosphodiesterase | T |
| OG0000059 | K01271 | pepQ; Xaa-Pro dipeptidase | E |
| OG0000079 | K06222 | dkgB; 2,5-Diketo-D-gluconate reductase B | S |
| OG0000175 | K02083 | allC; allantoate deiminase | E |
| OG0000214 | K03307 | TC.SSS; solute:Na+ symporter, SSS family | S |
| OG0000263 | K03406 | mcp; methyl-accepting chemotaxis protein | T |
| OG0000549 | K07089 | Uncharacterized protein | S |
| OG0000752 | K02030 | ABC.PA.S; polar amino acid transport system substrate-binding protein | ET |
| OG0001437 | K03406 | mcp; methyl-accepting chemotaxis protein | NT |
| OG0001606 | K04065 | osmY; hyperosmotically inducible periplasmic protein | S |
| OG0001699 | K02429 | fucP; MFS transporter, FHS family, L-fucose permease | G |
| OG0001868 | K03885 | ndh; NADH:quinone reductase (non-electrogenic) | C |
| OG0001942 | K02014 | TC.FEV.OM; iron complex outermembrane recepter protein | P |
| OG0002072 | K01785 | galM; aldose 1-epimerase | G |
| OG0002073 | K09781 | Uncharacterized protein | S |
| OG0002091 | K02426 | sufE; cysteine desulfuration protein | S |
| OG0002103 | K13924 | cheBR; two-component system, chemotaxis family, CheB/CheR fusion protein | T |
| OG0002106 | K03314 | nhaB; Na+:H+ antiporter, NhaB family | P |
| OG0002119 | K03409 | cheX; chemotaxis protein CheX | N |
| OG0002126 | K03585 | mexA; membrane fusion protein, multidrug efflux system | M |
| OG0002127 | K18288 | ict-Y; itaconate CoA-transferase | C |
| OG0002139 | K09954 | Uncharacterized protein | S |
| OG0002142 | K00569 | tpmT; thiopurine S-methyltransferase | Q |
| OG0002165 | K08234 | yaeR; glyoxylase I family protein | E |
| OG0002170 | K06886 | glbN; hemoglobin | S |
| OG0002173 | K02529 | galR; LacI family transcriptional regulator, galactose operon repressor | K |
| OG0002185 | K01487 | guaD; guanine deaminase | F |
| OG0002186 | K13482 | xdhB; xanthine dehydrogenase large subunit | F |
| OG0002187 | K06901 | adeQ; adenine/guanine/hypoxanthine permease | S |
| OG0002188 | K20920 | vpsM; polysaccharide biosynthesis protein VpsM | S |
| OG0002207 | K06200 | cstA; carbon starvation protein | T |
| OG0002216 | K04761 | oxyR; LysR family transcriptional regulator, hydrogen peroxide-inducible genes activator | K |
| OG0002218 | K13481 | xdhA; xanthine dehydrogenase small subunit | F |
| OG0002223 | K01090 | Protein phosphatase | IT |
| OG0002228 | K09897 | Uncharacterized protein | S |
| OG0002235 | K03969 | pspA; phage shock protein A | KT |
| OG0002238 | K20444 | rfbC; O-antigen biosynthesis protein | M |
| OG0002267 | K09929 | Uncharacterized protein | S |
| OG0002277 | K03088 | rpoE; RNA polymerase sigma-70 factor, ECF subfamily | K |
| OG0002278 | K07107 | ybgC; acyl-CoA thioester hydrolase | S |
| OG0002284 | K08990 | ycjF; putative membrane protein | S |
| OG0002285 | K06889 | Uncharacterized protein | S |
| OG0002291 | K09689 | kpsT; capsular polysaccharide transport system ATP-binding protein | GM |
| OG0002299 | K16840 | hpxQ; 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase | S |
| OG0002300 | K00681 | ggt; gamma-glutamyltranspeptidase/glutathione hydrolase | E |
| OG0002319 | K16088 | fhuE; outer-membrane receptor for ferric coprogen and ferric-Rhodotorulic acid | P |
| OG0002320 | K06918 | Uncharacterized protein | S |
| OG0002323 | K01834 | gpmA; 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | G |
| OG0002329 | K07127 | hiuH; 5-hydroxyisourate hydrolase | S |
| OG0002330 | K07402 | xdhC; xanthine dehydrogenase accessory factor | O |
| OG0002334 | K11811 | arsH; arsenical resistance protein ArsH | S |
| OG0002340 | K14153 | thiDE; hydroxymethylpyrimidine kinase/phosphomethylpyrimidine kinase/thiamine-phosphate diphosphorylase | H |
| OG0002351 | K00262 | gdhA; glutamate dehydrogenase (NADP+) | E |
| OG0002363 | K02055 | ABC.SP.S; putative spermidine/putrescine transport system substrate-binding protein | E |
| OG0002369 | K00362 | nirB; nitrite reductase (NADH) large subunit | C |
| OG0002370 | K00372 | nasA; assimilatory nitrate reductase catalytic subunit | C |
| OG0002371 | K07023 | YGK1; 5′-deoxynucleotidase | S |
| OG0002373 | K10107 | kpsE; capsular polysaccharide transport system permease protein | M |
| OG0002374 | K12990 | rfbF; rhamnosyltransferase | S |
| OG0002393 | K03669 | mdoH; membrane glycosyltransferase | M |
| OG0002394 | K03670 | mdoG; periplasmic glucans biosynthesis protein | P |
| OG0002397 | K03149 | thiG; thiazole synthase | H |
| OG0002403 | K09688 | kpsM; capsular polysaccharide transport system permease protein | GM |
| OG0002412 | K06149 | uspA; universal stress protein A | T |
| OG0002427 | K00363 | nirD; nitrite reductase (NADH) small subunit | P |
| OG0002430 | K05782 | benE; benzoate membrane transport protein | Q |
| OG0002442 | K01759 | gloA; lactoylglutathione lyase | E |
| OG0002448 | K00847 | scrK; fructokinase | G |
| OG0002483 | K02303 | cobA; uroporphyrin-III C-methyltransferase | H |
| OG0002496 | K00344 | qor; NADPH:quinone reductase | C |
| OG0002515 | K03793 | PTR1; pteridine reductase | IQ |
| OG0002516 | K01104 | Protein-tyrosine phosphatase | GM |
| OG0002523 | K09797 | Uncharacterized protein | S |
| OG0002534 | K07238 | zupT; zinc transporter, ZIP family | P |
| OG0002543 | K01425 | glsA; glutaminase | E |
| OG0002555 | K15977 | Putative oxidoreductase | S |
| OG0002594 | K01083 | 3-Phytase | I |
| OG0002688 | K16044 | iolW; scyllo-inositol 2-dehydrogenase (NADP+) | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-L.; Li, J.-X.; Wang, X.-C.; Li, Y.; Cao, Y.-F.; Duan, X.-W.; Sun, C.; Chen, C.; Xu, L. Alteromonas nitratireducens sp. nov., a Novel Nitrate-Reducing Bacterium Isolated from Marine Sediments, and the Evolution of Nitrate-Reducing Genes in the Genus Alteromonas. Microorganisms 2025, 13, 1888. https://doi.org/10.3390/microorganisms13081888
Chang Y-L, Li J-X, Wang X-C, Li Y, Cao Y-F, Duan X-W, Sun C, Chen C, Xu L. Alteromonas nitratireducens sp. nov., a Novel Nitrate-Reducing Bacterium Isolated from Marine Sediments, and the Evolution of Nitrate-Reducing Genes in the Genus Alteromonas. Microorganisms. 2025; 13(8):1888. https://doi.org/10.3390/microorganisms13081888
Chicago/Turabian StyleChang, Ying-Li, Jia-Xi Li, Xing-Chen Wang, Yang Li, Yun-Fei Cao, Xiang-Wen Duan, Cong Sun, Can Chen, and Lin Xu. 2025. "Alteromonas nitratireducens sp. nov., a Novel Nitrate-Reducing Bacterium Isolated from Marine Sediments, and the Evolution of Nitrate-Reducing Genes in the Genus Alteromonas" Microorganisms 13, no. 8: 1888. https://doi.org/10.3390/microorganisms13081888
APA StyleChang, Y.-L., Li, J.-X., Wang, X.-C., Li, Y., Cao, Y.-F., Duan, X.-W., Sun, C., Chen, C., & Xu, L. (2025). Alteromonas nitratireducens sp. nov., a Novel Nitrate-Reducing Bacterium Isolated from Marine Sediments, and the Evolution of Nitrate-Reducing Genes in the Genus Alteromonas. Microorganisms, 13(8), 1888. https://doi.org/10.3390/microorganisms13081888







