Molecular Surveillance of ESBL and Carbapenemase Genes in Gram-Negative Bacterial Pathogens Isolated from Various Clinical Samples Collected from Northern Region of United Arab Emirates
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Design
2.3. Data Collection and Clinical Sample Processing
2.4. Study Populations
2.5. Bacterial Isolation and Staining
2.6. Bacterial Identification and Antibiotic Sensitivity Test
2.7. Detection of ESBL and Carbapenem-Resistant Genes
2.8. Partial Genomic Sequencing of blaCTX-M
2.9. Statistical Analysis
3. Results
3.1. Isolation of Bacterial Pathogens
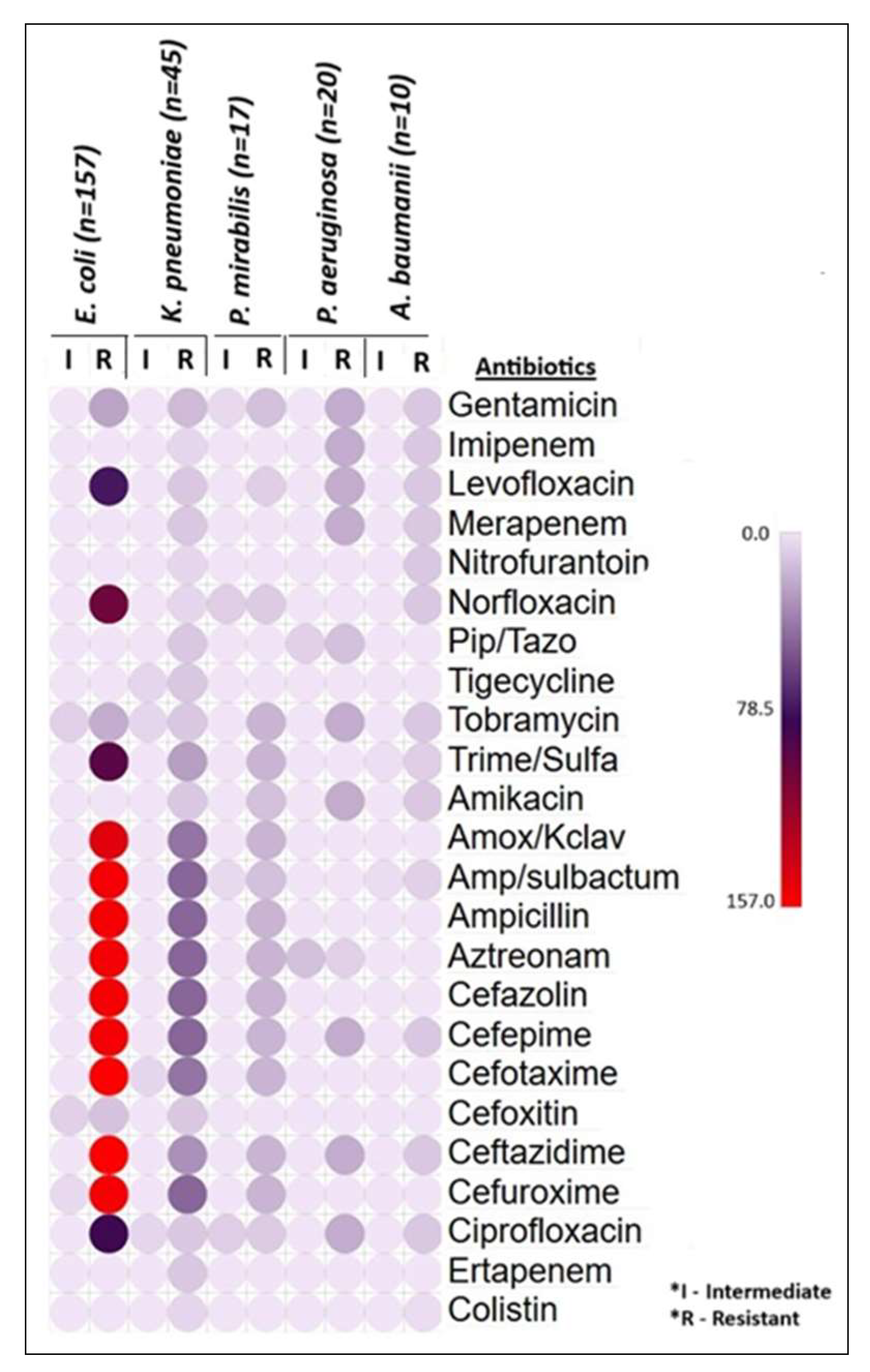
3.2. Bacterial Identification and Antibiotic Susceptibility Test
3.3. Unveiling the ESBL and Carbapenem-Resistant Genes in Gram-Negative Bacterial Pathogens
3.4. Genomic Sequencing and Phylogenetic Tree Relationships of the blaCTX-M
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| MDR | Multidrug resistant |
| ESBL | Extended Spectrum of Beta-lactamase |
| GMU | Gulf Medical University |
| TUH | Thumbay University Hospital |
| GCC | Gulf Cooperation Council |
| UAE | United Arab Emirates |
References
- Aljeldah, M.M. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics 2022, 11, 1082. [Google Scholar] [CrossRef]
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 26, 1340. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control 2017, 12, 1–24. [Google Scholar]
- ElAila, N.A.; Al-Laham, N.A.; Ayesh, B.M. Prevalence of extended spectrum beta lactamase and molecular detection of blaTEM, blaSHV and blaCTX-M genotypes among Gram negative bacilli isolates from pediatric patient population in Gaza strip. BMC Infect. Dis. 2023, 23, 99. [Google Scholar]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 2020, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, A.A.; Abdelaziz, N.A.; Amin, M.A.; Aziz, R.K. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci. Rep. 2019, 9, 4224. [Google Scholar] [CrossRef]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef]
- Mahony, M.; McMullan, B.; Brown, J.; Kennedy, S.E. Multidrug-resistant organisms in urinary tract infections in children. Pediatr. Nephrol. 2020, 35, 1563–1573. [Google Scholar] [CrossRef]
- Tsilipounidaki, K.; Athanasakopoulou, Z.; Müller, E.; Burgold-Voigt, S.; Florou, Z.; Braun, S.D.; Monecke, S.; Gatselis, N.K.; Zachou, K.; Stefos, A.; et al. Plethora of Resistance Genes in Carbapenem-Resistant Gram-Negative Bacteria in Greece: No End to a Continuous Genetic Evolution. Microorganisms 2022, 10, 159. [Google Scholar] [CrossRef]
- Cantón, R.; Akóva, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Tufa, T.B.; Mackenzie, C.R.; Orth, H.M.; Wienemann, T.; Nordmann, T.; Abdissa, S.; Hurissa, Z.; Schönfeld, A.; Bosselmann, M.; Häussinger, D.; et al. Prevalence and characterization of antimicrobial resistance among gram-negative bacteria isolated from febrile hospitalized patients in central Ethiopia. Antimicrob. Resist. Infect. Control 2022, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.E.; Algak, T.B.; Abbas, M.; Elamin, B.K. Emergence of bla TEM, bla CTX-M, bla SHV and bla OXA genes in multidrug-resistant Enterobacteriaceae and Acinetobacter baumannii in Saudi Arabia. Exp. Ther. Med. 2021, 22, 1450. [Google Scholar] [CrossRef]
- Moghnieh, R.A.; Kanafani, Z.A.; Tabaja, H.Z.; Sharara, S.L.; Awad, L.S.; Kanj, S.S. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. Lancet Infect. Dis. 2018, 18, e379–e394. [Google Scholar] [CrossRef]
- Al-Musawi, T.; Al-Agha, R.; Al-Khiami, S.; Al-Shamari, H.; Baghdadi, M.; Bosaeed, M.; Hadi, H.A.; Mady, A.; Sabra, N. Bacteremia in the Gulf Cooperation Council Region: A Review of the Literature 2013–2023. Infect. Drug Resist. 2025, 18, 2329–2355. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, J.; Najiba, M.; Abdulrazzaq, N.M.; AlRand, H. The UAE AMR Surveillance Consortium. Surveillance of antimicrobial resistance in the United Arab Emirates: The early implementation phase. Front. Public Health 2023, 11, 1247627. [Google Scholar] [CrossRef]
- Kebede, B.; Yihunie, W.; Abebe, D.; Tegegne, B.A.; Belayneh, A. Gram-negative bacteria isolates and their antibiotic-resistance patterns among pediatrics patients in Ethiopia: A systematic review. SAGE Open Med. 2022, 10, 20503121221094191. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, U.; Ponnaluri, S.; Villareal, L.; Gillespie, B.; Wen, A.; Miles, A.; Bucholz, B.; Marrs, C.F.; Iyer, R.K.; Misra, D.; et al. Gram Stains: A Resource for Retrospective Analysis of Bacterial Pathogens in Clinical Studies. PLoS ONE 2012, 7, e42898. [Google Scholar] [CrossRef]
- Beckman Coulter. MicroScan Neg Breakpoint Combo 50. Beckman Coulter, Inc. Brea, California. Available online: https://www.beckmancoulter.com/search#q=MicroScan%20Neg%20Breakpoint%20Combo%2050%20panel%20for%20Gram-negative%20organism&t=techdocs&sort=relevancy&numberOfResults=25&f:@td_parenttaxonomy=[Microbiology] (accessed on 16 October 2021).
- CLSI M100; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2020. Available online: https://clsi.org/shop/standards/m100/ (accessed on 16 October 2021).
- Beckman Coulter. DxM MicroScan WalkAway ID/AST System. Beckman Coulter, Inc. Brea, California. Available online: https://www.beckmancoulter.com/en/products/microbiology/dxm-microscan-walkaway-system?id=B1018-496 (accessed on 16 October 2021).
- Beckman Coulter. Beckman Coulter MicroScan LabPro Software WalkAway 9631049. Information Management Software LabPro Connect. Beckman Coulter, Inc. Brea, California. Available online: https://www.beckmancoulter.com/search#q=Beckman%20Coulter%20MicroScan%20LabPro%20software%20WalkAway%209631049%20&t=products&sort=relevancy&numberOfResults=25&f:@td_parenttaxonomy=[Microbiology] (accessed on 16 October 2021).
- G-Spin. Genomic DNA Extraction Kit for Bacteria. Available online: https://intronbio.com:6001/intronbioen/product/product_view.php?PRDT_ID=9 (accessed on 1 November 2021).
- Mahalingam, N.; Manivannan, B.; Khamari, B.; Siddaramappa, S.; Adak, S.; Bulagonda, E.P. Detection of Antibiotic Resistance Determinants and Their Transmissibility among Clinically Isolated Carbapenem-Resistant Escherichia coli from South India. Med. Princ. Pract. 2018, 8, 428–435. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Aljallal, A.; Radwan, H.H.; Shibl, A.M. Characterization of carbapenemases, ESBLs, and plasmid-mediated quinolone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J. Infect. Pub. Health 2018, 11, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Sonnevend, Á.; Ghazawi, A.A.; Hashmey, R.; Jamal, W.; Rotimi, V.O.; Shibl, A.M.; Al-Jardani, A.; Al-Abri, S.S.; Tariq, W.U.Z.; Weber, S.; et al. Characterization of carbapenem-resistant Enterobacteriaceae with high rate of autochthonous transmission in the Arabian Peninsula. PLoS ONE 2015, 10, e0131372. [Google Scholar] [CrossRef]
- Badger-Emeka, L.I.; Kausar, N.; Estrella, E.; Angeles, G.B. A Three-Year Look at the Phylogenetic Profile, Antimicrobial Resistance, and Associated Virulence Genes of Uropathogenic Escherichia coli. Pathogens 2022, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, J. National AMR Surveillance Report. United Arab Emirates Ministry of Health and Prevention. 2022. Available online: https://mohap.gov.ae/en/w/national-amr-surveillance-report-2022 (accessed on 2 December 2022).
- Ardati, K.O.; Murdeshwar, S.R.; Chacko, S.T.; Jagtap, A.; Jacob, S. Prevalence of Bacterial Pathogens and Antimicrobial Susceptibility Pattern in Bahrain Tertiary Care Hospital. J. Bahrain Med. Soc. 2019, 31, 5–16. [Google Scholar]
- Kabrah, A. Extended-Spectrum Beta-Lactamase and Carbapenem-Resistant Gram-Negative Pathogens in Makkah, Saudi Arabia. Ethiop. J. Health Sci. 2022, 36, 1221–1230. [Google Scholar]
- Shaaban, O.A.; Mahmoud, N.A.; Zeidan, A.A.; Kumar, N.; Finan, A.C. Prevalence and Resistance Patterns of Pediatric Urinary Tract Infections in Bahrain. Cureus 2021, 3, e20859. [Google Scholar] [CrossRef]
- Perez-Lopez, A.; Sundararaju, S.; Al-Mana, H.; Tsui, K.M.; Hasan, M.R.; Suleiman, M.; Janahi, M.; Al Maslamani, E.; Tang, P. Molecular Characterization of Extended-Spectrum β-Lactamase–Producing Escherichia coli and Klebsiella pneumoniae Among the Pediatric Population in Qatar. Front. Microbiol. 2020, 11, 581711. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Rabaan, A.A.; Saunar, J.V.; Bazzi, A.M. Antimicrobial resistance of gram-negative bacteria: A six-year longitudinal study in a hospital in Saudi Arabia. J. Infect. Public Health 2020, 3, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Brune, J.E.; Dickenmann, M.; Sidler, D.; Walti, L.N.; Golshayan, D.; Manuel, O.; Haidar, F.; Neofytos, D.; Schnyder, A.; Boggian, K.; et al. Frequency and impact on renal transplant outcomes of urinary tract infections due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species. Front. Med. 2024, 11, 1329778. [Google Scholar] [CrossRef]
- Alfaresi, M.; Sing, G.K.; Senok, A. First Report of blaCTX-M-28 in Enterobacteriaceae Isolates in the United Arab Emirates. J. Pathog. 2018, 2018, 1304793. [Google Scholar] [CrossRef]
- Dash, N.R.; Albataineh, M.T.; Alhourani, N.; Khoudeir, A.M.; Ghanim, M.; Wasim, M.; Mahmoud, I. Community-acquired urinary tract infections due to extended-spectrum β-lactamase-producing organisms in United Arab Emirates. Travel Med. Infect. Dis. 2018, 22, 46–50. [Google Scholar] [CrossRef]
- Azim, N.S.A.; Nofal, M.Y.; Nofal, M.Y.; Al-Harbi, M.A.; Al-Zaban, M.I.; Somily, A.M. Molecular-diversity, Prevalence and Antibiotic Susceptibility of Pathogenic Klebsiella Pneumoniae under Saudi Condition. Pak. J. Biol. Sci. 2019, 22, 174–179. [Google Scholar] [CrossRef]
- Al-Sweih, N.; Jamal, W.; Mokaddas, E.; Habashy, N.; Kurdi, A.; Mohamed, N. Evaluation of the in vitro activity of ceftaroline, ceftazidime/avibactam and comparator antimicrobial agents against clinical isolates from paediatric patients in Kuwait: ATLAS data 2012–19. JAC Antimicrob. Resist. 2021, 3, dlab159. [Google Scholar] [CrossRef] [PubMed]
- Brek, T.; Alghamdi, A.K.; Abujamel, T.S.; Yasir, M.; Alattas, E.M.; Hazazi, M.S.; Al-Zahrani, I.A. Prevalence and molecular determinants of carbapenemase-producing Klebsiella pneumoniae isolated from Jazan, Saudi Arabia. J. Infect. Dev. Ctries. 2023, 17, 1420–1429. [Google Scholar] [CrossRef]
- Hafiz, T.A.; Alghamdi, G.S.; Alkudmani, Z.S.; Alyami, A.S.; AlMazyed, A.; Alhumaidan, O.S.; Mubaraki, M.A.; Alotaibi, F.E. Multidrug-Resistant Proteus mirabilis Infections and Clinical Outcome at Tertiary Hospital in Riyadh, Saudi Arabia. Infect. Drug Resist. 2024, 17, 571–581. [Google Scholar] [CrossRef]
- Omer, T.H.S.; Mustafa, S.A.M.; Mohamed, S.O.O. Extended Spectrum β-Lactamase-Mediated Resistance and Antibiogram of Pseudomonas aeruginosa Isolates from Patients Attending Two Public Hospitals in Khartoum, Sudan. Int. J. Microb. 2020, 2020, 2313504. [Google Scholar] [CrossRef] [PubMed]
- Elhariri, M.; Hamza, D.; Elhelw, R.; Dorgham, S.M. Extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa in camel in Egypt: Potential human hazard. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 21. [Google Scholar] [CrossRef]
- Sannathimmappa, M.B.; Nambiar, V.; Aravindakshan, R. Antibiotic Resistance Pattern of Acinetobacter baumannii Strains: A Retrospective Study from Oman. Saudi. J. Med. Med. Sci. 2021, 9, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Basatian-Tashkan, B.; Niakan, M.; Khaledi, M.; Afkhami, H.; Sameni, F.; Bakhti, S.; Mirnejad, R. Antibiotic resistance assessment of Acinetobacter baumannii isolates from Tehran hospitals due to the presence of efflux pumps encoding genes adeA and adeS genes by molecular method. BMC Res. Notes 2020, 13, 543. [Google Scholar] [CrossRef]
- Bostanghadiri, N.; Narimisa, N.; Mirshekar, M.; Dadgar-Zankbar, L.; Taki, E.; Navidifar, T.; Darban-Sarokhalil, D. Prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2024, 13, 24. [Google Scholar] [CrossRef]
- Khuntayaporn, P.; Thirapanmethee, K.; Traidej Chomnawang, M.T. An Update of Mobile Colistin Resistance in Non-Fermentative Gram-Negative Bacilli. Front. Cell Infect. Microbiol. 2022, 12, 1–15. [Google Scholar] [CrossRef]
- Pishtiwan, A.H.; Khadija, K.M. Prevalence of blaTEM, blaSHV and blaCTX-M Genes among ESBL-Producing Klebsiella pneumoniae and Escherichia coli Isolated from Thalassemia Patients in Erbil, Iraq. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019041. [Google Scholar]
- Bajpai, T.; Pandey, M.; Varma, M.; Bhatambare, G.S. Prevalence of TEM, SHV, and CTX-M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna J. Med. 2017, 7, 12–16. [Google Scholar] [CrossRef]
- Zaki, M.; El-Halaby, H.; Elmansoury, E.; Zeid, M.; Khaled, K.; Nomir, M. Genetic Study of Extended Spectrum Beta-Lactamase and Carbapenemase Producing Escherichia coli Causing Sepsis among Egyptian Children. Open Microbiol. J. 2019, 13, 128–137. [Google Scholar] [CrossRef]
- Al-Rashed, N.; Bindayna, K.M.; Shahid, M.; Saeed, N.K.; Darwish, A.; Joji, R.M.; Al-Mahmeed, A. Prevalence of Carbapenemases in Carbapenem-Resistant Acinetobacter baumannii Isolates from the Kingdom of Bahrain. Antibiotics 2023, 12, 1198. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Reuter, S.; Wille, J.; Xanthopoulou, K.; Stefanik, D.; Grundmann, H.; Higgins, P.G.; Seifert, H. A global view on carbapenem-resistant Acinetobacter baumannii. mBio 2023, 14, e0226023. [Google Scholar] [CrossRef] [PubMed]
- Belouad, E.M.; Benaissa, E.; El-Mrimar, N.; Bssaibis, F.; Maleb, A.; Elouennass, M. Predominance of OXA-48 Carbapenemase-Producing Enterobacterales in a Moroccan Hospital. Int. J. Microbiol. 2023, 2023, 8581883. [Google Scholar] [CrossRef]
- Moglad, E.; Alanazi, N.; Altayb, H.N. Genomic Study of Chromosomally and Plasmid-Mediated Multidrug Resistance and Virulence Determinants in Klebsiella Pneumoniae Isolates Obtained from a Tertiary Hospital in Al-Kharj, KSA. Antibiotics 2022, 11, 1564. [Google Scholar] [CrossRef]
- Sid Ahmed, M.A.; Bansal, D.; Acharya, A.; Elmi, A.A.; Hamid, J.M.; Sid Ahmed, A.M.; Chandra, P.; Ibrahim, E.; Sultan, A.A.; Doiphode, S.; et al. Antimicrobial susceptibility and molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae from intensive care units at Hamad Medical Corporation, Qatar. Antimicrob. Resist. Infect. Cont. 2016, 5, 4. [Google Scholar] [CrossRef]
- Elbadawi, H.S.; Elhag, K.M.; Mahgoub, E.; Altayb, H.N.; Ntoumi, F.; Elton, L.; McHugh, T.D.; Tembo, J.; Ippolito, G.; Osman, A.Y.; et al. Detection and characterization of carbapenem resistant Gram-negative bacilli isolates recovered from hospitalized patients at Soba University Hospital, Sudan. BMC Microbiol. 2021, 21, 136. [Google Scholar] [CrossRef] [PubMed]
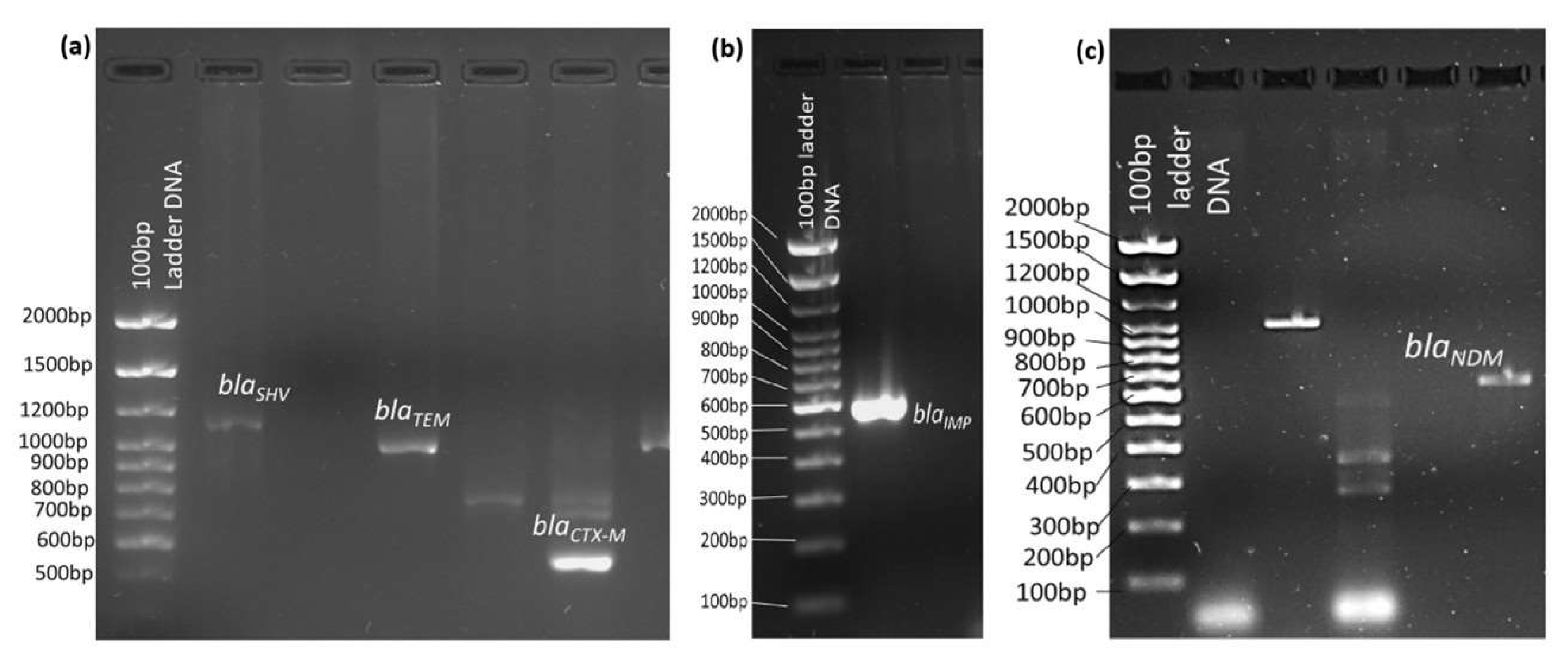





| Gene Type | Amplicon | Primer Sequence (5′-3′) | Amplicon Size (bp) | Annealing Temperature °C | Reference |
|---|---|---|---|---|---|
| ESBL gene primers | blaCTX-M | F-ATGTGCAGYACCAGTAARGTKAT GCC R-TGGGTRARRTARGTSACCAGAAY CAGCGG | 593 | 58 | [15] |
| blaTEM | F-TCGCCGCATACACTATTCTCAGAA TGA R-ACGCTCACCGGCTCCAGATTTAT | 445 | 50 | ||
| blaSHV | F-GGGTTATTCTT ATTTGTCGC, R-TTAGCGTTGCCAGTGCTC | 747 | 58 | ||
| Carbapenem-resistant gene primers | blaNDM | F-GGTTTG GCGATCTGGTTTTC R-CGGAATGGCTCATCACGATC | 621 | 57 | [27] |
| blaIMP | F-CTACCGCAGCAGAGTCTTTG R-AACCAGTTTTGCCTTACCAT | 587 | 56 | ||
| blaOXA48 | F-GCGTGGTTAAGGATGAACAC R-CATCAAGTTCAACCCAACCG | 438 | 57 | [15] | |
| Control primer | 16S rRNA gene | F-AGAGTTTGATCMTGGCTCAG R-ACGGHTACCTTGTTACGACTT | 1500 | 55 |
| Clinical Samples n = 3670 (%) | No. of Bacterial Pathogens Isolated n = 1098 (29.9%) | No. of Samples for No Bacterial Growth n = 2572 (70.1%) | p-Value/ Chi-Square Test | |
|---|---|---|---|---|
| Gram-Negative Bacteria n = 833 (22.7%) | Gram-Positive Bacteria n = 265 (7.2%) | |||
| Urine n = 1970 (53.7%) | 503 (13.7%) | 37 (1.0%) | 1430 (39%) | <0.001/335 * |
| Blood n = 879 (23.9%) | 48 (1.3%) | 71 (1.9%) | 760 (20.7%) | |
| Pus n = 469 (12.8%) | 111 (3.0%) | 142 (3.9%) | 216 (5.9%) | |
| Sputum n = 352 (9.6%) | 171 (4.7%) | 15 (0.4%) | 166 (4.5%) | |
| No. of Samples Positive for Gram-Ve Bacteria n (%) | No. of Gram-Ve Bacteria Isolated n = 833 | ||||
|---|---|---|---|---|---|
| E. coli n = 394 (47.2%) | K. pneumoniae n = 200 (24%) | P. aeruginosa n = 129 (15.5%) | P. mirabilis n = 40 (4.8%) | A. baumannii n = 70 (8.4%) | |
| Urine n = 503 (60.3%) | 354 (42.5%) | 116 (13.9%) | 15 (1.8%) | 12 (1.4%) | 6 (0.7%) |
| Blood n = 48 (5.7%) | 8 (0.9%) | 15 (1.8%) | 9 (1.1%) | 0 | 16 (1.9%) |
| Pus n = 111 (13.3%) | 25 (3.0%) | 35 (4.2%) | 19 (2.3%) | 24 (2.9%) | 8 (1.0%) |
| Sputum n = 171 (20.5%) | 7 (0.8%) | 34 (4.1%) | 86 (10.3%) | 4 (0.5%) | 40 (4.8%) |
| Antibiotics | E. coli n = 157 (%) | K. pneumoniae n = 45 (%) | P. mirabilis n = 17 (%) | P. aeruginosa n = 20 (%) | A. baumannii n = 10 (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I | R | I | R | I | R | I | R | I | R | |
| Gentamicin | 0 | 23 (15%) | 0 | 15 (33%) | 4 (23%) | 13 (77%) | 0 | 20 (100%) | 0 | 10 (100%) |
| Imipenem | 0 | 0 | 0 | 5 (11%) | 0 | 0 | 0 | 20 (100%) | 0 | 10 (100%) |
| Levofloxacin | 0 | 73 (46%) | 0 | 10 (22%) | 0 | 8 (47%) | 0 | 20 (100%) | 0 | 10 (100%) |
| Meropenem | 0 | 0 | 0 | 10 (22%) | 0 | 0 | 0 | 20 (100%) | 0 | 10 (100%) |
| Nitrofurantoin | 0 | 0 | 0 | 5 (11%) | 0 | 0 | 0 | 0 | 0 | 10 (100%) |
| Norfloxacin | 0 | 100 (64%) | 0 | 5 (11%) | 8 (47%) | 9 (53%) | 0 | 0 | 0 | 10 (100%) |
| Piperacillin/Tazobactam | 0 | 0 | 0 | 10 (22%) | 0 | 0 | 7 (35%) | 13 (65%) | 0 | 0 |
| Tigecycline | 0 | 0 | 5 (11%) | 10 (22%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Tobramycin | 7 (5%) | 20 (13%) | 5 (11%) | 10 (22%) | 0 | 17 (100%) | 0 | 20 (100%) | 0 | 10 (100%) |
| Trimethoprim/ Sulfamethoxazole | 0 | 92 (59%) | 0 | 25 (55%) | 0 | 17 (100%) | 0 | 0 | 2 (20%) | 8 (80%) |
| Amikacin | 0 | 0 | 0 | 10 (22%) | 0 | 13 (77%) | 0 | 20 (100%) | 0 | 10 (100%) |
| Amoxicillin + Clavulanate | 0 | 145 (92%) | 0 | 40 (89%) | 0 | 17 (100%) | 0 | 0 | 0 | 0 |
| Ampicillin/ sulbactam | 0 | 153 (97%) | 0 | 45 (100%) | 4 (23%) | 13 (77%) | 0 | 0 | 3 (30%) | 7 (70%) |
| Ampicillin | 0 | 153 (97%) | 0 | 45 (100%) | 0 | 17 (100%) | 0 | 0 | 0 | 0 |
| Aztreonam | 0 | 153 (97%) | 0 | 31 (69%) | 0 | 17 (100%) | 13 (65%) | 7 (35%) | 0 | 0 |
| Cefazolin | 0 | 153 (97%) | 0 | 45 (100%) | 0 | 17 (100%) | 0 | 0 | 0 | 0 |
| Cefepime | 0 | 153 (97%) | 0 | 45 (100%) | 0 | 17 (100%) | 0 | 20 (100%) | 0 | 10 (100%) |
| Cefotaxime | 0 | 157 (100%) | 5 (11%) | 40 (89%) | 0 | 17 (100%) | 0 | 0 | 0 | 0 |
| Cefoxitin | 7 (5%) | 12 (7.6%) | 0 | 10 (22%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Ceftazidime | 0 | 157 (100%) | 0 | 30 (67%) | 0 | 17 (100%) | 0 | 20 (100%) | 0 | 10 (100%) |
| Cefuroxime | 4 (3%) | 153 (97%) | 0 | 45 (100%) | 0 | 17 (100%) | 0 | 0 | 0 | 0 |
| Ciprofloxacin | 0 | 81 (52%) | 5 (11%) | 10 (22%) | 8 (47%) | 9 (53%) | 0 | 20 (100%) | 0 | 10 (100%) |
| Ertapenem | 0 | 0 | 0 | 10 (22%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Colistin | 0 | 0 | 0 | 5 (11%) | 0 | 0 | 0 | 0 | 0 | 3 (30%) |
| No. of Resistant Bacteria in Clinical Samples n = 249 (%) | Single Gene Detection | Multiple Gene Detection | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaCTX-M n = 111 | blaTEM n = 85 | blaSHV n = 45 | blaNDM n = 3 | blaIMP n = 6 | blaCTX-M +TEM n = 33 | blaCTX-M +SHV n = 26 | blaTEM +SHV n = 3 | blaCTX-M+TEM +SHV n = 6 | blaTEM +NDM n = 2 | blaTEM +IMP n = 1 | blaCTX-M+TEM+SHV+IMP n = 2 | ||
| Urine n = 156 | E. coli n = 120 (77%) | 56 (46.7%) | 44 (36.7%) | 20 (16.7%) | 0 | 0 | 11 (9%) | 11 (9%) | 0 | 6 (5%) | 0 | 0 | 0 |
| K. pnuemoniae n = 28 (18%) | 8 (28.6%) | 10 (35.7%) | 7 (35.7%) | 0 | 3 (21.4%) | 8 (28.6%) | 7 (25%) | 3 (10.7%) | 0 | 0 | 0 | 0 | |
| P. aeruginosa n = 5 (3%) | 2 (40%) | 2 (40%) | 1 (20%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P. mirabilis n = 3 (2%) | 2 (66%) | 1 (33%) | 0 | 0 | 0 | 1 (33%) | 0 | 0 | 0 | 0 | 0 | 0 | |
| A. baumannii n = 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Blood n = 14 | E. coli n = 7 (50%) | 5 (71.4%) | 1 (14.3%) | 1 (14.3%) | 0 | 0 | 1 (14.3%) | 0 | 0 | 0 | 0 | 0 | 0 |
| K. pnuemoniae n = 2 (14.3%) | 1 (50%) | 1 (50%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P. aeruginosa n = 2 (14.3%) | 1 (50%) | 0 | 1 (50%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P. mirabilis n = 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A. baumannii n = 3 (2%) | 2 (66%) | 1 (33.3%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pus n = 49 | E. coli n = 25 (51%) | 8 (32%) | 10(40%) | 7 (28%) | 0 | 0 | 3 (12%) | 3 (12%) | 0 | 0 | 0 | 0 | 0 |
| K. pnuemoniae n = 7 (14.3%) | 5 (71.4%) | 1 (14.3%) | 1 (14.3%) | 0 | 0 | 3 (43%) | 2 (28.6%) | 0 | 0 | 0 | 0 | 0 | |
| P. aeruginosa n = 3 (6%) | 1 (33%) | 1 (33%) | 0 | 0 | 1 (33%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P. mirabilis n = 14 (28.6%) | 8 (57%) | 4 (43%) | 0 | 0 | 0 | 2 (14.3%) | 0 | 0 | 0 | 0 | 0 | 0 | |
| A. baumannii n = 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sputum n = 30 | E. coli n = 5 (16.7%) | 3 (60%) | 1 (20%) | 1 (20%) | 0 | 0 | 0 | 1 (20%) | 0 | 0 | 0 | 0 | 0 |
| K. pnuemoniae n = 8 (26.6%) | 4 (50%) | 2 (25%) | 1 (12.5%) | 1 (12.5%) | 0 | 3 (37.5%) | 1 (12.5%) | 0 | 0 | 1 (12.5%) | 0 | 0 | |
| P. aeruginosa n = 10 (33%) | 3 (42.9%) | 5 (28.6%) | 1 (14.3%) | 0 | 1 (14.3%) | 1 (14.3%) | 1 (14.3%) | 0 | 0 | 0 | 0 | 1 (14.3%) | |
| P. mirabilis n = 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| A. baumannii n = 7 (23%) | 2 (28.6%) | 1 (14.3%) | 1 (14.3%) | 2 (28.6%) | 1 (14.3%) | 0 | 0 | 0 | 0 | 1 (14.3%) | 1 (14.3%) | 1 (14.3%) | |
| Detected Genes | No. of Bacteria Carrying the Resistant Genes n = 249 | |||||
|---|---|---|---|---|---|---|
| E. coli n = 157 (63%) | K. pnuemoniae n = 45 (18%) | P. aeruginosa n = 20 (8%) | P. mirabilis n = 17 (6.8%) | A. baumannii n = 10 (4%) | ||
| Single gene | blaCTX-M n = 111 | 72 (29%) | 18 (40%) | 7 (35%) | 10 (59%) | 4 (40%) |
| blaTEM n = 86 | 56 (35.7%) | 14 (31%) | 8 (40%) | 6 (35.3%) | 2 (20%) | |
| blaSHV n = 43 | 29 (18.5%) | 9 (20%) | 3 (15%) | 1 (5.9%) | 1 (10%) | |
| blaNDM n = 3 | 0 | 1 (2.2%) | 0 | 0 | 2 (20%) | |
| blaIMP n = 6 | 0 | 3 (6.6%) | 2 (10%) | 0 | 1 (10%) | |
| Mixed genes | blaCTX-M+TEM n = 33 | 15 (9.5%) | 14 (31%) | 1 (5%) | 3 (17.7%) | 0 |
| blaCTX-M+SHV n = 26 | 15 (9.5%) | 10 (22.2%) | 1 (5%) | 0 | 0 | |
| blaTEM+SHV n = 3 | 0 | 3 (6.6%) | 0 | 0 | 0 | |
| blaTEM+NDM n = 2 | 0 | 1 (2.2%) | 0 | 0 | 1 (10%) | |
| blaTEM+IMP n = 1 | 0 | 0 | 0 | 0 | 1 (10%) | |
| blaCTX-M+TEM+SHV n = 6 | 6 (4%) | 0 | 0 | 0 | ||
| blaCTX-M+TEM+SHV+IMP n = 2 | 0 | 0 | 1 (5%) | 0 | 1 (10%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragupathi, P.; Khamisani, V.; Sadiq, A.F.; Mobiddo, M.A.; Parwaiz, N.; Bagchi, S.; Rahamathullah, N. Molecular Surveillance of ESBL and Carbapenemase Genes in Gram-Negative Bacterial Pathogens Isolated from Various Clinical Samples Collected from Northern Region of United Arab Emirates. Microorganisms 2025, 13, 1880. https://doi.org/10.3390/microorganisms13081880
Ragupathi P, Khamisani V, Sadiq AF, Mobiddo MA, Parwaiz N, Bagchi S, Rahamathullah N. Molecular Surveillance of ESBL and Carbapenemase Genes in Gram-Negative Bacterial Pathogens Isolated from Various Clinical Samples Collected from Northern Region of United Arab Emirates. Microorganisms. 2025; 13(8):1880. https://doi.org/10.3390/microorganisms13081880
Chicago/Turabian StyleRagupathi, Premalatha, Vaneezeh Khamisani, Aisha Fadila Sadiq, Mariam Aliyu Mobiddo, Nasir Parwaiz, Sovan Bagchi, and Nazeerullah Rahamathullah. 2025. "Molecular Surveillance of ESBL and Carbapenemase Genes in Gram-Negative Bacterial Pathogens Isolated from Various Clinical Samples Collected from Northern Region of United Arab Emirates" Microorganisms 13, no. 8: 1880. https://doi.org/10.3390/microorganisms13081880
APA StyleRagupathi, P., Khamisani, V., Sadiq, A. F., Mobiddo, M. A., Parwaiz, N., Bagchi, S., & Rahamathullah, N. (2025). Molecular Surveillance of ESBL and Carbapenemase Genes in Gram-Negative Bacterial Pathogens Isolated from Various Clinical Samples Collected from Northern Region of United Arab Emirates. Microorganisms, 13(8), 1880. https://doi.org/10.3390/microorganisms13081880







