The Role of Genomic Islands in the Pathogenicity and Evolution of Plant-Pathogenic Gammaproteobacteria
Abstract
1. Introduction
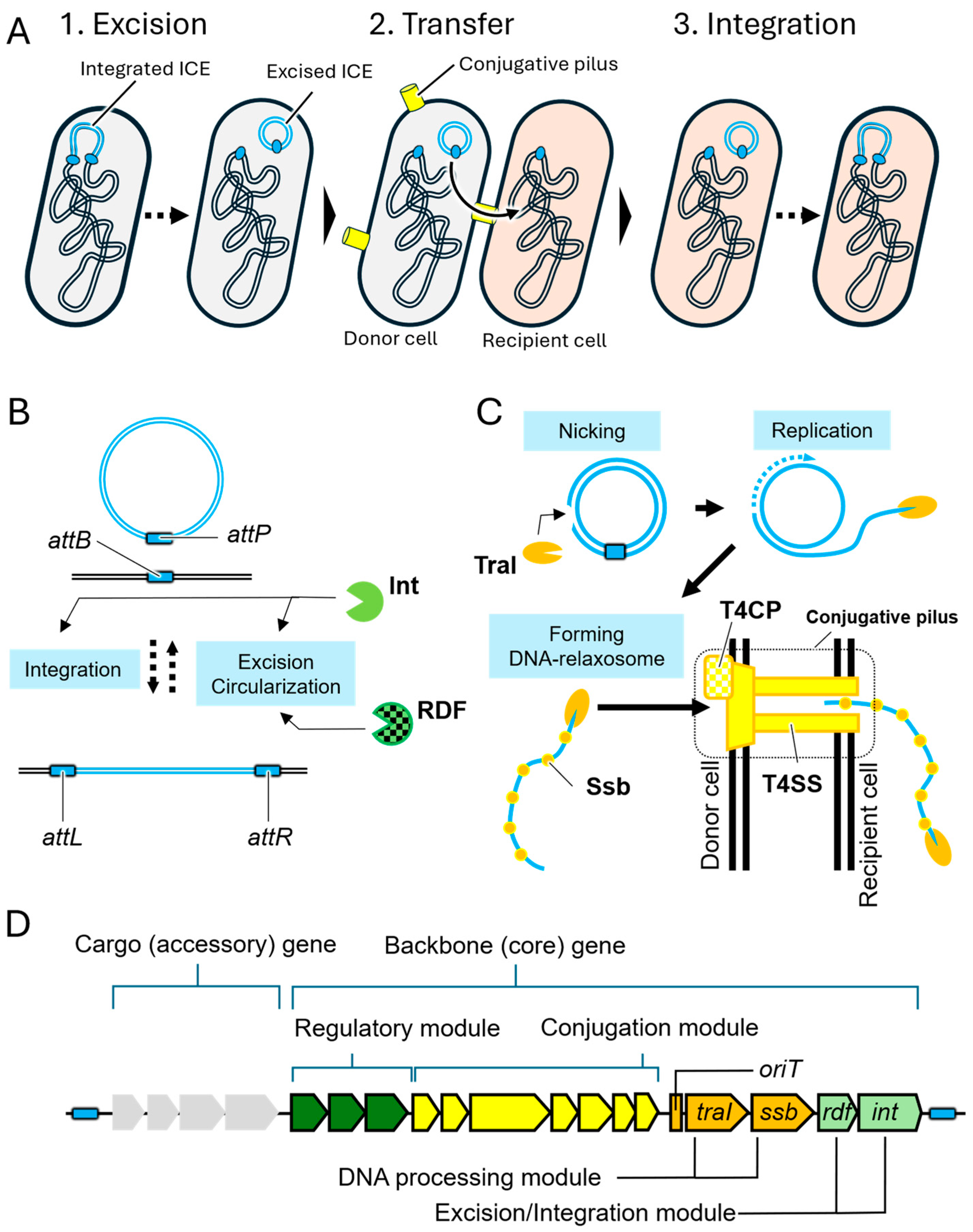
2. Structure and Transfer Mechanisms of ICEs
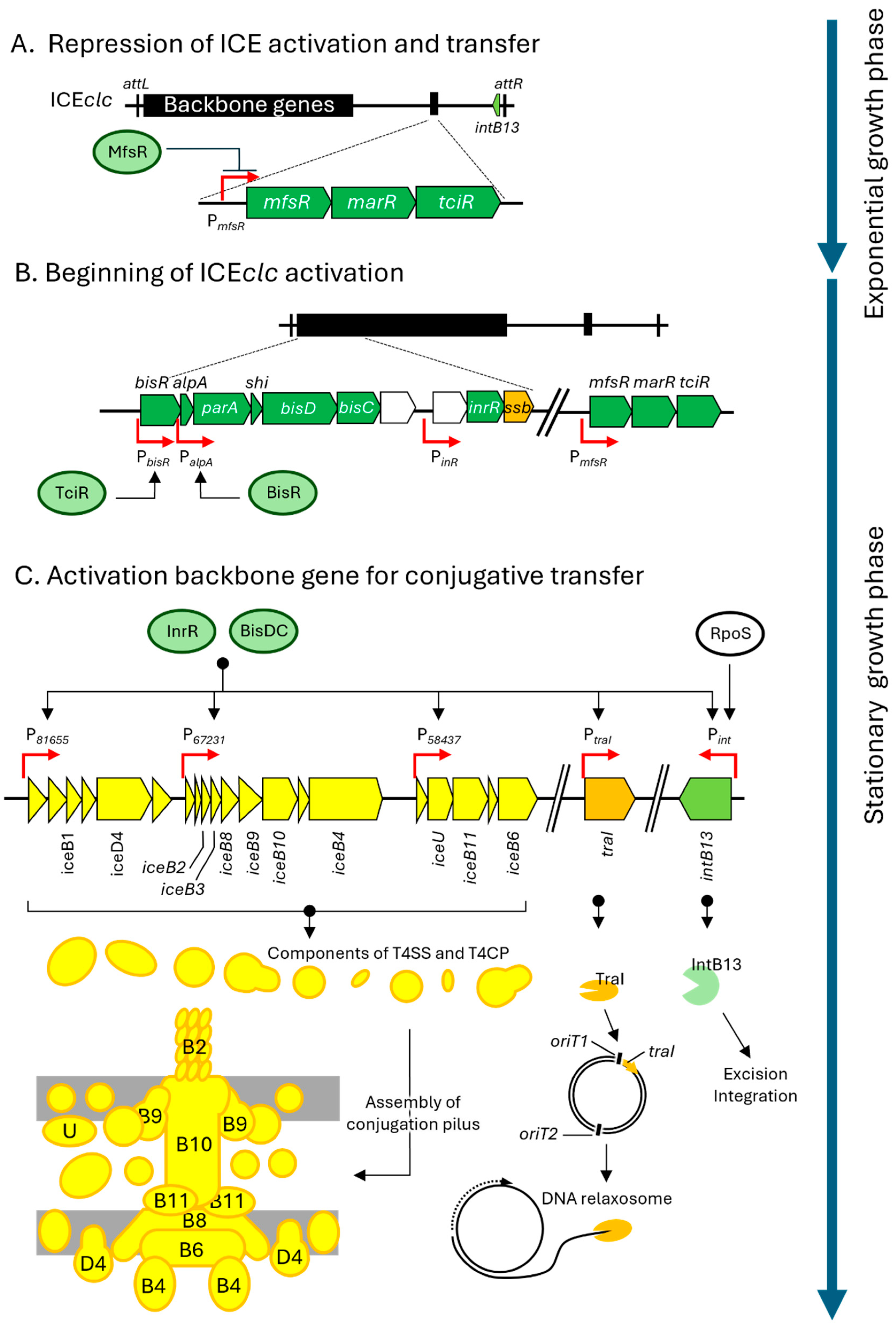
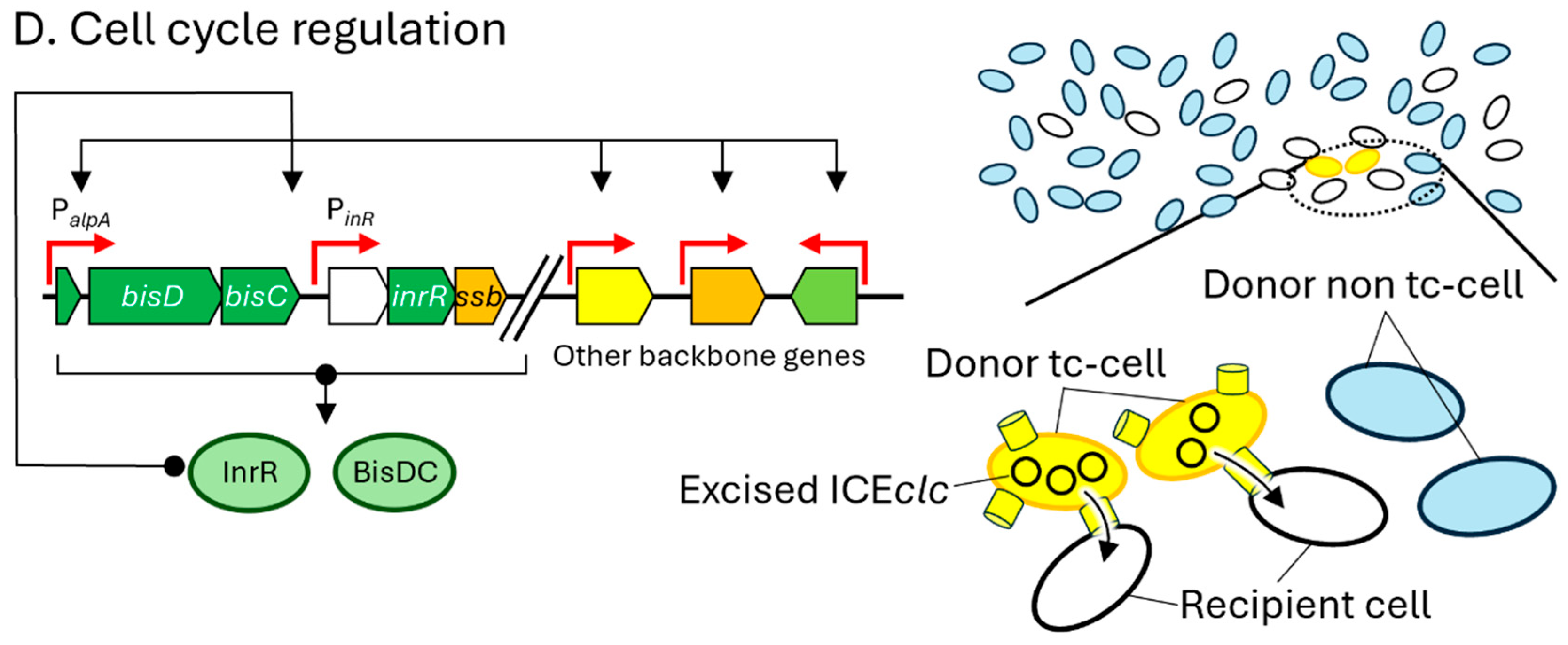
3. Specific Mechanisms of ICEclc for Excision and Transfer
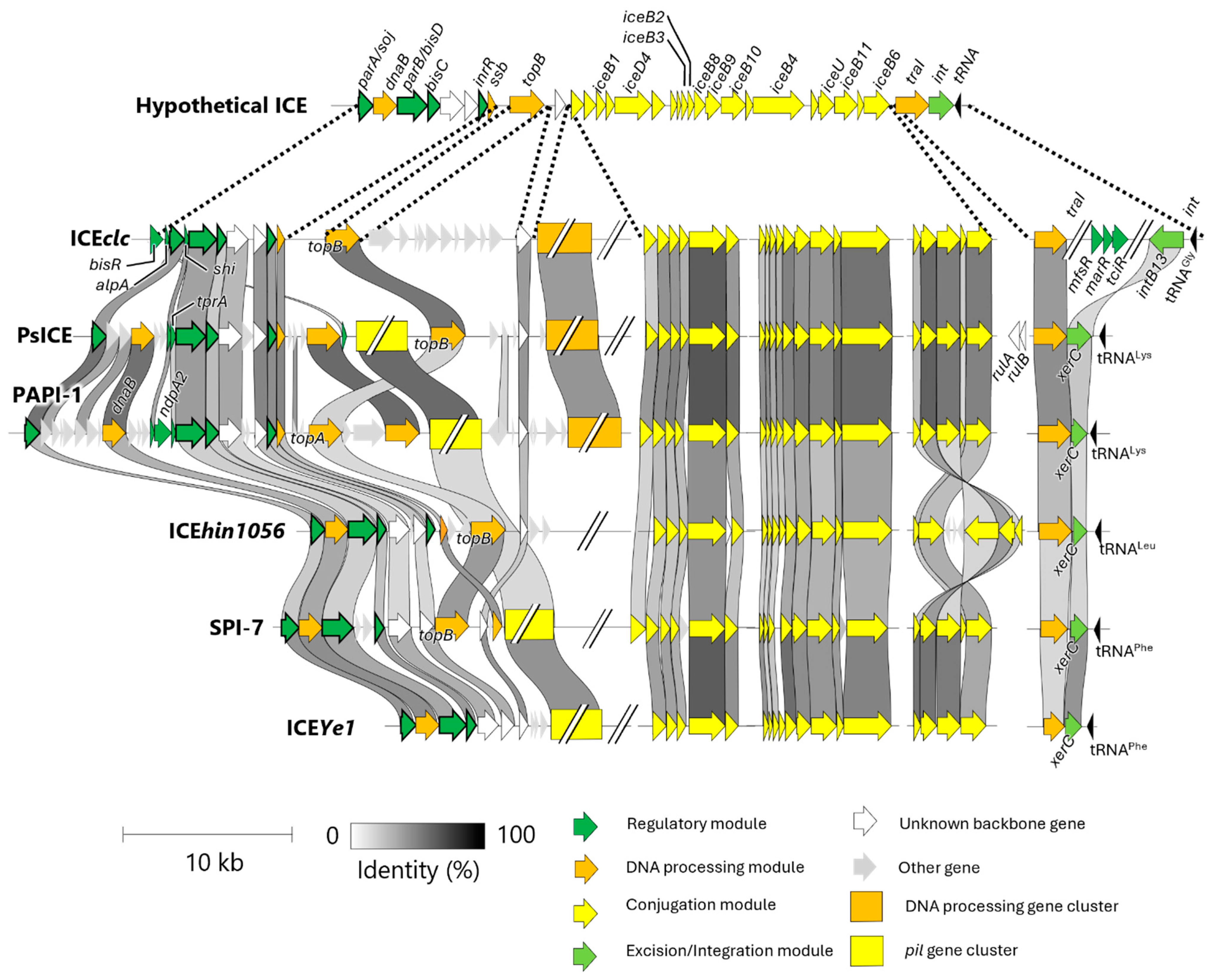
4. Classification of ICEclc and Its Homologous
5. Possible Mechanisms of PsICE Excision and Transfer
6. Role of Cargo Genes in Pseudomonas syringae
7. Role of Genomic Island in Other Plant-Pathogenic Bacteria
8. Future Research Direction
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HGT | Horizontal gene transfer |
| GI | Genomic island |
| ICE | Integrative and conjugative element |
| RDF | Recombination directionality factor |
| ssDNA | Single-strand DNA |
| T4CP | Type IV coupling protein |
| T4SS | Type IV secretion system |
| Tc cell | Transfer-competent cell |
| MLSA | Multilocus sequence analysis |
| H-NS | Histone-like nucleotide structuring protein |
| PG | Phylogroup |
| TCA | Tricarboxylic acid |
| CFA | Coronafacic acid |
| T3SS | Type III secretion system |
| aCGH | array comparative genomic hybridization |
References
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef]
- Mwangi, R.W.; Mustafa, M.; Charles, K.; Wagara, I.W.; Kappel, N. Selected emerging and reemerging plant pathogens affecting the food basket: A threat to food security. J. Agric. Food Res. 2023, 14, 100827. [Google Scholar] [CrossRef]
- Sundin, G.W.; Castiblanco, L.F.; Yuan, X.; Zeng, Q.; Yang, C.-H. Bacterial disease management: Challenges, experience, innovation and future prospects: Challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 2016, 17, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- Gowtham, H.G.; Murali, M.; Shilpa, N.; Amruthesh, K.N.; Gafur, A.; Antonius, S.; Sayyed, R.Z. Harnessing abiotic elicitors to bolster plant’s resistance against bacterial pathogens. Plant Stress 2024, 11, 100371. [Google Scholar] [CrossRef]
- Hacker, J.; Kaper, J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Hsiao, W.W.L.; Brinkman, F.S.L. Detecting genomic islands using bioinformatics approaches. Nat. Rev. Microbiol. 2010, 8, 373–382. [Google Scholar] [CrossRef]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef]
- Kiss, J.; Szabó, M.; Hegyi, A.; Douard, G.; Praud, K.; Nagy, I.; Olasz, F.; Cloeckaert, A.; Doublet, B. Identification and characterization of oriT and two mobilization genes required for conjugative transfer of Salmonella genomic island 1. Front. Microbiol. 2019, 10, 457. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Finlay, B.B. Pathogenicity islands: A molecular toolbox for bacterial virulence. Cell. Microbiol. 2006, 8, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Arnold, D.L.; Pitman, A.; Jackson, R.W. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 2003, 4, 407–420. [Google Scholar] [CrossRef]
- Ho Sui, S.J.; Fedynak, A.; Hsiao, W.W.L.; Langille, M.G.I.; Brinkman, F.S.L. The association of virulence factors with genomic islands. PLoS ONE 2009, 4, e8094. [Google Scholar] [CrossRef]
- Bertelli, C.; Tilley, K.E.; Brinkman, F.S.L. Microbial genomic island discovery, visualization and analysis. Brief Bioinform. 2019, 20, 1685–1698. [Google Scholar] [CrossRef]
- Sundin, G.W.; Wang, N. Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef]
- Wozniak, R.A.F.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar] [CrossRef]
- Johnson, C.M.; Grossman, A.D. Integrative and conjugative elements (ICEs): What they do and how they work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Xu, Z.; Harrison, E.M.; Tai, C.; Wei, Y.; He, X.; Jia, S.; Deng, Z.; Rajakumar, K.; Ou, H.-Y. ICEberg: A web-based resource for integrative and conjugative elements found in bacteria. Nucleic Acids Res. 2012, 40, D621–D626. [Google Scholar] [CrossRef]
- Burrus, V. Mechanisms of stabilization of integrative and conjugative elements. Curr. Opin. Microbiol. 2017, 38, 44–50. [Google Scholar] [CrossRef]
- Rudy, C.; Taylor, K.L.; Hinerfeld, D.; Scott, J.R.; Churchward, G. Excision of a conjugative transposon in vitro by the Int and Xis proteins of Tn916. Nucleic Acids Res. 1997, 25, 4061–4066. [Google Scholar] [CrossRef][Green Version]
- Lee, C.A.; Auchtung, J.M.; Monson, R.E.; Grossman, A.D. Identification and characterization of Int (integrase), Xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 2007, 66, 1356–1369. [Google Scholar] [CrossRef] [PubMed]
- Haskett, T.L.; Terpolilli, J.J.; Ramachandran, V.K.; Verdonk, C.J.; Poole, P.S.; O’Hara, G.W.; Ramsay, J.P. Sequential induction of three recombination directionality factors directs assembly of tripartite integrative and conjugative elements. PLoS Genet. 2018, 14, e1007292. [Google Scholar] [CrossRef] [PubMed]
- Bean, E.L.; Herman, C.; Anderson, M.E.; Grossman, A.D. Biology and engineering of integrative and conjugative elements: Construction and analyses of hybrid ICEs reveal element functions that affect species-specific efficiencies. PLoS Genet. 2022, 18, e1009998. [Google Scholar] [CrossRef] [PubMed]
- Dorn, E.; Hellwig, M.; Reineke, W.; Knackmuss, H.J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 1974, 99, 61–70. [Google Scholar] [CrossRef]
- Ravatn, R.; Studer, S.; Springael, D.; Zehnder, A.J.; van der Meer, J.R. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 1998, 180, 4360–4369. [Google Scholar] [CrossRef]
- Mohd-Zain, Z.; Turner, S.L.; Cerdeño-Tárraga, A.M.; Lilley, A.K.; Inzana, T.J.; Duncan, A.J.; Harding, R.M.; Hood, D.W.; Peto, T.E.; Crook, D.W. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 2004, 186, 8114–8122. [Google Scholar] [CrossRef]
- Pitman, A.R.; Jackson, R.W.; Mansfield, J.W.; Kaitell, V.; Thwaites, R.; Arnold, D.L. Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr. Biol. 2005, 15, 2230–2235. [Google Scholar] [CrossRef]
- Colombi, E.; Straub, C.; Künzel, S.; Templeton, M.D.; McCann, H.C.; Rainey, P.B. Evolution of copper resistance in the kiwifruit pathogen Pseudomonas syringae pv. actinidiae through acquisition of integrative conjugative elements and plasmids. Environ. Microbiol. 2017, 19, 819–832. [Google Scholar] [CrossRef]
- Baltrus, D.A.; Feng, Q.; Kvitko, B.H. Genome context influences evolutionary flexibility of nearly identical type III effectors in two phytopathogenic Pseudomonads. Front. Microbiol. 2022, 13, 826365. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Kunishi, K.; Matsui, H.; Sakata, N.; Noutoshi, Y.; Toyoda, K.; Ichinose, Y. Genomic islands of Pseudomonas syringae pv. tabaci 6605: Identification of PtaGI-1 as a pathogenicity island with effector genes and a tabtoxin cluster. Mol. Plant Pathol. 2025, 26, e70087. [Google Scholar] [CrossRef]
- Colombi, E.; Bertels, F.; Doulcier, G.; Mc Connell, E.; Pichugina, T.; Sohn, K.H.; Straub, C.; Mc Cann, H.C.; Rainey, P.B. Rapid dissemination of host metabolism–manipulating genes via integrative and conjugative elements. Proc. Natl. Acad. Sci. USA 2024, 11, e2309263121. [Google Scholar] [CrossRef]
- Pradervand, N.; Sulser, S.; Delavat, F.; Miyazaki, R.; Lamas, I.; van der Meer, J.R. An operon of three transcriptional regulators controls horizontal gene transfer of the integrative and conjugative element ICEclc in Pseudomonas knackmussii B13. PLoS Genet. 2014, 10, e1004441. [Google Scholar] [CrossRef] [PubMed]
- Minoia, M.; Gaillard, M.; Reinhard, F.; Stojanov, M.; Sentchilo, V.; van der Meer, J.R. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 20792–20797. [Google Scholar] [CrossRef]
- Carraro, N.; Richard, X.; Sulser, S.; Delavat, F.; Mazza, C.; van der Meer, J.R. An analog to digital converter controls bistable transfer competence development of a widespread bacterial integrative and conjugative element. eLife 2020, 9, e57915. [Google Scholar] [CrossRef] [PubMed]
- Sulser, S.; Vucicevic, A.; Bellini, V.; Moritz, R.; Delavat, F.; Sentchilo, V.; Carraro, N.; van der Meer, J.R. A bistable prokaryotic differentiation system underlying development of conjugative transfer competence. PLoS Genet. 2022, 18, e1010286. [Google Scholar] [CrossRef]
- Daveri, A.; Benigno, V.; van der Meer, J.R. Characterization of an atypical but widespread type IV secretion system for transfer of the integrative and conjugative element (ICEclc) in Pseudomonas putida. Nucleic Acids Res. 2023, 51, 2345–2362. [Google Scholar] [CrossRef]
- Miyazaki, R.; van der Meer, J.R. A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol. Microbiol. 2011, 79, 743–758. [Google Scholar] [CrossRef]
- Sentchilo, V.; Ravatn, R.; Werlen, C.; Zehnder, A.J.B.; van der Meer, J.R. Unusual integrase gene expression on the clc genomic island in Pseudomonas sp. strain B13. J. Bacteriol. 2003, 185, 4530–4538. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, R.; Minoia, M.; Pradervand, N.; Sulser, S.; Reinhard, F.; van der Meer, J.R. Cellular variability of rpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 2012, 8, e1002818. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, F.; Miyazaki, R.; Pradervand, N.; van der Meer, J.R. Cell differentiation to “mating bodies” induced by an integrating and conjugative element in free-living bacteria. Curr. Biol. 2013, 23, 255–259. [Google Scholar] [CrossRef]
- Carter, M.Q.; Chen, J.; Lory, S. The Pseudomonas aeruginosa pathogenicity island PAPI-1 is transferred via a novel type IV pilus. J. Bacteriol. 2010, 192, 3249–3258. [Google Scholar] [CrossRef]
- Shimoda, E.; Muto, T.; Horiuchi, T.; Furuya, N.; Komano, T. Novel class of mutations of pilS mutants, encoding plasmid R64 type IV prepilin: Interface of PilS-PilV interactions. J. Bacteriol. 2008, 190, 1202–1208. [Google Scholar] [CrossRef]
- Vos, S.M.; Tretter, E.M.; Schmidt, B.H.; Berger, J.M. All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011, 12, 827–841. [Google Scholar] [CrossRef]
- Qiu, X.; Gurkar, A.U.; Lory, S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19830–19835. [Google Scholar] [CrossRef]
- Kim, J.J.; Sundin, G.W. Regulation of the rulAB mutagenic DNA repair operon of Pseudomonas syringae by UV-B (290 to 320 nanometers) radiation and analysis of rulAB-mediated mutability in vitro and in planta. J. Bacteriol. 2000, 182, 6137–6144. [Google Scholar] [CrossRef]
- Beaber, J.W.; Hochhut, B.; Waldor, M.K. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004, 427, 72–74. [Google Scholar] [CrossRef]
- Takano, S.; Fukuda, K.; Koto, A.; Miyazaki, R. A novel system of bacterial cell division arrest implicated in horizontal transmission of an integrative and conjugative element. PLoS Genet. 2019, 15, e1008445. [Google Scholar] [CrossRef] [PubMed]
- Dangla-Pélissier, G.; Roux, N.; Schmidt, V.; Chambonnier, G.; Ba, M.; Sebban-Kreuzer, C.; de Bentzmann, S.; Giraud, C.; Bordi, C. The horizontal transfer of Pseudomonas aeruginosa PA14 ICE PAPI-1 is controlled by a transcriptional triad between TprA, NdpA2 and MvaT. Nucleic Acids Res. 2021, 49, 10956–10974. [Google Scholar] [CrossRef] [PubMed]
- Neale, H.C.; Jackson, R.W.; Preston, G.M.; Arnold, D.L. Supercoiling of an excised genomic island represses effector gene expression to prevent activation of host resistance. Mol. Microbiol. 2018, 110, 444–454. [Google Scholar] [CrossRef]
- de Assis, J.C.S.; Gonçalves, O.S.; Fernandes, A.S.; de Queiroz, M.V.; Bazzolli, D.M.S.; Santana, M.F. Genomic analysis reveals the role of integrative and conjugative elements in plant pathogenic bacteria. Mob. DNA 2022, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, O.S.; de Queiroz, M.V.; Santana, M.F. Potential evolutionary impact of integrative and conjugative elements (ICEs) and genomic islands in the Ralstonia solanacearum species complex. Sci. Rep. 2020, 10, 12498. [Google Scholar] [CrossRef]
- Botelho, J.; Schulenburg, H. The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 2021, 29, 8–18. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “Pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Dillon, M.M.; Thakur, S.; Almeida, R.N.D.; Wang, P.W.; Weir, B.S.; Gutman, D. Recombination of ecologically and evolutionarily significant loci maintains genetic cohesion in the Pseudomonas syringae species complex. Genome Biol. 2019, 20, 3. [Google Scholar] [CrossRef]
- Ranjit, S.; Deblais, L.; Poelstra, J.; Bhandari, M.; Rotondo, F.; Scaria, J.; Miller, S.A.; Rajashekara, G. In vitro, in planta, and comparative genomic analyses of Pseudomonas syringae pv. syringae strains of pepper (Capsicum annuum var. annuum). Microbiol. Spectr. 2024, 12, e0006424. [Google Scholar] [CrossRef]
- Scortichini, M.; Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G. Pseudomonas syringae pv. actinidiae: A re-emerging, multi-faceted, pandemic pathogen. Mol. Plant Pathol. 2012, 13, 631–640. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; de Vicente, A.; Carrión, V.J.; Sundin, G.W.; Cazorla, F.M. Recruitment and rearrangement of three different genetic determinants into a conjugative plasmid increase copper resistance in Pseudomonas syringae. Appl. Environ. Microbiol. 2013, 79, 1028–1033. [Google Scholar] [CrossRef]
- Hemara, L.M.; Hoyte, S.M.; Arshed, S.; Schipper, M.M.; Wood, P.N.; Marshall, S.L.; Andersen, M.T.; Patterson, H.R.; Vanneste, J.L.; Peacock, L.; et al. Genomic biosurveillance of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae biovar 3 reveals adaptation to selective pressures in New Zealand orchards. Mol. Plant Pathol. 2025, 26, e70056. [Google Scholar] [CrossRef]
- Jackson, R.W.; Mansfield, J.W.; Arnold, D.L.; Sesma, A.; Paynter, C.D.; Murillo, J.; Taylor, J.D.; Vivian, A. Excision from tRNA genes of a large chromosomal region, carrying avrPphB, associated with race change in the bean pathogen, Pseudomonas syringae pv. phaseolicola. Mol. Microbiol. 2000, 38, 186–197. [Google Scholar] [CrossRef]
- Lovell, H.C.; Jackson, R.W.; Mansfield, J.W.; Godfrey, S.A.C.; Hancock, J.T.; Desikan, R.; Arnold, D.L. In planta conditions induce genomic changes in Pseudomonas syringae pv. phaseolicola. Mol. Plant Pathol. 2011, 12, 167–176. [Google Scholar] [CrossRef]
- Wei, C.-F.; Kvitko, B.H.; Shimizu, R.; Crabill, E.; Alfano, J.R.; Lin, N.-C.; Martin, G.B.; Huang, H.-C.; Collmer, A. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007, 51, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.S.; Sebaihia, M.; Pritchard, L.; Holden, M.T.G.; Hyman, L.J.; Holeva, M.C.; Thomson, N.R.; Bentley, S.D.; Churcher, L.J.C.; Mungall, K.; et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 2004, 101, 11105–11110. [Google Scholar] [CrossRef]
- Vanga, B.R.; Butler, R.C.; Toth, I.K.; Ronson, C.W.; Pitman, A.R. Inactivation of PbTopo IIIβ causes hyper-excision of the Pathogenicity Island HAI2 resulting in reduced virulence of Pectobacterium atrosepticum. Mol. Microbiol. 2012, 84, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Liu, H.; Booth, C.; Douglas, E.; François, P.; Schrenzel, J.; Hedley, P.E.; Birch, P.R.J.; Toth, I.K. Microarray comparative genomic hybridisation analysis incorporating genomic organisation, and application to enterobacterial plant pathogens. PLoS Comput. Biol. 2009, 5, e1000473. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.-S.; Kim, J.F.; Beer, S.V. The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infectiondouble dagger. Mol. Plant Pathol. 2005, 6, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Aull, H.G.; Jacobs-Sera, D.; Garlena, R.A.; Russell, D.A.; Smith, B.E.; Mahalingam, V.; Abad, L.; Gauthier, C.H.; Hatfull, G.F. The prophage and plasmid mobilome as a likely driver of Mycobacterium abscessus diversity. mBio 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- Jimenez, D.; Beltran, D.; Castillo, J.A. Mobile genetic elements of Xylella fastidiosa and their contribution to pathogenicity. Plant Pathol. 2024, 73, 2490–2499. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Loper, J.E.; Paulsen, I.T.; Thomashow, L.S. Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol. 2009, 9, 8. [Google Scholar] [CrossRef]
- Jackson, R.W.; Vinatzer, B.; Arnold, D.L.; Dorus, S.; Murillo, J. The influence of the accessory genome on bacterial pathogen evolution. Mob. Genet. Elem. 2011, 1, 55–65. [Google Scholar] [CrossRef]
- Durrant, M.G.; Li, M.M.; Siranosian, B.A.; Montgomery, S.B.; Bhatt, A.S. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 2020, 27, 140–153.e9. [Google Scholar] [CrossRef]
- Hulin, M.T.; Rabiey, M.; Zeng, Z.; Vadillo Dieguez, A.; Bellamy, S.; Swift, P.; Mansfield, J.W.; Jackson, R.W.; Harrison, R.J. Genomic and functional analysis of phage-mediated horizontal gene transfer in Pseudomonas syringae on the plant surface. New Phytol. 2023, 237, 959–973. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Gillings, M.R. New perspectives on mobile genetic elements: A paradigm shift for managing the antibiotic resistance crisis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20200462. [Google Scholar] [CrossRef] [PubMed]
- Olatunji, A.O.; Olaboye, J.A.; Maha, C.C.; Kolawole, T.O.; Abdul, S. Next-generation strategies to combat antimicrobial resistance: Integrating genomics, CRISPR, and novel therapeutics for effective treatment. Eng. Sci. Technol. J. 2024, 5, 2284–2303. [Google Scholar] [CrossRef]
- Peters, J.M.; Koo, B.-M.; Patino, R.; Heussler, G.E.; Hearne, C.C.; Qu, J.; Inclan, Y.F.; Hawkins, J.S.; Lu, C.H.S.; Silvis, M.R.; et al. Enabling genetic analysis of diverse bacteria with mobile-CRISPRi. Nat. Microbiol. 2019, 4, 244–250. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Cabezón, E.; Ripoll-Rozada, J.; Peña, A.; de la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Westermann, A.J.; Barquist, L.; Vogel, J. Resolving host-pathogen interactions by dual RNA-seq. PLoS Pathog. 2017, 13, e1006033. [Google Scholar] [CrossRef]
- Islam, T. Genomic surveillance for tackling emerging plant diseases, with special reference to wheat blast. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2024, 19, 1. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef]
- Stockdale, J.E.; Liu, P.; Colijn, C. The potential of genomics for infectious disease forecasting. Nat. Microbiol. 2022, 7, 1736–1743. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, G.; Schneider, M.V.; Moya, A. Live genomics for pathogen monitoring in public health. Pathogens 2014, 3, 93–108. [Google Scholar] [CrossRef]
- Prasanna, N.L.; Choudhary, S.; Kumar, S.; Choudhary, M.; Meena, P.K.; Samreen; Saloni, S.; Ghanghas, R. Advances in plant disease diagnostics and surveillance—A review. Plant Cell Biotechnol. Mol. Biol. 2024, 25, 137–150. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.-Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, R.; Chaudhuri, K.; Chaudhuri, P. On detection and assessment of statistical significance of genomic islands. BMC Genom. 2008, 9, 150. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, X.; Guo, Y.; Chen, H.; Liu, X.; He, P.; Li, W.; Zhang, M.Q.; Dai, Q. Systematic comparison of genome information processing and boundary recognition tools used for genomic island detection. Comput. Biol. Med. 2023, 166, 107550. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-Db: A manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. 2022, 88, e0099122. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, Y.; Ishiga, Y.; Sakata, N. The Role of Genomic Islands in the Pathogenicity and Evolution of Plant-Pathogenic Gammaproteobacteria. Microorganisms 2025, 13, 1803. https://doi.org/10.3390/microorganisms13081803
Watanabe Y, Ishiga Y, Sakata N. The Role of Genomic Islands in the Pathogenicity and Evolution of Plant-Pathogenic Gammaproteobacteria. Microorganisms. 2025; 13(8):1803. https://doi.org/10.3390/microorganisms13081803
Chicago/Turabian StyleWatanabe, Yuta, Yasuhiro Ishiga, and Nanami Sakata. 2025. "The Role of Genomic Islands in the Pathogenicity and Evolution of Plant-Pathogenic Gammaproteobacteria" Microorganisms 13, no. 8: 1803. https://doi.org/10.3390/microorganisms13081803
APA StyleWatanabe, Y., Ishiga, Y., & Sakata, N. (2025). The Role of Genomic Islands in the Pathogenicity and Evolution of Plant-Pathogenic Gammaproteobacteria. Microorganisms, 13(8), 1803. https://doi.org/10.3390/microorganisms13081803





