Degradation of Synthetic and Natural Textile Materials Using Streptomyces Strains: Model Compost and Genome Exploration for Potential Plastic-Degrading Enzymes
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Textile Materials
2.3. Strain Selection and Characterization
2.4. Textile Degradation in Model Compost
2.5. Characterization of Textile Materials
2.5.1. The Fourier-Transform Infrared Spectroscopy (FTIR)
2.5.2. Field Emission Scanning Electron Microscopy (FESEM) Analysis
2.6. Biological Evaluations
2.6.1. Aliivibrio fisheri Toxicity Test
2.6.2. Cytotoxicity Assay
2.7. Degradation Products Analysis Using Liquid Chromatography—Mass Spectrometry
2.8. Streptomyces Genome Sequencing and Analysis
3. Results
3.1. Characterization of Selected Streptomyces Strains
3.2. Textile Degradation in Compost
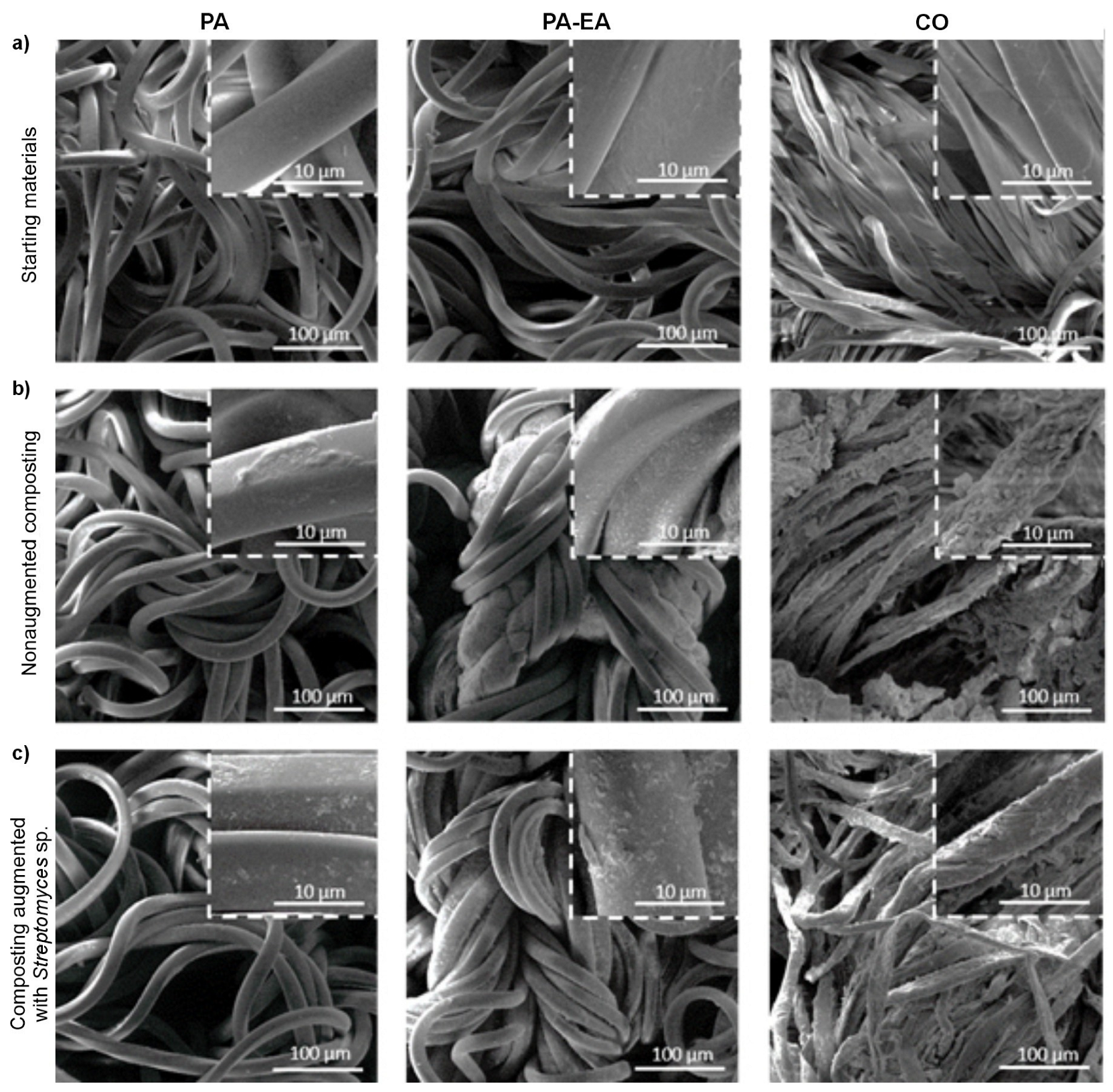
3.3. Textile Surface Analysis
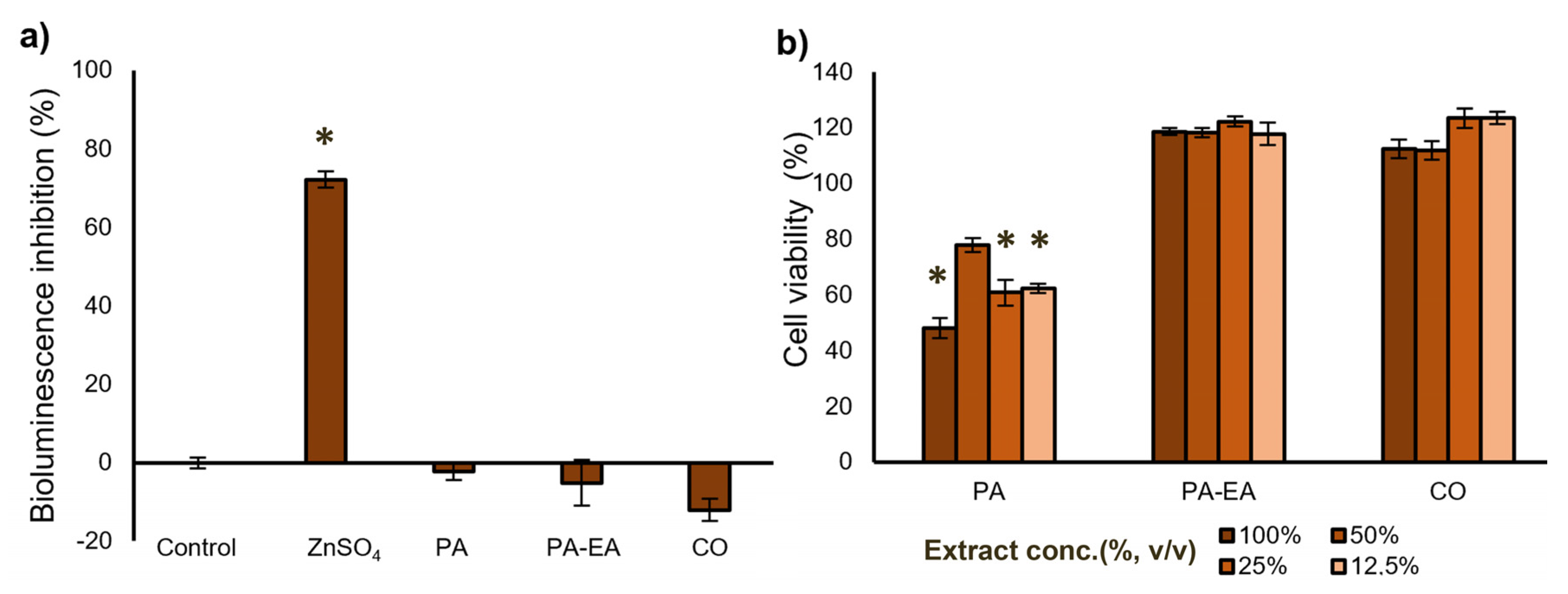
3.4. Eco- and Cytotoxicity of the Textile Material Extracts
3.5. LC-MS Analysis of Degradation Products
3.6. Genomic Exploration of Three Streptomyces
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Foundation, C.E. The Circularity Gap Report. Available online: https://www.circularity-gap.world/2024 (accessed on 24 January 2024).
- Exchange, T. 11th Edition of Textile Exchange’s Annual Materials Market Report. Available online: https://textileexchange.org/app/uploads/2024/09/Materials-Market-Report-2024.pdf (accessed on 26 September 2024).
- Roundup, T. 17 Most Worrysing Textile Waste Statistics & Facts. Available online: https://theroundup.org/textile-waste-statistics/ (accessed on 18 March 2024).
- Yadav, P.; Mishra, V. Comprehending microplastic pollution in diverse environment: Assessing fate, impacts, and remediation approaches. Int. Biodeterior. Biodegrad. 2025, 196, 105953. [Google Scholar] [CrossRef]
- UNEP. Sustainable Fashion to Take Centre Stage on Zero Waste Day. Available online: https://www.unep.org/technical-highlight/sustainable-fashion-take-centre-stage-zero-waste-day (accessed on 7 February 2025).
- Arioli, M.; Puiggalí, J.; Franco, L. Nylons with applications in energy generators, 3D printing and biomedicine. Molecules 2024, 29, 2443. [Google Scholar] [CrossRef]
- Pietroluongo, M.; Padovano, E.; Frache, A.; Badini, C. Mechanical recycling of an end-of-life automotive composite component. Sustain. Mater. Technol. 2020, 23, e00143. [Google Scholar] [CrossRef]
- Shah, Z.; Gulzar, M.; Hasan, F.; Shah, A.A. Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polym. Degrad. Stab. 2016, 134, 349–356. [Google Scholar] [CrossRef]
- Jovanović, T.; Penava, Ž.; Vrljičak, Z. Impact of the elastane percentage on the elastic properties of knitted fabrics under cyclic loading. Materials 2022, 15, 6512. [Google Scholar] [CrossRef]
- Shahriari-Khalaji, M.; Alassod, A.; Nozhat, Z. Cotton-based health care textile: A mini review. Polym. Bull. 2022, 79, 10409–10432. [Google Scholar] [CrossRef]
- Murthy, N.; Wilson, S.; Sy, J. 9.28-biodegradation of polymers. Polym. Sci. A Compr. Ref. 2012, 9, 547–560. [Google Scholar] [CrossRef]
- Zheng, L.; Wang, M.; Li, Y.; Xiong, Y.; Wu, C. Recycling and degradation of polyamides. Molecules 2024, 29, 1742. [Google Scholar] [CrossRef]
- Sun, S. Enzymatic depolymerization of polyamides (nylons): Current challenges and future directions. Polym. Degrad. Stab. 2025, 238, 111341. [Google Scholar] [CrossRef]
- Friedrich, J.; Zalar, P.; Mohorčič, M.; Klun, U.; Kržan, A. Ability of fungi to degrade synthetic polymer nylon-6. Chemosphere 2007, 67, 2089–2095. [Google Scholar] [CrossRef]
- Tomita, K.; Ikeda, N.; Ueno, A. Isolation and characterization of a thermophilic bacterium, Geobacillus thermocatenulatus, degrading nylon 12 and nylon 66. Biotechnol. Lett. 2003, 25, 1743–1746. [Google Scholar] [CrossRef]
- Baxi, N.; Shah, A. Biological treatment of the components of solid oligomeric waste from a nylon-6 production plant. World J. Microbiol. Biotechnol. 2000, 16, 835–840. [Google Scholar] [CrossRef]
- Bhavsar, P.; Bhave, M.; Webb, H.K. Solving the plastic dilemma: The fungal and bacterial biodegradability of polyurethanes. World J. Microbiol. Biotechnol. 2023, 39, 122. [Google Scholar] [CrossRef]
- Ji, J.; Pei, J.; Ding, F.; Zeng, C.; Zhou, J.; Dong, W.; Cui, Z.; Yan, X. Isolation and characterization of polyester polyurethane-degrading bacterium Bacillus sp. YXP1. Environ. Res. 2024, 249, 118468. [Google Scholar] [CrossRef]
- Najam, M.; Javaid, S.; Iram, S.; Pasertsakoun, K.; Oláh, M.; Székács, A.; Aleksza, L. Microbial Biodegradation of Synthetic Polyethylene and Polyurethane Polymers by Pedospheric Microbes: Towards Sustainable Environmental Management. Polymers 2025, 17, 169. [Google Scholar] [CrossRef]
- Kim, J.-H.; Choi, S.H.; Park, M.G.; Park, D.H.; Son, K.-H.; Park, H.-Y. Polyurethane biodegradation by Serratia sp. HY-72 isolated from the intestine of the Asian mantis Hierodula patellifera. Front. Microbiol. 2022, 13, 1005415. [Google Scholar] [CrossRef] [PubMed]
- Arshad, K.; Skrifvars, M.; Vivod, V.; Valh, J.; Voncina, B. Biodegradation of natural textile materials in soil. Tekstilec 2014, 57, 118–132. [Google Scholar] [CrossRef]
- Li, L.; Frey, M.; Browning, K.J. Biodegradability study on cotton and polyester fabrics. J. Eng. Fibers Fabr. 2010, 5, 155892501000500406. [Google Scholar] [CrossRef]
- Alam, K.; Mazumder, A.; Sikdar, S.; Zhao, Y.-M.; Hao, J.; Song, C.; Wang, Y.; Sarkar, R.; Islam, S.; Zhang, Y. Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 2022, 13, 968053. [Google Scholar] [CrossRef]
- Gonzalez-Silva, A.; San Juan-Mendo, M.; Delgado-Prudencio, G.; Hernández-García, J.A.; Larios-Serrato, V.; Aguilar, C.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils. Microorganisms 2024, 12, 2637. [Google Scholar] [CrossRef]
- Vojnovic, S.; Aleksic, I.; Ilic-Tomic, T.; Stevanovic, M.; Nikodinovic-Runic, J. Bacillus and Streptomyces spp. as hosts for production of industrially relevant enzymes. Appl. Microbiol. Biotechnol. 2024, 108, 185. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, H.; Liu, Y.; Zhang, J.; Yu, H. Chitin biodegradation by lytic polysaccharide monooxygenases from Streptomyces coelicolor in vitro and in vivo. Int. J. Mol. Sci. 2022, 24, 275. [Google Scholar] [CrossRef] [PubMed]
- Herrera, D.A.G.; Mojicevic, M.; Pantelic, B.; Joshi, A.; Collins, C.; Batista, M.; Torres, C.; Freitas, F.; Murray, P.; Nikodinovic-Runic, J. Exploring microorganisms from plastic-polluted sites: Unveiling plastic degradation and PHA production potential. Microorganisms 2023, 11, 2914. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; Keller, M.B.; Paul, B.; Schubert, S.W.; Clausen, K.S.; Jensen, K.; Clarke, D.J.; Westh, P.; Dobson, A.D. Purification and biochemical characterization of SM14est, a PET-hydrolyzing enzyme from the marine sponge-derived Streptomyces sp. SM14. Front. Microbiol. 2023, 14, 1170880. [Google Scholar] [CrossRef]
- Verschoor, J.-A.; Croese, M.R.; Lakemeier, S.E.; Mugge, A.; Burgers, C.M.; Innocenti, P.; Willemse, J.; Crooijmans, M.E.; van Wezel, G.P.; Ram, A.F. Polyester degradation by soil bacteria: Identification of conserved BHETase enzymes in Streptomyces. Commun. Biol. 2024, 7, 725. [Google Scholar] [CrossRef]
- Pantelic, B.; Skaro Bogojevic, S.; Milivojevic, D.; Ilic-Tomic, T.; Lončarević, B.; Beskoski, V.; Maslak, V.; Guzik, M.; Makryniotis, K.; Taxeidis, G.; et al. Set of Small Molecule Polyurethane (PU) Model Substrates: Ecotoxicity Evaluation and Identification of PU Degrading Biocatalysts. Catalysts 2023, 13, 278. [Google Scholar] [CrossRef]
- Ilic-Tomic, T.; Genčić, M.S.; Živković, M.Z.; Vasiljevic, B.; Djokic, L.; Nikodinovic-Runic, J.; Radulović, N.S. Structural diversity and possible functional roles of free fatty acids of the novel soil isolate Streptomyces sp. NP10. Appl. Microbiol. Biotechnol. 2015, 99, 4815–4833. [Google Scholar] [CrossRef]
- Schneider, O.; Ilic-Tomic, T.; Rückert, C.; Kalinowski, J.; Genčić, M.S.; Živković, M.Z.; Stankovic, N.; Radulović, N.S.; Vasiljevic, B.; Nikodinovic-Runic, J. Genomics-based insights into the biosynthesis and unusually high accumulation of free fatty acids by Streptomyces sp. NP10. Front. Microbiol. 2018, 9, 1302. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.; Buttner, M.; Chater, K.; Hopwood, D. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000; pp. 44–61. [Google Scholar]
- Meddeb-Mouelhi, F.; Moisan, J.K.; Beauregard, M. A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzyme Microb. Technol. 2014, 66, 16–19. [Google Scholar] [CrossRef]
- Sourkouni, G.; Jeremić, S.; Kalogirou, C.; Höfft, O.; Nenadovic, M.; Jankovic, V.; Rajasekaran, D.; Pandis, P.; Padamati, R.; Nikodinovic-Runic, J. Study of PLA pre-treatment, enzymatic and model-compost degradation, and valorization of degradation products to bacterial nanocellulose. World J. Microbiol. Biotechnol. 2023, 39, 161. [Google Scholar] [CrossRef]
- Marković, D.; Tseng, H.-H.; Nunney, T.; Radoičić, M.; Ilic-Tomic, T.; Radetić, M. Novel antimicrobial nanocomposite based on polypropylene non-woven fabric, biopolymer alginate and copper oxides nanoparticles. Appl. Surf. Sci. 2020, 527, 146829. [Google Scholar] [CrossRef]
- Pantelic, B.; Araujo, J.A.; Jeremic, S.; Azeem, M.; Attallah, O.A.; Siaperas, R.; Mojicevic, M.; Chen, Y.; Fournet, M.B.; Topakas, E. A novel Bacillus subtilis BPM12 with high bis (2 hydroxyethyl) terephthalate hydrolytic activity efficiently interacts with virgin and mechanically recycled polyethylene terephthalate. Environ. Technol. Innov. 2023, 32, 103316. [Google Scholar] [CrossRef]
- Mutsuga, M.; Yamaguchi, M.; Kawamura, Y. Quantification of isocyanates and amines in polyurethane foams and coated products by liquid chromatography–tandem mass spectrometry. Food Sci. Nutr. 2014, 2, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Mutsuga, M.; Ohno, H.; Kawamura, Y.; Akiyama, H. Isolation and quantification of polyamide cyclic oligomers in kitchen utensils and their migration into various food simulants. PLoS ONE 2016, 11, e0159547. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Buchholz, P.C.; Feuerriegel, G.; Zhang, H.; Perez-Garcia, P.; Nover, L.L.; Chow, J.; Streit, W.R.; Pleiss, J. Plastics degradation by hydrolytic enzymes: The plastics-active enzymes database—PAZy. Proteins Struct. Funct. Bioinf. 2022, 90, 1443–1456. [Google Scholar] [CrossRef]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. PlasticDB: A database of microorganisms and proteins linked to plastic biodegradation. Database 2022, 2022, baac008. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C. Open-access bacterial population genomics: BIGSdb software, the PubMLST. org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Feeney, M.A.; Newitt, J.T.; Addington, E.; Algora-Gallardo, L.; Allan, C.; Balis, L.; Birke, A.S.; Castaño-Espriu, L.; Charkoudian, L.K.; Devine, R. ActinoBase: Tools and protocols for researchers working on Streptomyces and other filamentous actinobacteria. Microb. Genom. 2022, 8, 000824. [Google Scholar] [CrossRef] [PubMed]
- Consortium, C. Ten years of CAZypedia: A living encyclopedia of carbohydrate-active enzymes. Glycobiology 2018, 28, 3–8. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, X.; Duan, J.; Koide, R.T.; Xu, L.; Chu, J. Variation in microbial CAZyme families across degradation severity in a steppe grassland in northern China. Front. Environ. Sci. 2023, 11, 1080505. [Google Scholar] [CrossRef]
- Spasic, J.; Mandic, M.; Djokic, L.; Nikodinovic-Runic, J. Streptomyces spp. in the biocatalysis toolbox. Appl. Microbiol. Biotechnol. 2018, 102, 3513–3536. [Google Scholar] [CrossRef]
- Sintim, H.Y.; Bary, A.I.; Hayes, D.G.; Wadsworth, L.C.; Anunciado, M.B.; English, M.E.; Bandopadhyay, S.; Schaeffer, S.M.; DeBruyn, J.M.; Miles, C.A. In situ degradation of biodegradable plastic mulch films in compost and agricultural soils. Sci. Total Environ. 2020, 727, 138668. [Google Scholar] [CrossRef]
- Sülar, V.; Devrim, G. Biodegradation behaviour of different textile fibres: Visual, morphological, structural properties and soil analyses. Fibres Text. East. Eur. 2019, 27, 100–111. [Google Scholar] [CrossRef]
- Tomšič, B.; Marković, D.; Janković, V.; Simončič, B.; Nikodinovic-Runic, J.; Ilic-Tomic, T.; Radetić, M. Biodegradation of cellulose fibers functionalized with CuO/Cu 2 O nanoparticles in combination with polycarboxylic acids. Cellulose 2022, 29, 287–302. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Z.; Zhang, D.; Qiu, Y.; Wang, Y.; Quan, S.; Zhao, L. Exploring microbial degradation of polyamide 4 in soils: Unveiling degradation mechanisms, pathways, and the contribution of strain NR4. J. Clean. Prod 2023, 429, 139535. [Google Scholar] [CrossRef]
- Krasowska, K.; Janik, H.; Gradys, A.; Rutkowska, M. Degradation of polyurethanes in compost under natural conditions. J. Appl. Polym. Sci. 2012, 125, 4252–4260. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Z.; Ling, M.; Zhang, K.; Xu, W.; Xu, Z.; Huang, X.; Qiao, Y.; Luo, Y.; Zhang, W. Dynamics of plastisphere microbial communities in mangrove sediments and their potential impact on N-cycling. Int. Biodeterior. Biodegrad. 2025, 196, 105929. [Google Scholar] [CrossRef]
- Collie, S.; Brorens, P.; Hassan, M.; Fowler, I. Biodegradation behavior of wool and other textile fibers in aerobic composting conditions. Int. J. Environ. Sci. Technol. 2025, 22, 2113–2125. [Google Scholar] [CrossRef]
- Tiwari, N.; Santhiya, D.; Sharma, J.G. Significance of landfill microbial communities in biodegradation of polyethylene and nylon 6, 6 microplastics. J. Hazard. Mater. 2024, 462, 132786. [Google Scholar] [CrossRef]
- Wang, J.; Qiu, J.; Xu, S.; Li, J.; Shen, L. Electron beam irradiation influencing the mechanical properties and water absorption of polycaprolactam (PA6) and polyhexamethylene adipamide (PA66). RSC Adv. 2020, 10, 21481–21486. [Google Scholar] [CrossRef]
- Wang, H.; Feng, J.; Lu, J.; Li, R.; Lu, Y.; Liu, S.; Cavaco-Paulo, A.; Fu, J. Surface modification of polyamide fabric based on a multi-enzyme system. Fibers Polym. 2024, 25, 2585–2596. [Google Scholar] [CrossRef]
- Yang, I.-H.; Wu, X.-Y.; Chou, Y.-N. One-Step Zwitterionic Modification of Polyamide–Polyurethane Mixed Textile through Acidic Catalyzation. Langmuir 2025, 41, 8106–8116. [Google Scholar] [CrossRef]
- Kanelli, M.; Vasilakos, S.; Ladas, S.; Symianakis, E.; Christakopoulos, P.; Topakas, E. Surface modification of polyamide 6.6 fibers by enzymatic hydrolysis. Process Biochem. 2017, 59, 97–103. [Google Scholar] [CrossRef]
- Abidi, N.; Cabrales, L.; Haigler, C.H. Changes in the cell wall and cellulose content of developing cotton fibers investigated by FTIR spectroscopy. Carbohydr. Polym. 2014, 100, 9–16. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Zhang, S.; Gao, Q.; Lu, Q.; Peng, R.; Xu, P.; Shang, H.; Yuan, Y.; Zou, H. Micro-FTIR combined with curve fitting method to study cellulose crystallinity of developing cotton fibers. Anal. Bioanal. Chem. 2021, 413, 1313–1320. [Google Scholar] [CrossRef]
- Liu, Y. Recent progress in fourier transform infrared (FTIR) spectroscopy study of compositional, structural and physical attributes of developmental cotton fibers. Materials 2013, 6, 299–313. [Google Scholar] [CrossRef]
- Klun, U.; Friedrich, J.; Kržan, A. Polyamide-6 fibre degradation by a lignolytic fungus. Polym. Degrad. Stab. 2003, 79, 99–104. [Google Scholar] [CrossRef]
- Marjo, C.E.; Gatenby, S.; Rich, A.M.; Gong, B.; Chee, S. ATR-FTIR as a tool for assessing potential for chemical ageing in Spandex/Lycra®/elastane-based fabric collections. Stud. Conserv. 2017, 62, 343–353. [Google Scholar] [CrossRef]
- Smith, S.; Ozturk, M.; Frey, M. Soil biodegradation of cotton fabrics treated with common finishes. Cellulose 2021, 28, 4485–4494. [Google Scholar] [CrossRef]
- Abbas, M.; Adil, M.; Ehtisham-ul-Haque, S.; Munir, B.; Yameen, M.; Ghaffar, A.; Shar, G.A.; Tahir, M.A.; Iqbal, M. Vibrio fischeri bioluminescence inhibition assay for ecotoxicity assessment: A review. Sci. Total Environ. 2018, 626, 1295–1309. [Google Scholar] [CrossRef]
- Kennedy, G.L., Jr. Toxicity of Hexamethylenediamine (HMDA). Drug Chem. Toxicol. 2005, 28, 15–33. [Google Scholar] [CrossRef]
- Kennedy, G.L., Jr. Toxicity of adipic acid. Drug Chem. Toxicol. 2002, 25, 191–202. [Google Scholar] [CrossRef]
- Wiegand, C.; Hipler, U.-C. Evaluation of biocompatibility and cytotoxicity using keratinocyte and fibroblast cultures. Skin Pharmacol. Physiol. 2009, 22, 74–82. [Google Scholar] [CrossRef]
- de Witt, J.; Molitor, R.; Gätgens, J.; Ortmann de Percin Northumberland, C.; Kruse, L.; Polen, T.; Wynands, B.; van Goethem, K.; Thies, S.; Jaeger, K.E. Biodegradation of poly (ester-urethane) coatings by Halopseudomonas formosensis. Microb. Biotechnol. 2024, 17, e14362. [Google Scholar] [CrossRef]
- Schmidt, J.; Wei, R.; Oeser, T.; Dedavid e Silva, L.A.; Breite, D.; Schulze, A.; Zimmermann, W. Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers 2017, 9, 65. [Google Scholar] [CrossRef]
- de Witt, J.; Ostheller, M.-E.; Jensen, K.; van Slagmaat, C.A.; Polen, T.; Seide, G.; Thies, S.; Wynands, B.; Wierckx, N. Increasing the diversity of nylonases for poly (ester amide) degradation. Green Chem. 2024, 26, 9911–9922. [Google Scholar] [CrossRef]
- Heumann, S.; Eberl, A.; Fischer-Colbrie, G.; Pobeheim, H.; Kaufmann, F.; Ribitsch, D.; Cavaco-Paulo, A.; Guebitz, G.M. A novel aryl acylamidase from Nocardia farcinica hydrolyses polyamide. Biotechnol. Bioeng. 2009, 102, 1003–1011. [Google Scholar] [CrossRef]
- Bell, E.L.; Rosetto, G.; Ingraham, M.A.; Ramirez, K.J.; Lincoln, C.; Clarke, R.W.; Gado, J.E.; Lilly, J.L.; Kucharzyk, K.H.; Erickson, E. Natural diversity screening, assay development, and characterization of nylon-6 enzymatic depolymerization. Nat. Commun 2024, 15, 1217. [Google Scholar] [CrossRef]
- Branson, Y.; Söltl, S.; Buchmann, C.; Wei, R.; Schaffert, L.; Badenhorst, C.P.; Reisky, L.; Jäger, G.; Bornscheuer, U.T. Urethanasen für die enzymatische Hydrolyse niedermolekularer Carbamate und das Recycling von Polyurethanen. Angew. Chem. 2023, 135, e202216220. [Google Scholar] [CrossRef]
- Kanokratana, P.; Eurwilaichitr, L.; Pootanakit, K.; Champreda, V. Identification of glycosyl hydrolases from a metagenomic library of microflora in sugarcane bagasse collection site and their cooperative action on cellulose degradation. J. Biosci. Bioeng. 2015, 119, 384–391. [Google Scholar] [CrossRef]
- Lehninger, A.L.; Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry; Macmillan: London, UK, 2005. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janković, V.; Pantelic, B.; Ponjavic, M.; Marković, D.; Radetić, M.; Nikodinovic-Runic, J.; Ilic-Tomic, T. Degradation of Synthetic and Natural Textile Materials Using Streptomyces Strains: Model Compost and Genome Exploration for Potential Plastic-Degrading Enzymes. Microorganisms 2025, 13, 1800. https://doi.org/10.3390/microorganisms13081800
Janković V, Pantelic B, Ponjavic M, Marković D, Radetić M, Nikodinovic-Runic J, Ilic-Tomic T. Degradation of Synthetic and Natural Textile Materials Using Streptomyces Strains: Model Compost and Genome Exploration for Potential Plastic-Degrading Enzymes. Microorganisms. 2025; 13(8):1800. https://doi.org/10.3390/microorganisms13081800
Chicago/Turabian StyleJanković, Vukašin, Brana Pantelic, Marijana Ponjavic, Darka Marković, Maja Radetić, Jasmina Nikodinovic-Runic, and Tatjana Ilic-Tomic. 2025. "Degradation of Synthetic and Natural Textile Materials Using Streptomyces Strains: Model Compost and Genome Exploration for Potential Plastic-Degrading Enzymes" Microorganisms 13, no. 8: 1800. https://doi.org/10.3390/microorganisms13081800
APA StyleJanković, V., Pantelic, B., Ponjavic, M., Marković, D., Radetić, M., Nikodinovic-Runic, J., & Ilic-Tomic, T. (2025). Degradation of Synthetic and Natural Textile Materials Using Streptomyces Strains: Model Compost and Genome Exploration for Potential Plastic-Degrading Enzymes. Microorganisms, 13(8), 1800. https://doi.org/10.3390/microorganisms13081800






