Abstract
Contaminated poultry is one of the major sources of food-borne non-typhoidal Salmonella (NTS). The aim of this study was to evaluate the presence of Salmonella along the slaughter process in low- and high-throughput poultry abattoirs in South Africa and to determine their characteristics. Samples were collected from 500 chicken carcass rinsates at various processing stages in three abattoirs. Salmonella detection and identification was conducted in accordance with the ISO 6579 methodology. NTS serotyping was performed with serotype-specific PCRs. The Kirby–Bauer disk diffusion method was used to determine antimicrobial resistance in Salmonella. PCR was used to analyze thirteen antimicrobial genes and four virulence genes. Salmonella spp. was detected in 11.8% (59/500; CI: 9.5–15) of the samples tested. The predominant serovars were Salmonella Enteritidis (n = 21/59; 35.59%) and Salmonella Typhimurium (n = 35; 59.32%). Almost all Salmonella isolates were susceptible to all tested antimicrobials except three. Despite the low resistance to tetracyclines at the phenotypic level, approximately half of the strains carried tetA genes, which may be due to “silent” antimicrobial resistance genes. Diverse virulence genes were detected among the confirmed NTS serotypes. We found a predominance of S. Enteritidis and S. Typhimurium from chicken carcasses with diverse virulence and resistance genes. As we detected differences between the slaughterhouses, an in-depth study should be performed on the risk of Salmonella in low- and high-throughput abattoirs. The integrated monitoring and surveillance of NTS in poultry is warranted in South Africa to aid in the design of mitigation strategies.
1. Introduction
The Salmonella genus is divided into two species: Salmonella bongori and Salmonella enterica. Salmonella enterica is further categorized into six subspecies: S. enterica, S. salamae, S. arizonae, S. diarizonae, S. indica, and S. houtenae [1]. Approximately 99% of Salmonella human infections are caused by a small number of serotypes from the Salmonella enterica subspecies enterica group, even though over 2610 Salmonella enterica serotypes have been identified [2,3]. According to the World Health Organization, as well as preliminary analysis from The Institute for Health Metrics and Evaluation (IHME), diarrheal illnesses due to NTS accounted for approximately 73.9 million cases, resulting in 61,600 fatalities in 2019 [4]. Although African countries have fewer reported instances of NTS gastroenteritis than other regions, probably due to underreporting, the frequency of invasive non-typhoidal Salmonella (iNTS) remains high [5]. Especially in Sub-Saharan Africa, NTS causes invasive salmonellosis and bacteraemia [6,7,8]. In Africa, the most common serovars are Salmonella Enteritidis and Salmonella Typhimurium, accounting for 26% and 25% of all Salmonella infections, respectively [9,10]. From 2013 to 2015, six of seven Salmonella enterica Enteritidis outbreaks in South Africa were foodborne [10].
It was shown before that animal-derived food products are the primary carriers of NTS [11,12]. Poultry meat accounts for most of the foodborne salmonellosis cases [13,14,15]. Salmonella is found in chicken gastrointestinal tracts, as well as the oviducts, and may contaminate the carcass during slaughter [16,17]. In modern abattoirs, the rapid line speed during processing keeps the birds in close proximity, which increases the potential of microbial cross-contamination between infected and uninfected carcasses [18]. In South Africa, the Meat Safety Act of 2000 (Act No. 40 of 2000) regulates the hygiene and quality of meat, as well as the quantity of units that can be slaughtered [19]. The Foodstuffs, Cosmetics, and Disinfectants Act of 1972 (Act No. 54 of 1972) further regulates microbiological food monitoring [20].
Antibiotics are used to treat Salmonella infections. However, in mild infections, antibiotic therapy is not recommended as salmonellosis is a self-limiting disease [21]. There are numerous reports regarding antimicrobial resistance among Salmonella [22,23,24,25,26,27,28,29]. Drivers of antimicrobial resistance in Salmonella include horizontal gene transfer, environmental factors, and the use of antibiotics in human and veterinary medicine [30]. Generally, bacteria resist antimicrobial agents through the enzymatic inactivation of antimicrobials, the modification of target proteins receptors, and extrusion by the efflux pump [31,32,33,34]. Third-generation cephalosporin resistance is of major public health importance, which is generally the result of β-lactam ring disruption by enzymes encoded by mainly a multitude of variants of blaCMY, blaTEM, blaSHV, blaPSE, blaOXA, and blaCTX-M genes [35]. Another major antimicrobial resistance of public health importance is fluroquinolone resistance, which is mediated by point mutations in the Salmonella gyrA and/or parC genes [1,36]. Taking into consideration that the infection in humans originates mainly from food animals or foods contaminated by products of food animals, antimicrobial resistance in Salmonella from food animals is a major public health issue [37,38]. The resistance genes can be found on plasmids, gene cassettes in integrons, or other mobile genetic elements (MGE) [1,39]. MGEs can be transferred between serovars and to other bacterial genera through horizontal gene transfer, resulting in the spread of resistance [40,41].
The main virulence factors of Salmonella enterica are located on the chromosomal Salmonella pathogenicity islands (SPI), which are large mobile genetic elements (MGE), capable of acquiring genes as gene cassettes [42,43,44]. There are 24 Salmonella pathogenicity islands that have been identified [45,46]; however, it is worth noting that not all these SPIs have been scientifically validated to contribute to phenotypic virulence [47,48,49], with SPI-1, SPI-2, SPI-3, SPI-4, and SPI-5 being abundant in most Salmonella serovars [45,50,51,52]. SivH is a pathogenicity determinant implicated in the intracellular survival of Salmonella. The high prevalence of SivH genes can be attributed to their association with islands that are specific to Salmonella capable of infecting warm-blooded animals [53].
South Africa has legislation in place that makes it a contravention to obtain prescribed antimicrobial medicines without a prescription; however, the Farm Feeds, Agriculture Remedies, Fertilizers, and Stock Remedies Act (Act No. 36, 1947 [54]) permits the acquisition of listed stock remedies for livestock treatment without a prescription [55]. In low- and middle-income countries (LMICs) such as South Africa, there is little information regarding the occurrence and distribution of NTS on chicken carcasses during slaughtering process and its antimicrobial resistance patterns [11,56,57]. Therefore, the aim of this study was to evaluate the presence of Salmonella along the slaughter process in low- and high-throughput poultry abattoirs in South Africa and to determine their characteristics.
2. Materials and Methods
2.1. Ethical Approval
Ethical approval for this study was obtained from University of Zululand research ethics committee with reference number UZREC 171110-030 PGM 2022/61. This study was a part of an umbrella project for Prof. Evelyn Madoroba, with ethics approval reference UZREC 171110-030 Dept 2022/11.
2.2. Study Design, Study Site, and Sample Collection
This study was conducted on abattoirs that were randomly selected based on the 2019 European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDPC) [58] reports for monitoring foodborne pathogens. This study used the sample size determination equation for cross-sectional analysis to determine the appropriate sample size [59,60].
is standard normal variant (5% type 1 error, i.e., 1, 96), p is the estimated prevalence from previous studies, and d is the unconditional error (p < 0.05).
= 384 poultry samples
Assuming a prevalence of 50%, the required minimum sample size is 384. To increase the robustness of the conclusions, the poultry sample size for this study was 500.
In South Africa, abattoirs are divided into three categories: rural, high-throughput (HT), and low-throughput (LT) [19]. Between July and September 2022, 500 chicken rinsates were sampled from two low-throughput and one high-throughput abattoir. Rinsate samples were collected randomly at three processing stages (scalding, evisceration, final water). This was performed to have good representation of Salmonella prevalence at different critical stages in KZN abattoirs. There was no particular stratification. Scalding was conducted at 50–55 °C in order to loosen the chicken feathers and to ensure that the carcasses are not over scalded [19]. Prior to evisceration, the chicken carcasses were washed according to the provisions in South Africa’s Foodstuffs, Cosmetics, and Disinfectants Act, 1972 (Act No. 54 of 1972). Evisceration was conducted in a hanging position on the evisceration line. After evisceration, the chicken carcasses were washed with cold water according to the provisions of South Africa’s Foodstuffs, Cosmetics, and Disinfectants Ad, 1972 (Act No. 54 of 1972). The samples for scalding water were collected immediately from a channel. Evisceration samples were taken from water obtained from washing individual chicken carcasses. Chicken rinsates were collected by placing the sterile containers below the carcasses as they were washed on a line.
Chicken carcass rinsates were aseptically placed in sterile Stomacher bags and delivered to the University of Zululand main campus in Kwa-Dlangezwa on ice for microbiological testing and analysis within 12 h. Figure 1 illustrates the schematic representation of the workflow used for this study.
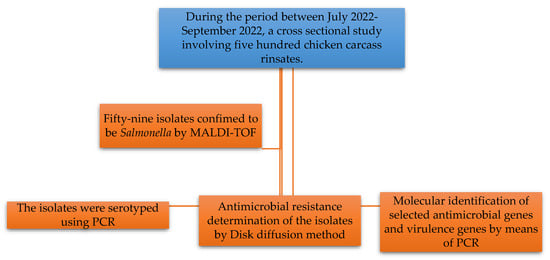
Figure 1.
Schematic representation of the workflow used for this study.
2.3. Microbiological Analysis and Cryopreservation of Isolates
Isolation and Identification of Salmonella
Detection and phenotypic confirmation of Salmonella was performed according to the ISO-6579-1:2017/Amd 1:2020 International Organization for Standardization (ISO-6579-1:2017/Amd 1:2020 [61] protocol with slight modification. Briefly, the method involved pre-enrichment whereby 10 mL of each sample was measured and aseptically transferred into 90 mL of the pre-enrichment broth, buffered peptone water (BPW) (Oxoid Ltd., Hampshire, UK), and incubated at 37 °C for 18–24 h. Selective enrichment was performed after incubation, whereby 0.1 mL and 1 mL aliquots of the samples were inoculated into 10 mL Rappaport–Vassiliadis soy broth (RVS) (Oxoid, UK) and Muller–Kauffmann tetrathionate-novobiocin broth (MKTTn), respectively (Oxoid, UK), followed by incubation at 37 ± 2 °C and 42 ± 2 °C for 24 ± 2 h, respectively. Loopfuls of the samples from the selective broths (MKTTn and RVS) were streaked onto xylose lysine deoxycholate (XLD) agar (Oxoid, UK) and Bismuth sulphide agar (BSA) (Oxoid, UK), followed by incubation at 37 ± 2 °C for 24 ± 2 h. Colonies that were pink and black centred on XLD and had black metallic sheen on BSA were considered presumptive Salmonella.
Presumptive isolates were purified on XLD and on nutrient agar (Oxoid, UK), followed by incubation at 37 ± 2 °C for 24 ± 2 h. All presumptive Salmonella were cryopreserved in stock cultures of nutrient broth supplemented with sterile glycerol to a final concentration of 50%, followed by storage at −80 °C until required. The presumptive isolates were also preserved in 1 mL of 10% skimmed milk and stored at −80 °C.
2.4. Salmonella Identification Using MALDI-TOF MS
All preserved Salmonella isolates were revived and sub-cultured on XLD to ensure purity. The revived presumptive Salmonella isolates were subjected to MALDI-TOF MS for species confirmation according to the manufacturer’s instruction [62,63].
2.5. PCR Serotyping of Salmonella Isolates
The predominant Salmonella serovars in poultry are usually Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium in Africa [6]; hence, this study focused on these two serovars. Conventional PCR was undertaken to screen for S. Enteritidis and S. Typhimurium. The forward and reverse primer sequences for each gene are shown in Table 1.

Table 1.
Primer sequences used to screen for S. Enteritidis and S. Typhimurium using PCR.
A 20 µL master mix consisting of 10 µL NEB OneTaq 2X MasterMix with Standard Buffer (Inqaba Biotec, Pretoria, South Africa; Catalogue No. M0482S), 1 µL of genomic DNA (10–30 ng/μL), 1 µL forward primer (10 μM), and 1 µL reverse primer (10 μM) was used. The volume was made up to 20 µL with nuclease-free water (Inqaba Biotec; Catalogue No. E476). Thermocycling of the PCR mixture was conducted under the following conditions: initial denaturation at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s; extension at 68 °C for 1 min; final elongation at 68 °C for 10 min; and the amplicons were kept at 4 °C for further analysis, as previously described [27].
2.6. Agarose Gel Electrophoresis of PCR Amplicons
The PCR amplicons were electrophoresed in a 1.5% agarose gel (CSL-AG100, Cleaver Scientific Ltd, Rugby, UK) and stained with ethidium bromide at 90 V for approximately 30 min. A 100 bp NEB Fast Ladder (N3238; (Inqaba Biotec, Pretoria, South Africa)) was used to estimate the size of the amplicons. The results were viewed and documented using the VILBER (E-Box; Vilber, Eberhardzell, Germany) gel documentation system.
2.7. Antimicrobial Susceptibility
The Kirby–Bauer disk diffusion method [66] was used to determine the antimicrobial susceptibility of confirmed Salmonella isolates. The strains were tested against commonly used antibiotics in poultry farming [58]: ampicillin (10 µg), chloramphenicol (30 µg), trimethoprim–sulfamethoxazole, oxytetracycline (10 µg), cefotaxime, gentamicin (10 µg), ciprofloxacin, and kanamycin (30 µg) (Davies Diagnostic, Randburg, South Africa). Briefly, four to five colonies were inoculated into 5 mL of sterile physiological saline solution, and the turbidity of the suspension was adjusted to match the 0.5 McFarland standard. Subsequently, the standardized Salmonella bacterial suspensions were swabbed onto Mueller–Hinton agar plates (Thermo-Fisher, Waltham, MA, USA) in three directions, rotating the plate by approximately 60° angles between each swabbing to ensure uniform distribution. Afterwards, the inoculated medium was allowed to dry for about five to ten minutes, and antibiotic disks were applied on the Mueller Hinton agar. Following a 16 h incubation period at 37 °C ± 2, the resulting zones of inhibition were measured in millimetres (mm). The obtained results were then classified as susceptible (S), intermediate (I), or resistant in accordance with the guidelines set by the Clinical Laboratory Standards Institute (CLSI) [66].
2.8. Evaluation of Antimicrobial Resistant Genes Using PCR
DNA Extraction
The conventional cell-lysis (boiling) method was used as previously described [25]. Briefly, the Salmonella isolates were resuscitated on nutrient agar and incubated at 37 ± 2 °C for 24 ± 2 h. A loopful of the culture medium was inoculated into an Eppendorf tube containing 1 mL of sterile distilled water. The inoculum was thoroughly mixed and then placed in a heating block at 99 °C for 10 min. It was then centrifuged at 13,000 rpm for 5 min. The pellet was discarded, and the DNA-containing supernatant was transferred to a sterile tube for PCR amplification of antimicrobial genes and virulence genes.
Multiplex PCR was used for the screening of selected antimicrobial resistance genes of Salmonella strains. Twelve antimicrobial resistance genes previously described by Lauteri and co-workers [67] were screened with slight modifications: blaTEM and blaPSE for beta-lactam resistance; aadA2, aac(3)IV, and aadB for aminoglycoside resistance; catA1 for chloramphenicol resistance; tetA, tetB, and tetC for tetracycline resistance; dfrA1 and dfrA14 for trimethoprim resistance. The forward and reverse primer sequences for each gene is shown in Table 2.

Table 2.
Primer sequences used to screen for each of the thirteen antimicrobial resistance genes in Salmonella spp. using multiplex PCR *.
The 25 µL PCR mixtures consisted of 12.5 μL NEB OneTaq 2X MasterMix with standard buffer (Inqaba Biotec, Pretoria, South Africa; Catalogue No. M0482S), 5 µL Genomic DNA (15–150 ng/μL), 1 μL of each of the forward and reverse primer, and the volume was topped up with molecular-grade water (Inqaba Biotec, Pretoria, South Africa; Catalogue No. E476). The thermocycling conditions consisted of 94 °C for 5 min of initial denaturation, 35 cycles at 94 °C for 30 s of denaturation, 50 °C for 30 s, 68 °C for 1 min of elongation, and 68 °C for 10 min of final extension. Agarose gel electrophoresis of PCR amplicons was conducted as described in Section 2.6.
2.9. Evaluation of Salmonella spp. Virulence Genes
The virulence genes screened in this study were previously described by Siddiky et al. (2021) [72] and included invasion proteinA (invA), aggregative fimbriae A (agfA), long polar fimbrial A (lpfA), and intimin-like inverse autotransporter protein H (SivH). The forward and reverse primer sequences for each gene is shown in Table 3.

Table 3.
Primer sequences to screen for each of the four virulence genes in Salmonella spp. using PCR *.
A 25 µL PCR master mixture was prepared consisting of 12.5 μL NEB OneTaq 2X MasterMix with standard buffer (Inqaba Biotec; Catalogue No. M0482S), 5 µL crude DNA (50–150 ng/μL), 5.5 µL molecular-grade water (Inqaba Biotec; Catalogue No. E476), and 1 μL of each of the forward and reverse primers. For amplification, the mixtures underwent thermocycling under the following conditions: for initial denaturation, 94 °C for 5 min; followed by 35 cycles of denaturation at 94 °C for 30 s; annealing at 50 °C for 30 s; elongation at 68 °C for 1 min; and final extension was at 68 °C for 10 min.
2.10. Agarose Gel Electrophoresis of PCR Amplicons
The PCR amplicons were electrophoresed on 1.5% agarose gels stained with ethidium bromide at 90 V for approximately 30 min. A 100 bp NEB Fast Ladder (N3238; (Inqaba Biotec, Pretoria, South Africa)) was used to estimate the size of the amplicon. The results were viewed and documented using VILBER (E-Box; Vilber, Eberhardzell, Germany) gel documentation.
2.11. Quality Control
To ensure the accuracy and validity of the results obtained in this study, reference strains Salmonella Typhimurium ATCC14028 and Escherichia coli ATCC 25922 were included in all experiments, as positive and negative controls, respectively [25]. Previous positive isolates obtained from Prof. Madoroba’s research group [23,27] were used as positive controls for virulence genes. Standard method validation approaches for all methods were undertaken.
2.12. Statistical Analysis
Confidence intervals (CI) were calculated using binomial distribution in Excel. The results of the prevalence of Salmonella spp. were compared between abattoirs using the chi square test. A p-value < 0.05 was considered statistically significant.
3. Results
3.1. MALDI-TOF Identification
From the eighty-one presumptive Salmonella isolates, fifty-nine Salmonella isolates were confirmed to be Salmonella. As a result, the overall prevalence of Salmonella in KZN abattoirs was 11.8% (59/500, CI: 9.5–15). Salmonella prevalence was reported to be 39.58% (19/48) in LT abattoir A rinsates, and 17.32% (40/231) in HT abattoir rinsates, with no Salmonella identified on chicken carcass rinsates in LT abattoir B. Salmonella occurrence was the highest during evisceration stage (21.01%), followed by final washing (10.28%), and no Salmonella was isolated during scalding. Table 4 summarizes the prevalence of Salmonella isolates from various processing stages in KZN abattoirs.

Table 4.
Prevalence of Salmonella in different chicken processing stages classified according to the abattoir category.
3.2. PCR Serotyping
Of the 59 Salmonella isolates, 35 were S. Typhimurium (59.3%, 95% CI 45.7–72), 21 were S. Enteritidis (35.6%, 95% CI 23.6–49), and only 3 were neither S. Typhimurium nor S. Enteritidis. The prevalences of S. Enteritidis and S. Typhimurium were found to be 22.03% (13/59) and 8.47% (5/59) in LT A, respectively. Similarly, the prevalences of S. Enteritidis, S. Typhimurium, and other serovars were represented as 13.56% (8/59), 51% (30/59), and 5.08% (3/59) in the HT abattoir, respectively (Figure 2). All genes that were screened in this study are illustrated in Figure 3 and Figure 4.
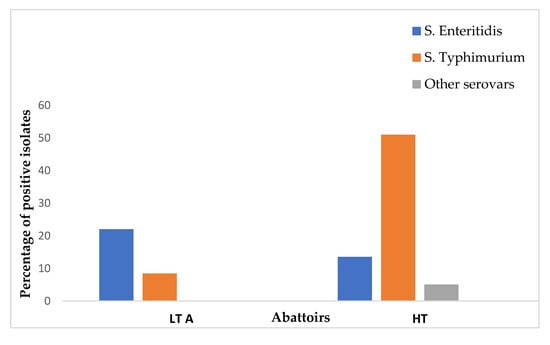
Figure 2.
Distribution of Salmonella serovars of chicken carcass rinsates from low-throughput and high-throughput abattoirs in KZN.
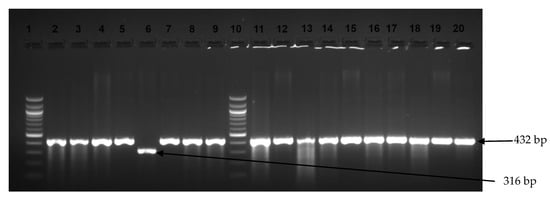
Figure 3.
Image showing examples of Salmonella Typhimurium and S. Enteritidis amplicons observed on an agarose gel. Lanes 1 and 10 show the 100 bp DNA ladder, while lanes 2–5, 7–9, and 11–20 display positive amplification for S. Typhimurium (432 bp). Lane 6 shows S. Enteritidis amplicons (316 bp).
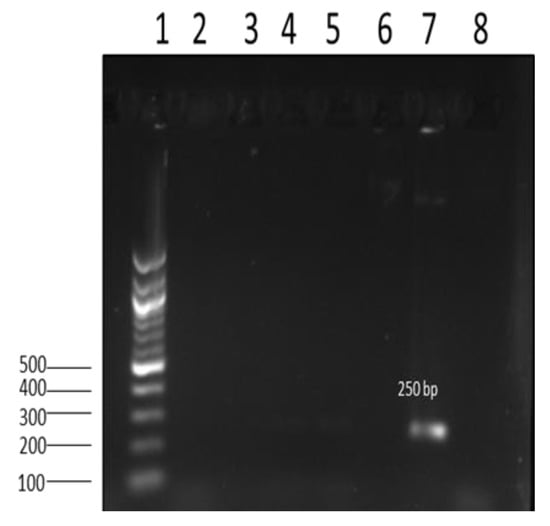
Figure 4.
Image showing amplicon of antimicrobial resistance gene aad2 (250 bp) from Salmonella isolates. Lane 1: 100 bp DNA ladder; lane 2–6; negative results for the aad2 gene; lane 7: positive 250 bp aad2 gene; lane 8: nuclease-free water (negative control).
3.3. Antimicrobial Susceptibility Testing
We found a low presence of antimicrobial resistance. All of the isolates from the LT slaughterhouse were susceptible. However, in the HT abattoir, a multi-resistant S. Enteritidis strain with the phenotypic-pattern ampicillin, oxytetracycline, cefotaxime, and trimethoprim/sulphonamide, as well as two S. Typhimurium strains, were resistant to oxytetracycline and ampicillin, while the remaining isolates were susceptible to all antibiotics.
3.4. Evaluation of Virulence Genes Using PCR
Table 5 shows the distribution of the screened virulence genes according to their serotypes and origin. All S. Enteriditis were positive for invA, while two S. Typhimurium strains were negative. Other virulence genes tested were present in 40-to-100% (sivH in S. Typhimurium) of the strains. The strains with an unknown serotype were positive for nearly all virulence genes except agfA.

Table 5.
Distribution of virulence genes in Salmonella enterica serotypes from chicken carcasses in LT A and HT abattoirs.
3.5. Evaluation of Antimicrobial Resistance Genes Using PCR
Three of the ten AMR genes tested were found in Salmonella isolates recovered from the two (LT A and HT) abattoirs. Overall, the most common AMR genes were tetA 54.2% (32/59), aadA2 (aminoglycosides) at 3.4%, and blaTEM (beta-lactam) at 1.7%. All of the isolates tested negative for the following AMR genes: aac(3)IV, aadB (aminoglycosides), blaPSE (beta-lactam), tetB, tetC (Tetracycline), catA1 (phenicol), and dfrA1 (trimethoprim) (Table 6).

Table 6.
Frequency of AMR genes in S. Typhimurium, S. Enteritidis, and other serotypes in different chicken abattoirs.
4. Discussion
Non-typhoidal Salmonella is a significant zoonotic pathogen posing a threat to human health [73]. Salmonella spp. was detected in 11.8% of the 500 rinsates from chicken carcasses collected during this study. The presence of NTS in chicken rinsates in this study is concerning because it can lead to the cross-contamination of poultry and the environment through contaminated water sources. These contaminated sources may lead to non-typhoidal salmonellosis, which is listed in the South African National Health Act, 2003 (Act No. 61 of 2003), under Category 3 of notifiable medical conditions. Furthermore, S. Enteritidis, which was one of the predominant serovars in this study, is listed in South Africa as a controlled disease on the List of Controlled and Notifiable Animal Diseases in Terms of the Animal Diseases Act, 1984 (Act No. 35 of 1984). Therefore, routine surveillance and strict hygiene should be practiced during the slaughter of chickens as abattoirs may serve as potential sources of Salmonella contamination in humans.
The rate of Salmonella occurrence in this study was found to be higher compared to a previous study in KwaZulu-Natal, which reported a prevalence 0.6% (1/162) Salmonella from carcass rinsates at an abattoir [11]. The difference in prevalence between our study and that of Ramtahal et al. (2022) [11] could be due to a variety of differences, including, but not limited to, the sample size (500 samples from this study compared to 162), the source of the samples, and the sampling strategy.
We found a higher prevalence in HT abattoirs, followed by the LT A abattoir, with no Salmonella occurrence in the LT B abattoir. In high-throughput slaughterhouses, contamination risks can be particularly high due to the large number of animals processed, the use of automation, and the high throughput rate. The speed of processing and high volume of animals can make it difficult to maintain adequate hygiene practices and to monitor for contamination effectively. In the low-throughput abattoir A, Salmonella contamination could be a result of a lack of resources, inadequate facilities, and limited regulatory oversight. No Salmonella was detected in the low-throughput B abattoir, which could suggest that good hygiene practices were observed during the processing of the flocks or that the chickens were not contaminated during production at the farm or during transportation to the abattoir. Further studies are necessary to verify the data as only a few slaughterhouses were included in this study, and there may be differences in operationality between the slaughterhouses. Studies accounting for these differences would be expected to determine better practices.
The reduction of Salmonella at the final carcass-washing stage is a clear indicator that interventions at the point of carcass washing can reduce the level of the bacterial contamination of carcasses to below detection. No Salmonella was isolated from the scalding water, which is perhaps an indication of the implementation of good hygiene measures in slaughtering and processing [25]. The temperature of the scalding water could be a hurdle that reduces the bacterial contamination of chicken carcasses.
The predominant Salmonella serovars identified in this study were S. Enteritidis and S. Typhimurium. These serovars have been shown to be predominant in different studies and have been associated with most cases of human salmonellosis. In South Africa, a retrospective study involving the incidence of Salmonella from different animal species from 1996 to 2006 revealed that out of the 183 serovars, S. Typhimurium was predominant, followed by S. Dublin and S. Enteritidis [74]. A study that was undertaken in Iran to determine Salmonella species in poultry markets revealed S. Typhimurium as the dominant and only serovar that was recovered [75]. A systematic review and meta-analysis in South Africa that used pooled prevalence to assess the “One Health” perspective of Salmonella serovars found S. Typhimurium and S. Enteritidis to constitute a relatively large proportion [9]. In Saudi Arabia, the prevalences of S. Enteritidis and S. Typhimurium associated with human infections in Riyadh were 40% and 13.5%, respectively [76].
While most of the Salmonella isolates from this study were susceptible to the antibiotics tested, one S. Enteritidis strain showed multi-drug resistance (MDR). Previously in KZN, all of the Salmonella isolates from wastewater, and 72.4% of carcass rinsates were susceptible to all of the tested antibiotics [11], while a study covering South Africa [56] found a high prevalence of resistance to tetracycline (93%), kanamycin (74%), trimethoprim–sulfamethoxazole (50%), gentamicin (48%), ampicillin (47%), and chloramphenicol (31%). The MDR S. Enteritidis strain from this study was also resistant to cefotaxime. The presence of resistance against third-generation cephalosporins is a worrisome observation as these strains may be selected by most beta-lactam antibiotics, as well as through co-resistance selection. Cefotaxime resistance is highly concerning as this antimicrobial is considered a potent treatment for septicaemic salmonellosis in adult humans. The emergence of resistant isolates highlights the urgent need for regular surveillance to prevent the spread of resistant strains.
Resistance to tetracyclines was low at the phenotypic level, but about half of the strains showed the presence of tetA genes. These are referred to as “silent” or “cryptic” antimicrobial resistance genes [77,78,79,80]. The reasons for this discrepancy in phenotypic and genotypic occurrence remain under investigation. Nevertheless, the presence of tetA genes does not always imply that the bacteria show phenotypic resistance [77,78,79,80]. However, “silent” antimicrobial resistance genes may have public health consequences because when the conditions are suitable, the “silent” genes may be expressed and regain resistance, which may lead to the failure of therapeutic agents [77]. Some of the resistance genes that were observed from this study were previously identified from other similar studies albeit from different sampling sites in South Africa [55,56]. It is important to use a holistic multidisciplinary “One Health” approach that takes into consideration the health of the animals, the environment, and the health of human beings to reduce microbial contamination and curb the spread of antimicrobial resistance [81,82].
Virulence genes play a crucial role in the pathogen’s ability to survive and multiply within the host cell. In this study, invA was the most prevalent virulence gene. This gene is located inside the Salmonella pathogenicity island-1 (SPI-1) [83] and is associated with the Type III Secretion System (T3SS), which aids in host cell invasion. The invA gene also serves as a biomarker for NTS Salmonella as it is present in nearly all Salmonella enterica subspecies enterica [83]. In this study, two isolates did not carry the invA gene. It has been reported that invA gene-negative Salmonella strains are rare and have been observed in few studies [23,84]. In South Africa, Naidoo et al. (2022) observed invA-negative Salmonella enterica Heidelberg from beef tripe [23]. As the invA gene plays a role in the invasion of epithelial cells during infection, Salmonella strains that are negative for the invA gene may invade epithelial cells using other mechanisms [85].
The agfA and IpfA genes are crucial to the infection process as they encode fimbriae, which play a vital role in the attachment process [72]. agfA is also involved in the process of biofilm formation, and the presence of this gene in Salmonella represents a risk of persistence on slaughterhouse equipment and other surfaces [86]. Other studies have also documented the occurrence of the agfA gene in S. Enteriditis and S. Typhimurium [23,27]. sivH (also known as sinH), an intimin-like inverse autotransporter protein, is involved in the internal colonization and persistence of Salmonella [72], indicating that most of the strains in this study are capable of maintaining their presence in the poultry population.
5. Conclusions
In this study, we determined the occurrence, serotypes, phenotypical antimicrobial resistance profiles, selected antibiotic resistance genes, and virulence factors of NTS in chicken rinsates from abattoirs located in selected areas of KwaZulu-Natal province, South Africa. Our findings show that chicken meat serves as a potential carrier of zoonotic Salmonella spp. With the exception of the identified S. Enteritidis MDR isolate, low antimicrobial resistance was observed in this study. However, despite the low resistance to tetracyclines at the phenotypic level, almost half of the strains showed the presence of tetA genes, which may be due to “silent” or “cryptic” antimicrobial resistance genes. The Salmonella harbored diverse virulence genes, which is important for pathogenicity and infectivity. Therefore, higher standards of basic hygiene, hygiene awareness, and proficient food management abilities in the abattoirs are recommended to reduce cross-contamination. Cross contamination was more pronounced in HT slaughterhouses and slaughter processes; therefore, hygiene and hazard analysis and critical control points should be reanalyzed and improved to reduce contamination levels. Further molecular epidemiology studies are necessary, including detailed characterization of NTS strains with whole-genome sequencing, which is critical for the prevention, reduction, and elimination of contaminants in the food chain.
Author Contributions
Conceptualization, E.M. and P.B.; methodology, B.B.M.; software, B.B.M.; validation, E.M. and P.B.; formal analysis, B.B.M.; investigation, B.B.M.; resources, E.M.; data curation, E.M. and P.B.; writing—original draft preparation, B.B.M.; writing—review and editing, E.M., P.B. and K.M.; visualization, E.M.; supervision, E.M. and P.B.; project administration, E.M.; funding acquisition, E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Foundation, South Africa, grant number reference SRUG190411429382, with unique grant (number 120838) awarded to Evelyn Madoroba. B.B.M was funded by Moses Kotane Institute.
Institutional Review Board Statement
This study was approved by University of Zululand Research Ethics Committee (UZREC). The reference number is UZREC 171110-030 Dept 2022/11, and the approval date is 20 September 2022.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors are indebted to all stakeholders who contributed to the success of this study. We are grateful to the University of Zululand for providing the facilities to undertake research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Eng, S.K.; Pusparajah, P.; Ab Mutalib, N.S.; Ser, H.L.; Chan, K.G.; Lee, L.H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Gupta, S.K.; Sharma, P.; McMillan, E.A.; Jackson, C.R.; Hiott, L.M.; Woodley, T.; Humayoun, S.B.; Barrett, J.B.; Frye, J.G.; McClelland, M. Genomic comparison of diverse Salmonella serovars isolated from swine. PLoS ONE 2019, 14, e0224518. [Google Scholar] [CrossRef] [PubMed]
- Pulford, C.V.; Wenner, N.; Redway, M.L.; Rodwell, E.V.; Webster, H.J.; Escudero, R.; Kröger, C.; Canals, R.; Rowe, W.; Lopez, J. The diversity, evolution and ecology of Salmonella in venomous snakes. PLoS Negl. Trop. Dis. 2019, 13, e0007169. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Nontyphoidal Salmonella Disease. Available online: https://www.who.int/teams/immunization-vaccines-and-biologicals/diseases/nontyphoidal-salmonella-disease (accessed on 15 September 2024).
- Crump, J.A.; Heyderman, R.S. A perspective on invasive Salmonella disease in Africa. Clin. Infect. Dis. 2015, 61, S235–S240. [Google Scholar] [CrossRef]
- Feasey, N.A.; Dougan, G.; Kingsley, R.A.; Heyderman, R.S.; Gordon, M.A. Invasive non-typhoidal Salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 2012, 379, 2489–2499. [Google Scholar] [CrossRef]
- Gilchrist, J.J.; MacLennan, C.A. Invasive nontyphoidal Salmonella disease in Africa. EcoSal Plus 2019, 8, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Uche, I.V.; MacLennan, C.A.; Saul, A. A systematic review of the incidence, risk factors and case fatality rates of invasive Nontyphoidal Salmonella (iNTS) disease in Africa (1966 to 2014). PLoS Negl. Trop. Dis. 2017, 11, e0005118. [Google Scholar] [CrossRef]
- Ramatla, T.; Lekota, K.E.; Tawana, M.; Onyiche, T.E.; Thekisoe, E. One Health Perspective of Salmonella Serovars in South Africa Using Pooled Prevalence: Systematic Review and Meta-Analysis. Int. J. Microbiol. 2022, 2022, 8952669. [Google Scholar] [CrossRef]
- Muvhali1, M.; Smith, A.M.; Moipone Rakgantso, A.M.; Keddy, K.H. Investigation of Salmonella Enteritidis outbreaks in South Africa using multi-locus variable-number tandem-repeats analysis, 2013–2015. BMC Infect. Dis. 2017, 17, 661. [Google Scholar] [CrossRef]
- Ramtahal, M.A.; Somboro, A.M.; Amoako, D.G.; Abia, A.L.K.; Perrett, K.; Bester, L.A.; Essack, S.Y. Molecular Epidemiology of Salmonella enterica in Poultry in South Africa Using the Farm-to-Fork Approach. Int. J. Microbiol. 2022, 2022, 5121273. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef]
- Gonçalves-Tenório, A.; Silva, B.N.; Rodrigues, V.; Cadavez, V.; Gonzales-Barron, U. Prevalence of pathogens in poultry meat: A Meta-Analysis of European Published Surveys. Foods 2018, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, B.; Mawad, A.M.M.; Saleh, M.; Kelley, W.G.; Harrington, P.J.; Lovestad, C.W.; Amezcua, J.; Sarhan, M.M.; El Zowalaty, M.E.; Ramadan, H. Salmonellosis: An overview of epidemiology, pathogenesis, and innovative approaches to mitigate the antimicrobial resistant infections. Antibiotics 2024, 13, 76. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar]
- Afema, J.A.; Byarugaba, D.K.; Shah, D.H.; Atukwase, E.; Nambi, M.; Sischo, W.M. Potential sources and transmission of Salmonella and antimicrobial resistance in Kampala, Uganda. PLoS ONE 2016, 11, e0152130. [Google Scholar] [CrossRef]
- Dione, M.M.; Geerts, S.M.S.; Antonio, M. Characterisation of novel strains of multiply antibiotic-resistant Salmonella recovered from poultry in Southern Senegal. J. Infect. Dev. Ctries 2012, 6, 436–442. [Google Scholar] [CrossRef]
- Akil, L.; Ahmad, H.A. Quantitative Risk Assessment Model of Human Salmonellosis Resulting from Consumption of Broiler Chicken. Diseases 2019, 7, 19. [Google Scholar] [CrossRef]
- Republic of South Africa Government Gazette 425 (21707). Meat Safety Act of 2000 (Act No. 40 of 2000). Available online: https://www.gov.za/sites/default/files/gcis_document/201409/a40-000.pdf (accessed on 1 March 2025).
- Republic of South Africa Government Gazette. Foodstuffs, Cosmetics, and Disinfectants Act of 1972 (Act No. 54 of 1972); Amended 2010. Available online: https://www.gov.za/sites/default/files/gcis_document/201504/act-54-1972.pdf (accessed on 1 March 2025).
- Hussain, M.A.; Wang, W.; Sun, C.; Gu, L.; Liu, Z.; Yu, T.; Ahmad, Y.; Jiang, Z.; Hou, J. Molecular Characterization of pathogenic Salmonella spp. from raw beef in Karachi, Pakistan. Antibiotics 2020, 9, 73. [Google Scholar] [CrossRef] [PubMed]
- Forgaciu, A.; Tabaran, A.; Colobatiu, L.; Mihaiu, R.; Dan, S.D.; Mihaiu, M. Concerning Increase in Antimicrobial Resistance Patterns of Pathogenic Strains of Salmonella Isolated in Poultry Meat Products. Antibiotics 2022, 11, 1469. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, S.; Butaye, P.; Maliehe, T.S.; Magwedere, K.; Basson, A.K.; Madoroba, E. Virulence Factors and Antimicrobial Resistance in Salmonella Species Isolated from Retail Beef in Selected KwaZulu-Natal Municipality Areas, South Africa. Appl. Sci. 2022, 12, 2843. [Google Scholar] [CrossRef]
- Naidoo, S.; Basson, A.K.; Butaye, P.; Madoroba, E. Salmonella enterica Subspecies enterica Serotypes Associated with Meat and Meat Products in African Countries: A Review. In Food Security and Safety; Babalola, O.O., Ed.; Springer Nature: Cham, Swizerland, 2021; pp. 763–789. [Google Scholar]
- Madoroba, E.; Gelaw, A.K.; Kapeta, D. Salmonella contamination, serovars and antimicrobial resistance profiles of cattle slaughtered in South Africa. Onderstepoort J. Vet. Res. 2016, 83, a1109. [Google Scholar] [CrossRef] [PubMed]
- Mathole, M.A.; Muchadeyi, F.C.; Mdladla, K.; Malatji, D.P.; Dzomba, E.F.; Madoroba, E. Presence, distribution, serotypes and antimicrobial resistance profiles of Salmonella among pigs, chickens and goats in South Africa. Food Control 2017, 72, 219–224. [Google Scholar] [CrossRef]
- Ndlovu, L.; Butaye, P.; Maliehe, T.S.; Magwedere, K.; Mankonkwana, B.B.; Basson, A.K.; Ngema, S.S.; Madoroba, E. Virulence and antimicrobial resistance profiling of Salmonella serovars recovered from retail poultry offal in KwaZulu-Natal province, South Africa. Pathogens 2023, 12, 641. [Google Scholar] [CrossRef] [PubMed]
- Elnekave, E.; Hong, S.L.; Lim, S.; Hayer, S.S.; Boxrud, D.; Taylor, A.J.; Lappi, V.; Noyes, N.; Johnson, T.J.; Rovira, A.; et al. Circulation of plasmids harboring resistance genes to quinolones and/or extended spectrum cephalosporins in multiple Salmonella enterica serotypes from swine in the United States. Antimicrob. Agents Chemother. 2019, 63, e02602-18. [Google Scholar] [CrossRef]
- Elnekave, E.; Hong, S.; Mather, A.E.; Boxrud, D.; Taylor, A.J.; Lappi, V.; Johnson, T.J.; Vannucci, F.; Davies, P.; Hedberg, C.; et al. Salmonella enterica Serotype 4,[5],12:i:- in Swine in the United States Midwest: An Emerging Multidrug-Resistant Clade. Clin. Infect. Dis. 2018, 66, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.K.; Fareeq, A.; Hussein, S.; Mahmood, K.L.; Qurbani, K.; Ibrahim, R.H.; Mohamed, M.G. Antimicrobial resistance: Impacts, challenges, and future prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar] [CrossRef]
- Saxena, D.; Maitra, R.; Bormon, R.; Czekanska, M.; Meiers, J.; Titz, A.; Verma, S.; Chopra, S. Tackling the outer membrane: Facilitating compound entry into Gram-negative bacterial pathogens. NPJ Antimicrob. Resist. 2023, 1, 17. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The challenge of efflux mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Mkize, N. Molecular Detection and Genetic Characterization of Antimicrobial Resistance Genes in Foodborne Pathogens Isolated from Slaughtered Broiler Chickens in Durban. Master’s Thesis, University of KwaZulu-Natal, Durban, South Africa, 2016. [Google Scholar]
- Acheampong, G.; Owusu, M.; Owusu-Ofori, A.; Osei, I.; Sarpong, N.; Sylverken, A.; Kung, H.-J.; Cho, S.-T.; Kuo, C.-H.; Park, S.E.; et al. Chromosomal and plasmid-mediated fluoroquinolone resistance in human Salmonella enterica infection in Ghana. BMC Infect. Dis. 2019, 19, 898. [Google Scholar] [CrossRef] [PubMed]
- Mthembu, T.P.; Zishiri, O.T.; El Zowalaty, M.E. Genomic characterization of antimicrobial resistance in food chain and livestock-associated Salmonella species. Animals 2021, 11, 872. [Google Scholar] [CrossRef]
- Dinagde, B.M.; Mumed, B.A. A Review on Antimicrobial Resistance of Bovine Salmonellosis and Its Public Health Importance: One Health Approach. Integr. J. Vet. Biosci. 2023, 6, 1–8. [Google Scholar]
- Bhat, B.A.; Mir, R.A.; Qadri, H.; Dhiman, R.; Almilaibary, A.; Alkhanani, M.; Mir, M.A. Integrons in the development of antimicrobial resistance: Critical review and perspectives. Front. Microbiol. 2023, 4, 1231938. [Google Scholar] [CrossRef]
- Nair, D.; Thomas, J.V.; Dewi, G.; Noll, S.; Brannon, J.; Johny, A.K. Reduction of multidrug-resistant Salmonella enterica serovar Heidelberg using a dairy-originated probiotic bacterium, Propionibacterium freudenreichii freudenreichii B3523, in growing turkeys. J. Appl. Poultry Res. 2019, 28, 356–363. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Wang, T.; Liang, W. The spread of antibiotic resistance genes in vivo model. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 3348695. [Google Scholar] [CrossRef] [PubMed]
- Coburn, B.; Li, Y.; Owen, D.; Vallance, B.A.; Finlay, B.B. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 2005, 73, 3219–3227. [Google Scholar] [CrossRef]
- Ibarra, J.A.; Steele-Mortimer, O. Salmonella—The ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009, 11, 1579–1586. [Google Scholar]
- van Duijkeren, E.; Schink, A.K.; Roberts, M.C.; Wang, Y.; Schwarz, S. Mechanisms of bacterial resistance to antimicrobial agents. Microbiol. Spectrum 2018, 6, 51–82. [Google Scholar] [CrossRef]
- Marcus, S.L.; Brumell, J.H.; Pfeifer, S.G.; Finlay, B.B. Salmonella pathogenicity islands: Big virulence in small packages. Microb. Infect. 2000, 2, 145–156. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; MacKenzie, K.D.; Cameron, A.D.S. Salmonella Pathogenicity Island 1 (SPI-1): The evolution and stabilization of a core genomic type three secretion system. Microorganisms 2020, 8, 576. [Google Scholar] [CrossRef] [PubMed]
- Fookes, M.; Schroeder, G.N.; Langridge, G.C.; Blondel, C.J.; Mammina, C.; Connor, T.R.; Seth-Smith, H.S.; Vernikos, G.S.; Robinson, K.S.; Sanders, M.; et al. Salmonella bongori Provides Insights into the Evolution of the Salmonellae. PLoS Pathogens 2011, 7, e1002191. [Google Scholar] [CrossRef]
- Hayward, M.R.; Petrovska, L.; Jansen, V.A.A.; Woodward, M.J. Population structure and associated phenotypes of Salmonella enterica serovars Derby and Mbandaka overlap with host range. BMC Microbiol. 2016, 16, 15. [Google Scholar] [CrossRef]
- Urrutia, I.M.; Fuentes, J.A.; Valenzuela, L.M.; Ortega, A.P.; Hidalgo, A.A.; Mora, G.C. Salmonella Typhi shdA: Pseudogene or allelic variant? Infect. Genet. Evol. 2014, 26, 146–152. [Google Scholar] [CrossRef]
- Mthembu, T.P.; Zishiri, O.T.; El Zowalaty, M.E. Detection and molecular identification of Salmonella virulence genes in livestock production systems in South Africa. Pathogens 2019, 8, 124. [Google Scholar] [CrossRef]
- Foley, S.L.; Johnson, T.J.; Ricke, S.C.; Nayak, R.; Danzeisen, J. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol. Mol. Biol. Rev. 2013, 77, 582–607. [Google Scholar] [CrossRef]
- Ashari, K.S.; Roslan, N.S.; Omar, A.R.; Bejo, M.H.; Ideris, A.; Mat Isa, N. Genome sequencing and analysis of Salmonella enterica subsp. enterica serovar Stanley UPM 517: Insights on its virulence-associated elements and their potentials as vaccine candidates. PeerJ 2019, 7, e6948. [Google Scholar] [CrossRef]
- Siddiky, N.A.; Sarker, S.; Khan, S.R.; Rahman, T.; Kafi, A.; Samad, M.A. Virulence and antimicrobial resistance profile of non-typhoidal Salmonella enterica serovars recovered from poultry processing environments at wet markets in Dhaka, Bangladesh. PLoS ONE 2022, 17, e0254465. [Google Scholar] [CrossRef]
- South African Government. Fertilizers, Farm Feeds, Seeds and Remedies Act 36 of 1947. Available online: https://www.gov.za/sites/default/files/gcis_document/201505/act-36-1947.pdf (accessed on 24 February 2023).
- Mokgophi, T.M.; Gcebe, N.; Fasina, F.; Adesiyun, A.A. Molecular characterization of virulence and resistance genes in Salmonella strains isolated from chickens sold at the informal chicken market in Gauteng Province, South Africa. J. Food Saf. 2024, 44, e13110. [Google Scholar] [CrossRef]
- Zishiri, O.T.; Mkhize, N.; Mukaratirwa, S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J. Vet. Res. 2016, 83, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Madoroba, E.; Magwedere, K.; Chaora, N.S.; Matle, I.; Muchadeyi, F.; Mathole, M.A.; Pierneef, R. Microbial communities of meat and meat products: An exploratory analysis of the product quality and safety at selected enterprises in South Africa. Microorganisms 2021, 9, 507. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef] [PubMed]
- Charan, J.; Biswas, C. How to Calculate Sample Size for Different Study Designs in Medical Research? Ind. J. Psychol. Med. 2013, 35, 121–126. [Google Scholar] [CrossRef]
- Käsbohrer, A.; Tenhangen, B.A.; Appel, B.; Fetsch, A. Development of Harmonised Survey Methods for Food-Borne Pathogens in Foodstuffs in the European Union; European Food Safety Authority: Parma, Italy, 2010. [Google Scholar]
- ISO 6579-1:2017/Amd 1:2020; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.—AMENDMENT 1: Broader Range of Incubation Temperatures, AMENDMENT to the Status of Annex D, and Correction of the Composition of MSRV and SC. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/obp/ui/en/#iso:std:iso:6579:-1:ed-1:v1:amd:1:v1:en (accessed on 20 January 2025).
- Dieckmann, R.; Helmuth, R.; Erhard, M.; Malorny, B. Rapid classification and identification of Salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 2008, 74, 7767–7778. [Google Scholar] [CrossRef]
- Dieckmann, R.; Malorny, B. Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars using Whole-Cell MALDI-TOF mass spectrometry. Appl. Environ. Microbiol. 2011, 77, 4136–4146. [Google Scholar]
- Wang, S.J.; Yeh, D.B. Designing of polymerase chain reaction primers for the detection of Salmonella Enteritidis in foods and faecal samples. Lett. Appl. Microbiol. 2002, 32, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Paião, F.G.; Arisitides, L.G.A.; Murate, L.S.; Vilas-Bôas, G.T.; Vilas-Boas, L.A.; Shimokomaki, M. Detection of Salmonella spp., Salmonella Enteritidis and Typhimurium in naturally infected broiler chickens by a multiplex PCR-based assay. Braz. J. Microbiol. 2013, 44, 37–41. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, M100, 33rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Lauteri, C.; Festino, A.R.; Conter, M.; Vergara, A. Prevalence and antimicrobial resistance profile in Salmonella spp. isolates from swine food chain. Ital. J. Food Saf. 2022, 11, 9980. [Google Scholar] [CrossRef]
- Prasertsee, T.; Nattakarn, K.; Panuwat, Y.; Pannita, S.; Chokesajjawatee, N.; Patchanee, P. Repetitive sequence-based PCR fingerprinting and the relationship of antimicrobial resistance characteristics and corresponding genes among Salmonella strains from pig production. Asian Pac. J. Trop. Dis. 2016, 6, 390–395. [Google Scholar] [CrossRef]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Smith, R.J.; Jardine, C. Antimicrobial Resistance in Escherichia coli Isolates from Swine and Wild Small Mammals in the Proximity of Swine Farms and in Natural Environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef]
- Kikuvi, G.M.; Ombui, J.N.; Mitema, E.S. Serotypes and antimicrobial residence profiles of Salmonella isolates from pigs at slaughter in Kenya. J. Infect. Dev. Ctries 2010, 4, 243–248. [Google Scholar] [CrossRef]
- El-Tayeb, M.; Ibrahim, A.S.S.; Al-Salamah, A.A.; Almaary, K.; Elbadawi, Y.B. Prevalence, serotyping and antimicrobials resistance mechanism of Salmonella enterica isolated from clinical and environmental samples in Saudi Arabia. Braz. J. Microbiol. 2017, 48, 499–508. [Google Scholar] [CrossRef]
- Siddiky, N.A.; Sarker, M.S.; Khan, M.; Begum, R.; Kabir, M.E.; Karim, M.R.; Rahman, M.T.; Mahmud, A.; Samad, M.A. Virulence and Antimicrobial Resistance Profiles of Salmonella enterica Serovars Isolated from Chicken at Wet Markets in Dhaka, Bangladesh. Microorganisms 2021, 9, 952. [Google Scholar] [CrossRef]
- Kumar, G.; Kumar, S.; Jangid, H.; Dutta, J.; Shidiki, A. The rise of non-typhoidal Salmonella: An emerging global public health concern. Front. Microbiol. 2025, 16, 1524287. [Google Scholar] [CrossRef]
- Kidanemariam, A.; Engelbrecht, M.; Picard, J. Retrospective study on the incidence of Salmonella isolations in animals in South Africa, 1996 to 2006. J. S. Afr. Vet. Assoc. 2010, 81, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Nazari Moghadam, M.; Rahimi, E.; Shakerian, A.; Momtaz, H. Prevalence of Salmonella Typhimurium and Salmonella Enteritidis isolated from poultry meat: Virulence and antimicrobial-resistant genes. BMC Microbiol. 2023, 23, 168. [Google Scholar] [CrossRef] [PubMed]
- Alghoribi, M.F.; Doumith, M.; Alrodayyan, M.; Al Zayer, M.; Köster, W.L.; Muhanna, A.; Aljohani, S.M.; Balkhy, H.H.; Desin, S.S. S. enteritidis and S. typhimurium Harboring SPI-1 and SPI-2 Are the Predominant serotypes associated with human salmonellosis in Saudi Arabia. Front. Cell. Infect. Microbiol. 2019, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Kime, L.; Randall, C.P.; Banda, F.I.; Coll, F.; Wright, J.; Richardson, J.; Empel, J.; Parkhill, J.; O’Neill, A.J. Transient silencing of antibiotic resistance by mutation represents a significant potential source of unanticipated therapeutic failure. mBio 2019, 10, e01755-19. [Google Scholar] [CrossRef]
- Deekshit, V.K.; Srikumar, S. To be, or not to be’—The dilemma of ‘silent’ antimicrobial resistance genes in bacteria. J. Appl. Microbiol. 2022, 133, 2902–2914. [Google Scholar] [CrossRef]
- Tamburini, E.; Mastromei, G. Do bacterial cryptic genes really exist? Res. Microbiol. 2000, 151, 179–182. [Google Scholar] [CrossRef]
- Stasiak, M.; Mackiw, E.; Kowalska, J.; Kucharek, K.; Postupolski, J. Silent genes: Antimicrobial resistance and antibiotic production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef]
- Charlier, J.; Barkema, H.W.; Becher, P.; De Benedictis, P.; Hansson, I.; Hennig-Pauka, I.; La Ragione, R.; Larsen, L.E.; Madoroba, E.; Maes, D.; et al. Disease Control Tools to Secure Animal and Public Health in a Densely Populated World. Lancet Planet. Health 2022, 6, e812–e824. [Google Scholar] [CrossRef]
- Wee, B.A.; Muloi, D.M.; van Bunnik, B.A.D. Quantifying the transmission of antimicrobial resistance at the human and livestock interface with genomics. Clin. Microbiol. Infect. 2020, 26, 1612–1616. [Google Scholar] [CrossRef]
- Wibisono, F.M.; Faridah, H.D.; Effendi, M.H.; Wibisono, F.J.; Witaningrum, A.M.; Tyasningsih, W.; Ugbo, E.N. Detection of invA virulence gene of multidrug-resistant Salmonella species isolated from the cloacal swab of broiler chickens in Blitar district, East Java, Indonesia. Vet. World 2021, 14, 3126–3131. [Google Scholar] [CrossRef]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; Curtiss, R.; Gyles, C.L. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell Probes 1992, 6, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Galán, J.E.; Curtiss, R., 3rd. Distribution of the invA, -B, -C, and -D genes of Salmonella Typhimurium among other Salmonella serovars: invA mutants of Salmonella Typhi are deficient for entry into mammalian cells. Infect. Immun. 1991, 59, 2901–2908. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, E.P.; Melo, R.T.; Oliveira, M.R.M.; Monteiro, G.P.; Peres, P.A.B.M.; Fonseca, B.B.; Giombelli, A.; Rossi, D.A. Characteristics of virulence, resistance and genetic diversity of strains of Salmonella Infantis isolated from broiler chicken in Brazil. Pesq. Vet. Bras. 2020, 40, 29–38. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).