Multi-Omics Reveals Aberrant Phenotypes of Respiratory Microbiome and Phospholipidomics Associated with Asthma-Related Inflammation
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Acquisition and Processing of Sputum Induction
2.3. Respiratory Microbiome Sequencing and Bioinformatics
2.4. Analysis of Serum Cytokines
2.5. EBC Collection
2.6. Lipid Extraction
2.7. Measurement of Phospholipids Using Liquid Chromatography–Mass Spectrometry (LC-MS)
2.8. Statistics
3. Results
3.1. Demographic and Clinical Characteristics of Study Participants
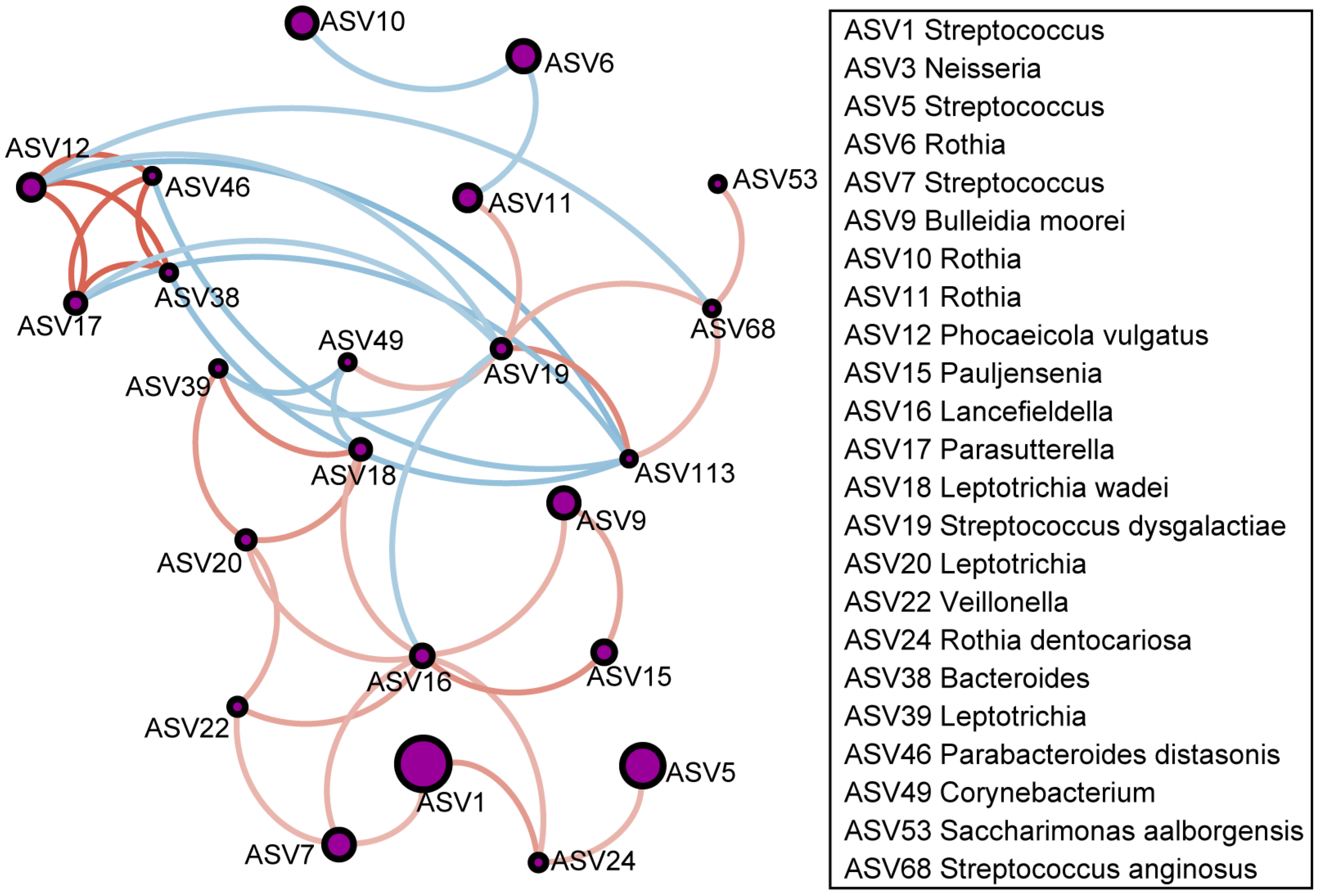
3.2. Differential Respiratory Microbiome in Patients with Asthma
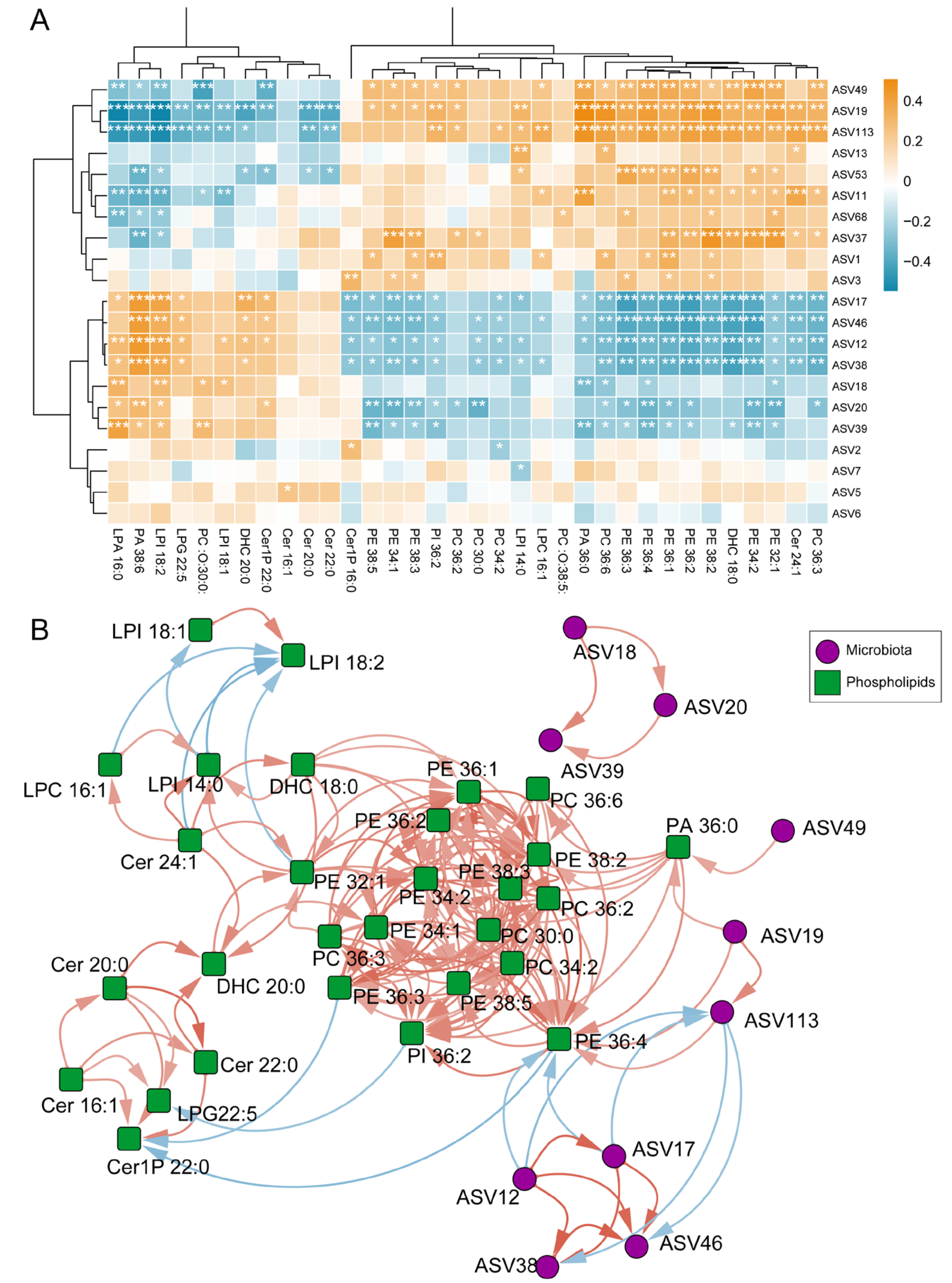
3.3. Differential EBC Phospholipid Levels in Patients with Asthma
3.4. Correlation Between Differential Phospholipids in EBC and Differential Microbes in Sputum
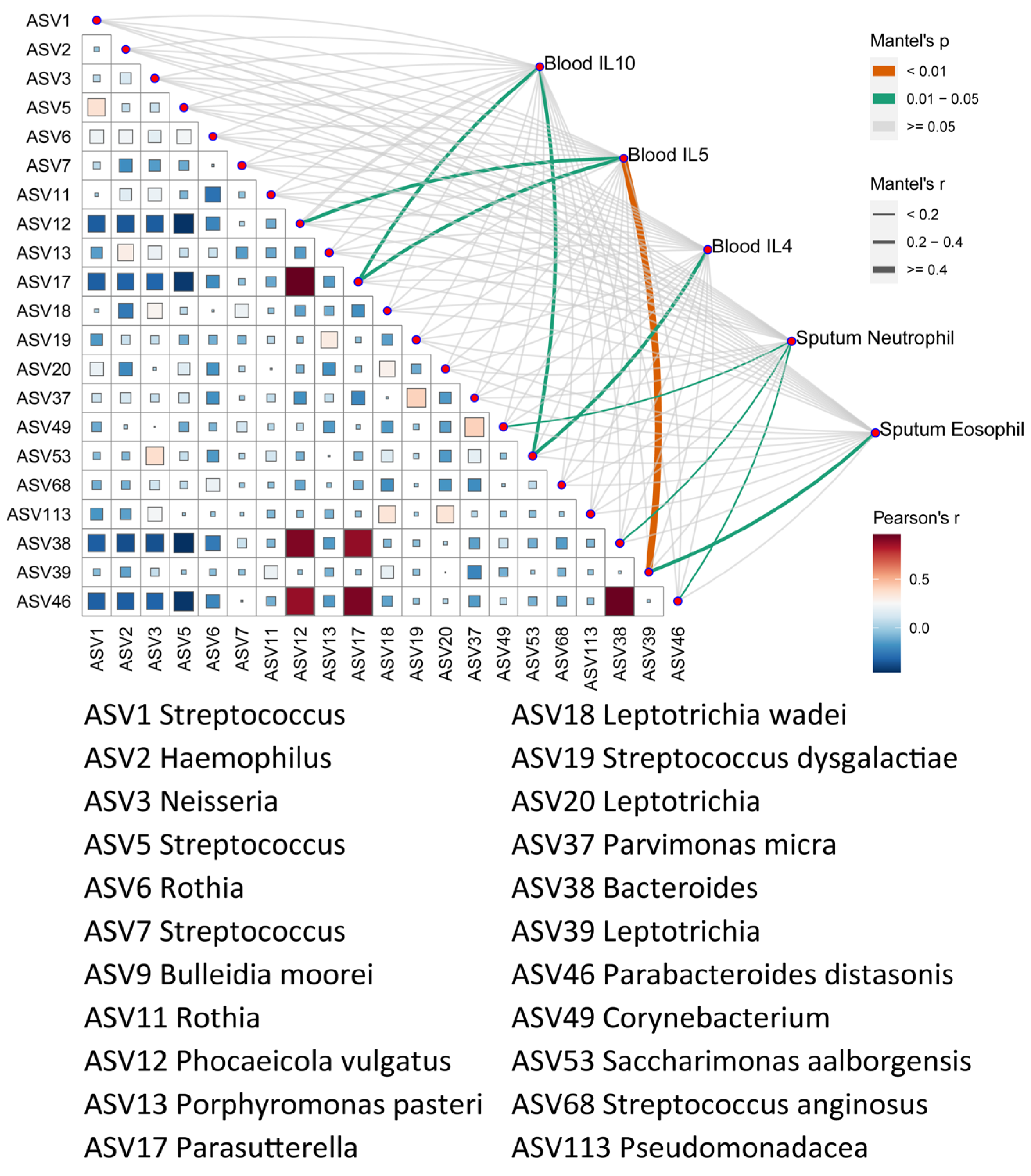
3.5. Correlation of Microbes with Clinical Characteristics
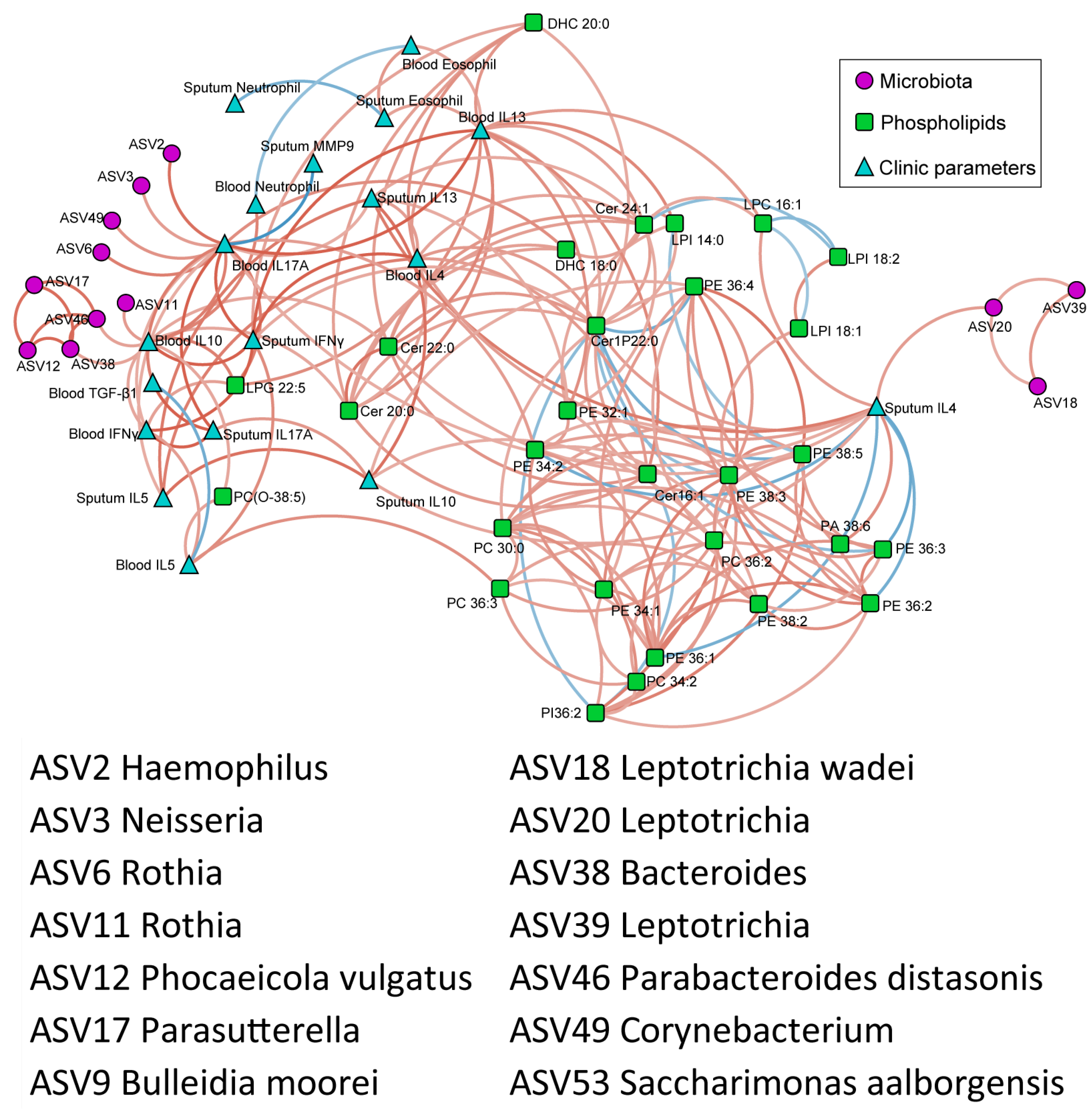
3.6. Correlation of Differential Phospholipids with Clinical Characteristics
3.7. Correlation of Phospholipids, Microbes, and Clinical Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Shi, G. Roles of sirtuins in asthma. Respir. Res. 2022, 23, 251. [Google Scholar] [CrossRef]
- Sze, E.; Bhalla, A.; Nair, P. Mechanisms and therapeutic strategies for non-T2 asthma. Allergy 2020, 75, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.S.; Hollins, F.; Sutcliffe, A.; Saunders, R.; Shah, S.; Siddiqui, S.; Gupta, S.; Haldar, P.; Green, R.; Pavord, I.; et al. Eosinophil protein in airway macrophages: A novel biomarker of eosinophilic inflammation in patients with asthma. J. Allergy Clin. Immunol. 2010, 126, 61.e63–69.e63. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.K.; Brunetti, T.; Quinn, K.; Doenges, K.; Campbell, M.; Arehart, C.; Taub, M.A.; Mathias, R.A.; Reisdorph, N.; Barnes, K.C.; et al. Discovering metabolite quantitative trait loci in asthma using an isolated population. J. Allergy Clin. Immunol. 2022, 149, 1807.e1816–1811.e1816. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, N.; Mauger, D.T.; Reisdorph, N.; Covar, R.; Malka, J.; Lemanske, R.F., Jr.; Morgan, W.J.; Guilbert, T.W.; Zeiger, R.S.; Bacharier, L.B.; et al. Predictors of asthma control and lung function responsiveness to step 3 therapy in children with uncontrolled asthma. J. Allergy Clin. Immunol. 2014, 133, 350–356. [Google Scholar] [CrossRef]
- Rabinovitch, N.; Reisdorph, N.; Silveira, L.; Gelfand, E.W. Urinary leukotriene E4 levels identify children with tobacco smoke exposure at risk for asthma exacerbation. J. Allergy Clin. Immunol. 2011, 128, 323–327. [Google Scholar] [CrossRef]
- Van Kruining, D.; Luo, Q.; van Echten-Deckert, G.; Mielke, M.M.; Bowman, A.; Ellis, S.; Oliveira, T.G.; Martinez-Martinez, P. Sphingolipids as prognostic biomarkers of neurodegeneration, neuroinflammation, and psychiatric diseases and their emerging role in lipidomic investigation methods. Adv. Drug Deliv. Rev. 2020, 159, 232–244. [Google Scholar] [CrossRef]
- Turi, K.N.; Michel, C.R.; Manke, J.; Doenges, K.A.; Reisdorph, N.; Bauer, A.K. Multi-Omics Analysis of Lung Tissue Demonstrates Changes to Lipid Metabolism during Allergic Sensitization in Mice. Metabolites 2023, 13, 406. [Google Scholar] [CrossRef]
- Yue, M.; Tao, S.; Gaietto, K.; Chen, W. Omics approaches in asthma research: Challenges and opportunities. Chin. Med. J. Pulm. Crit. Care Med. 2024, 2, 1–9. [Google Scholar] [CrossRef]
- Ver Heul, A.; Planer, J.; Kau, A.L. The Human Microbiota and Asthma. Clin. Rev. Allergy Immunol. 2019, 57, 350–363. [Google Scholar] [CrossRef]
- Denner, D.R.; Sangwan, N.; Becker, J.B.; Hogarth, D.K.; Oldham, J.; Castillo, J.; Sperling, A.I.; Solway, J.; Naureckas, E.T.; Gilbert, J.A.; et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J. Allergy Clin. Immunol. 2016, 137, 1398.e1393–1405.e1393. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Nelson, C.E.; Brodie, E.L.; Desantis, T.Z.; Baek, M.S.; Liu, J.; Woyke, T.; Allgaier, M.; Bristow, J.; Wiener-Kronish, J.P.; et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011, 127, 372.e3–381.e3. [Google Scholar] [CrossRef] [PubMed]
- Sverrild, A.; Kiilerich, P.; Brejnrod, A.; Pedersen, R.; Porsbjerg, C.; Bergqvist, A.; Erjefält, J.S.; Kristiansen, K.; Backer, V. Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J. Allergy Clin. Immunol. 2017, 140, 407.e411–417.e411. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V.; Nariya, S.; Bhakta, N.R.; Beigelman, A.; Castro, M.; Dyer, A.M.; Israel, E.; Kraft, M.; Martin, R.J.; et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 2017, 140, 63–75. [Google Scholar] [CrossRef]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef]
- Renz, H.; Skevaki, C. Early life microbial exposures and allergy risks: Opportunities for prevention. Nat. Rev. Immunol. 2021, 21, 177–191. [Google Scholar] [CrossRef]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef]
- Wang, J.; Chai, J.; Zhang, L.; Zhang, L.; Yan, W.; Sun, L.; Chen, Y.; Sun, Y.; Zhao, J.; Chang, C. Microbiota associations with inflammatory pathways in asthma. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2022, 52, 697–705. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2023, 42, 715–718. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191–e00216. [Google Scholar] [CrossRef]
- Horváth, I.; Hunt, J.; Barnes, P.J.; Alving, K.; Antczak, A.; Baraldi, E.; Becher, G.; van Beurden, W.J.; Corradi, M.; Dekhuijzen, R.; et al. Exhaled breath condensate: Methodological recommendations and unresolved questions. Eur. Respir. J. 2005, 26, 523–548. [Google Scholar] [CrossRef] [PubMed]
- Gans, M.D.; Gavrilova, T. Understanding the immunology of asthma: Pathophysiology, biomarkers, and treatments for asthma endotypes. Paediatr. Respir. Rev. 2020, 36, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, B.N.; Hammad, H.; Fahy, J.V. The Cytokines of Asthma. Immunity 2019, 50, 975–991. [Google Scholar] [CrossRef] [PubMed]
- Busse, W.W.; Viswanathan, R. What has been learned by cytokine targeting of asthma? J. Allergy Clin. Immunol. 2022, 150, 235–249. [Google Scholar] [CrossRef]
- Alamri, A. Diversity of Microbial Signatures in Asthmatic Airways. Int. J. Gen. Med. 2021, 14, 1367–1378. [Google Scholar] [CrossRef]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef]
- Valverde-Molina, J.; García-Marcos, L. Microbiome and Asthma: Microbial Dysbiosis and the Origins, Phenotypes, Persistence, and Severity of Asthma. Nutrients 2023, 15, 486. [Google Scholar] [CrossRef]
- Pfeiffer, S.; Jatzlauk, G.; Lund, J.V.; Boateng, E.; Kovacevic, D.; Hylkema, M.N.; Bartel, S.; Schloter, M.; Krauss-Etschmann, S. Oral application of vancomycin alters murine lung microbiome and pulmonary immune responses. Immun. Inflamm. Dis. 2022, 10, e675. [Google Scholar] [CrossRef]
- Ramos-Tapia, I.; Reynaldos-Grandón, K.L.; Pérez-Losada, M.; Castro-Nallar, E. Characterization of the upper respiratory tract microbiota in Chilean asthmatic children reveals compositional, functional, and structural differences. Front. Allergy 2023, 4, 1223306. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Chan, Y.L.; Tsai, Y.S.; Chen, S.A.; Wang, C.J.; Chen, K.F.; Chung, I.F. Airway Microbial Diversity is Inversely Associated with Mite-Sensitized Rhinitis and Asthma in Early Childhood. Sci. Rep. 2017, 7, 1820. [Google Scholar] [CrossRef]
- Chun, Y.; Do, A.; Grishina, G.; Grishin, A.; Fang, G.; Rose, S.; Spencer, C.; Vicencio, A.; Schadt, E.; Bunyavanich, S. Integrative study of the upper and lower airway microbiome and transcriptome in asthma. JCI Insight 2020, 5, e133707. [Google Scholar] [CrossRef]
- Zhou, Y.; Jackson, D.; Bacharier, L.B.; Mauger, D.; Boushey, H.; Castro, M.; Durack, J.; Huang, Y.; Lemanske, R.F.; Jr Storch, G.A.; et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat. Commun. 2019, 10, 5714. [Google Scholar] [CrossRef]
- Kim, Y.H.; Jang, H.; Kim, S.Y.; Jung, J.H.; Kim, G.E.; Park, M.R.; Hong, J.Y.; Kim, M.N.; Kim, E.G.; Kim, M.J.; et al. Gram-negative microbiota is related to acute exacerbation in children with asthma. Clin. Transl. Allergy 2021, 11, e12069. [Google Scholar] [CrossRef] [PubMed]
- Campanella, A.; De Summa, S.; Tommasi, S. Exhaled breath condensate biomarkers for lung cancer. J. Breath Res. 2019, 13, 044002. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Hu, M.; Chen, Y.; Liu, L.; Wu, L.; Hang, C.; Luo, X.; Xu, X. Metabolomics of bronchoalveolar lavage in children with persistent wheezing. Respir. Res. 2022, 23, 161. [Google Scholar] [CrossRef] [PubMed]
- Kertys, M.; Grendar, M.; Kosutova, P.; Mokra, D.; Mokry, J. Plasma based targeted metabolomic analysis reveals alterations of phosphatidylcholines and oxidative stress markers in guinea pig model of allergic asthma. Biochim. Et Biophys. Acta Mol. Basis Dis. 2020, 1866, 165572. [Google Scholar] [CrossRef]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Shi, F.; Zhang, X.; Zhang, Y.; Bi, K.; Chen, X.; Li, L.; Diao, H. Phospholipid metabolites of the gut microbiota promote hypoxia-induced intestinal injury via CD1d-dependent γδ T cells. Gut Microbes 2022, 14, 2096994. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Chan, Y.L.; Tsai, M.H.; Wang, C.J.; Chiang, M.H.; Chiu, C.C.; Su, S.C. Cross-talk between airway and gut microbiome links to IgE responses to house dust mites in childhood airway allergies. Sci. Rep. 2020, 10, 13449. [Google Scholar] [CrossRef]
- Jiang, T.; Dai, L.; Li, P.; Zhao, J.; Wang, X.; An, L.; Liu, M.; Wu, S.; Wang, Y.; Peng, Y.; et al. Lipid metabolism and identification of biomarkers in asthma by lipidomic analysis. Biochim. Et Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158853. [Google Scholar] [CrossRef]
- Nagasaki, T.; Schuyler, A.J.; Zhao, J.; Samovich, S.N.; Yamada, K.; Deng, Y.; Ginebaugh, S.P.; Christenson, S.A.; Woodruff, P.G.; Fahy, J.V.; et al. 15LO1 dictates glutathione redox changes in asthmatic airway epithelium to worsen type 2 inflammation. J. Clin. Investig. 2022, 132, e151685. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Ryan, E.; Gonzalez Pastor, B.; Gethings, L.A.; Clarke, D.J.; Joyce, S.A. Lipidomic Analysis Reveals Differences in Bacteroides Species Driven Largely by Plasmalogens, Glycerophosphoinositols and Certain Sphingolipids. Metabolites 2023, 13, 360. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- Brown, E.M.; Clardy, J.; Xavier, R.J. Gut microbiome lipid metabolism and its impact on host physiology. Cell Host Microbe 2023, 31, 173–186. [Google Scholar] [CrossRef]



| Characteristics | Asthma Patients | Healthy Controls | p Value |
|---|---|---|---|
| No. of subjects | 61 | 17 | |
| Age (years) | 47.3 ± 17.8 | 50.5 ± 6.4 | 0.475 |
| Sex (male/female), no. | 21/40 | 3/14 | 0.242 |
| BMI (kg/m2) | 24.5 ± 4.0 | 23.1 ± 3.3 | 0.193 |
| ACT score | 18.5 ± 5.8 | ND | |
| Smoking history (smoker/never), no. | 11/50 | 2/15 | 0.722 |
| Gastroesophageal reflux history | 0 | 0 | ND |
| ICS Dose | |||
| Low | 26 | ND | |
| Medium | 18 | ND | |
| High | 7 | ND | |
| FEV1 | 89.8 ± 18.6 | 106.7 ± 16.8 | 0.001 |
| FEV1/FVC | 72.3 ± 11.4 | 84.1 ± 7.0 | 0.000 |
| Allergy test positive, no. | 48 | ND | ND |
| T2-high asthma | 55 | ND | ND |
| Blood Eosinophil (×109/L) | 0.3 ± 0.2 | ND | ND |
| Blood neutrophil (×109/L) | 2.9 ± 1.2 | ND | ND |
| Sputum total cells count (×106/g) | 2.0 ± 1.6 | 1.3 ± 0.8 | 0.065 |
| Sputum Macrophage (%) | 16.1 ± 20.5 | 5.4 ± 6.0 | 0.034 |
| Sputum Lymphocyte (%) | 0.7 ± 3.2 | 0.5 ± 0.8 | 0.650 |
| Sputum Neutrophil (%) | 63.3 ± 30.0 | 87.9 ± 6.2 | 0.000 |
| Sputum Eosinophil (%) | 18.2 ± 24.7 | 0.3 ± 0.5 | 0.004 |
| Sputum IL-4 (pg/mL) | 2.4 ± 1.6 | ND | ND |
| Sputum IL-5 (pg/mL) | 7.5 ± 7.3 | ND | ND |
| Sputum IL-13 (pg/mL) | 7.1 ± 5.4 | ND | ND |
| Sputum IL-10 (pg/mL) | 2.2 ± 1.6 | ND | ND |
| Sputum IL-17 (pg/mL) | 1.0 ± 0.7 | ND | ND |
| Sputum IL-10 (pg/mL) | 2.2 ± 1.6 | ND | ND |
| Sputum IFN-γ (pg/mL) | 7.8 ± 10.3 | ND | ND |
| Sputum MMP-9 (pg/mL) | 51.5 ± 44.7 | ND | ND |
| Blood IL-4 (pg/mL) | 5.4 ± 4.2 | ND | ND |
| Blood IL-5 (pg/mL) | 8.1 ± 8.0 | ND | ND |
| Blood IL-13 (pg/mL) | 12.1 ± 8.9 | ND | ND |
| Blood IL-17 (pg/mL) | 32.1 ± 5.8 | ND | ND |
| Blood IL-10 (pg/mL) | 10.1 ± 5.1 | ND | ND |
| Blood IFN-γ (pg/mL) | 12.8 ± 5.1 | ND | ND |
| Blood TGF-β (pg/mL) | 846.2 ± 337.7 | ND | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Li, Z.; Zhang, X.; Zhao, J.-C.; Chai, J.; Chang, C. Multi-Omics Reveals Aberrant Phenotypes of Respiratory Microbiome and Phospholipidomics Associated with Asthma-Related Inflammation. Microorganisms 2025, 13, 1761. https://doi.org/10.3390/microorganisms13081761
Liu H, Li Z, Zhang X, Zhao J-C, Chai J, Chang C. Multi-Omics Reveals Aberrant Phenotypes of Respiratory Microbiome and Phospholipidomics Associated with Asthma-Related Inflammation. Microorganisms. 2025; 13(8):1761. https://doi.org/10.3390/microorganisms13081761
Chicago/Turabian StyleLiu, Huan, Zemin Li, Xu Zhang, Jiang-Chao Zhao, Jianmin Chai, and Chun Chang. 2025. "Multi-Omics Reveals Aberrant Phenotypes of Respiratory Microbiome and Phospholipidomics Associated with Asthma-Related Inflammation" Microorganisms 13, no. 8: 1761. https://doi.org/10.3390/microorganisms13081761
APA StyleLiu, H., Li, Z., Zhang, X., Zhao, J.-C., Chai, J., & Chang, C. (2025). Multi-Omics Reveals Aberrant Phenotypes of Respiratory Microbiome and Phospholipidomics Associated with Asthma-Related Inflammation. Microorganisms, 13(8), 1761. https://doi.org/10.3390/microorganisms13081761






