Community Structure, Growth-Promoting Potential, and Genomic Analysis of Seed-Endophytic Bacteria in Stipagrostis pennata
Abstract
1. Introduction
2. Materials and Methods
2.1. Material Pretreatment
2.2. Total DNA Extraction, PCR Amplification, and High-Throughput Sequencing
2.3. Statistical and Bioinformatics Analysis
2.4. Isolation and Purification of Seed-Endophytic Bacteria
2.5. Evaluation of PGP Activity of Seed-Endophytic Bacteria
2.6. Seed Endophyte-Mediated Growth Promotion Assay in Potted Wheat
2.7. Abiotic Stress Tolerance of L7 Strain
2.7.1. Osmotic Stress Tolerance
2.7.2. Salt Tolerance
2.7.3. pH Tolerance
2.8. Identification of Strain L7
2.9. L7 Sample Preparation and Genomic Sequencing
2.10. Gene Composition Analysis and Functional Annotation
2.11. Statistical Analysis
3. Results
3.1. Community Composition and Diversity Analysis
3.2. Isolation of Endophytic Bacteria from S. pennata Seeds
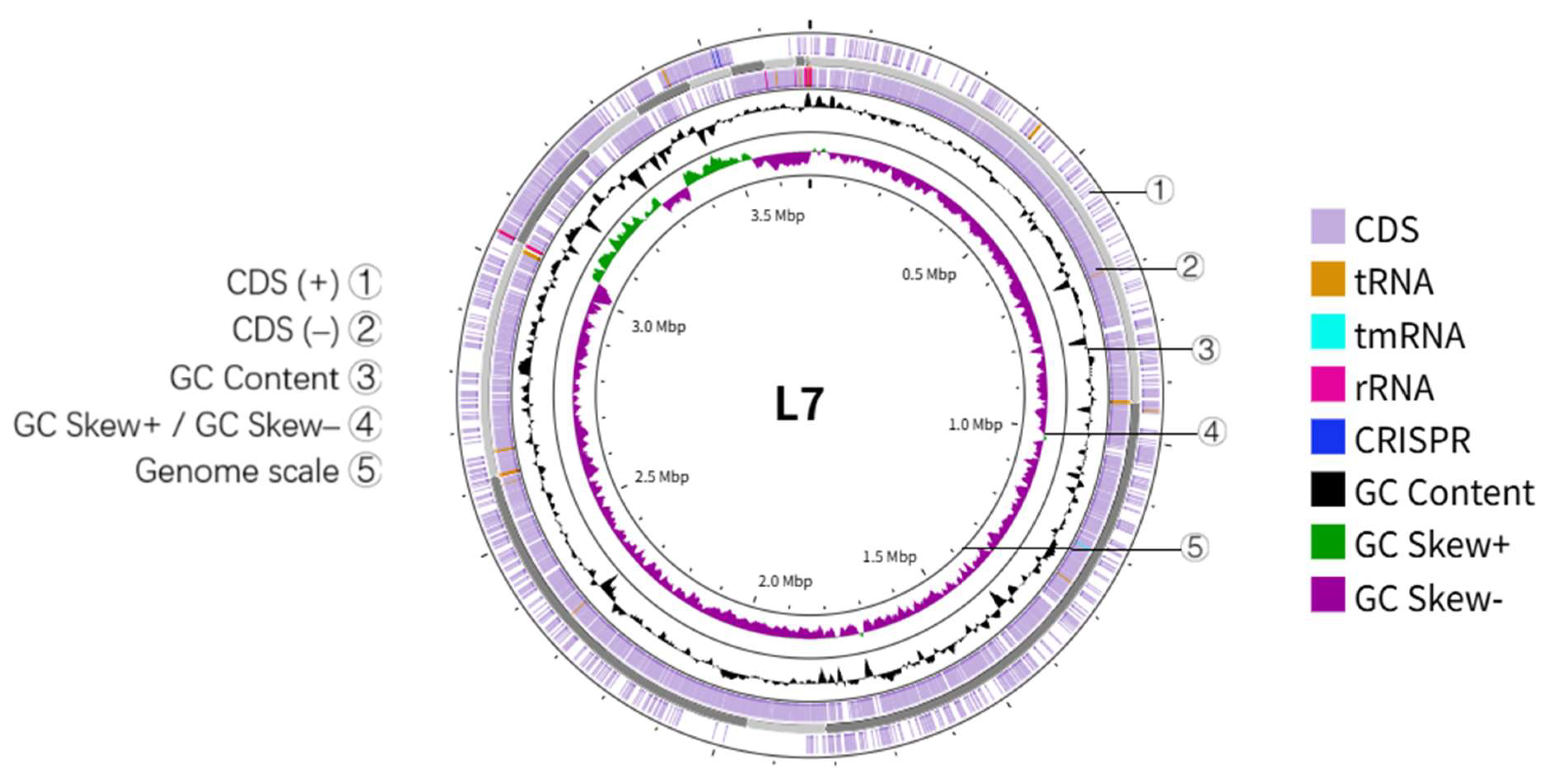
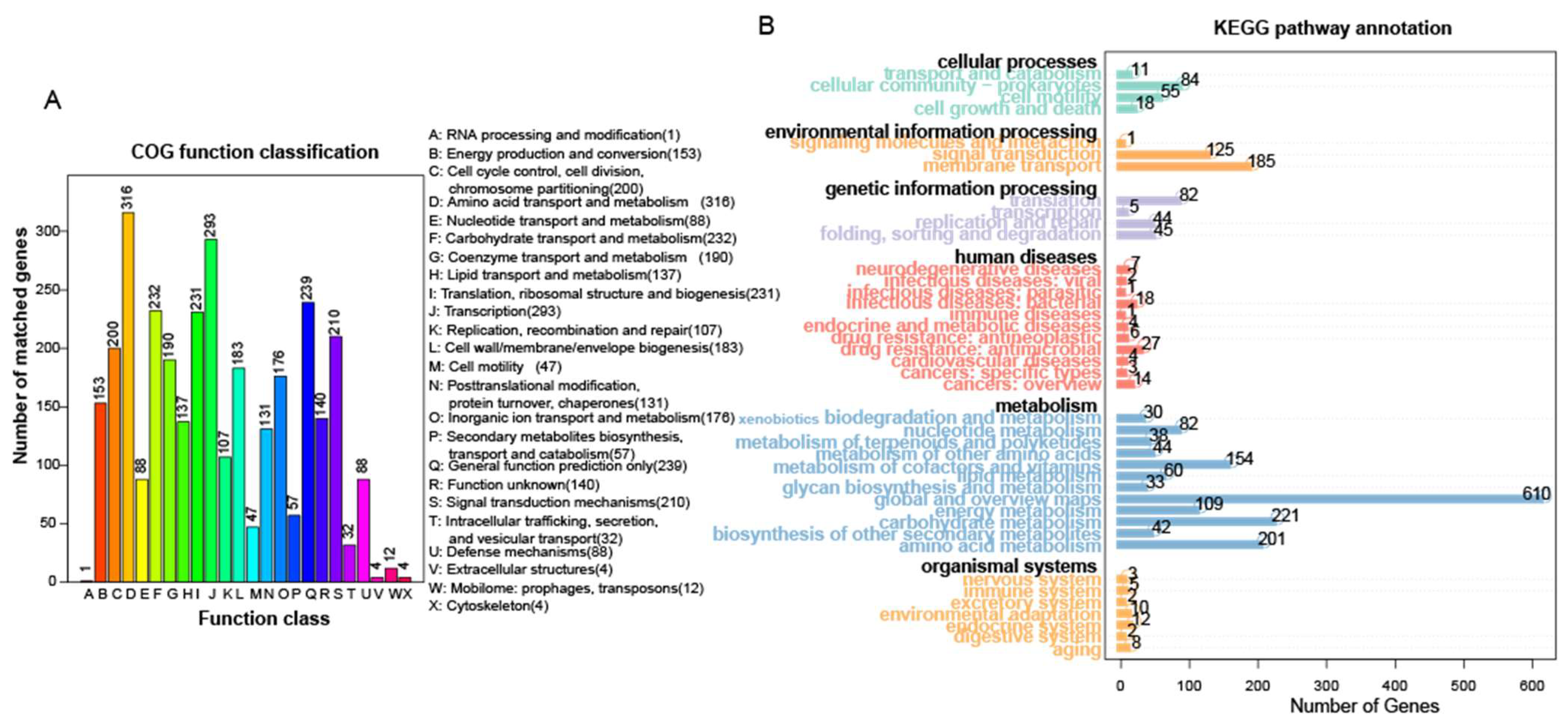
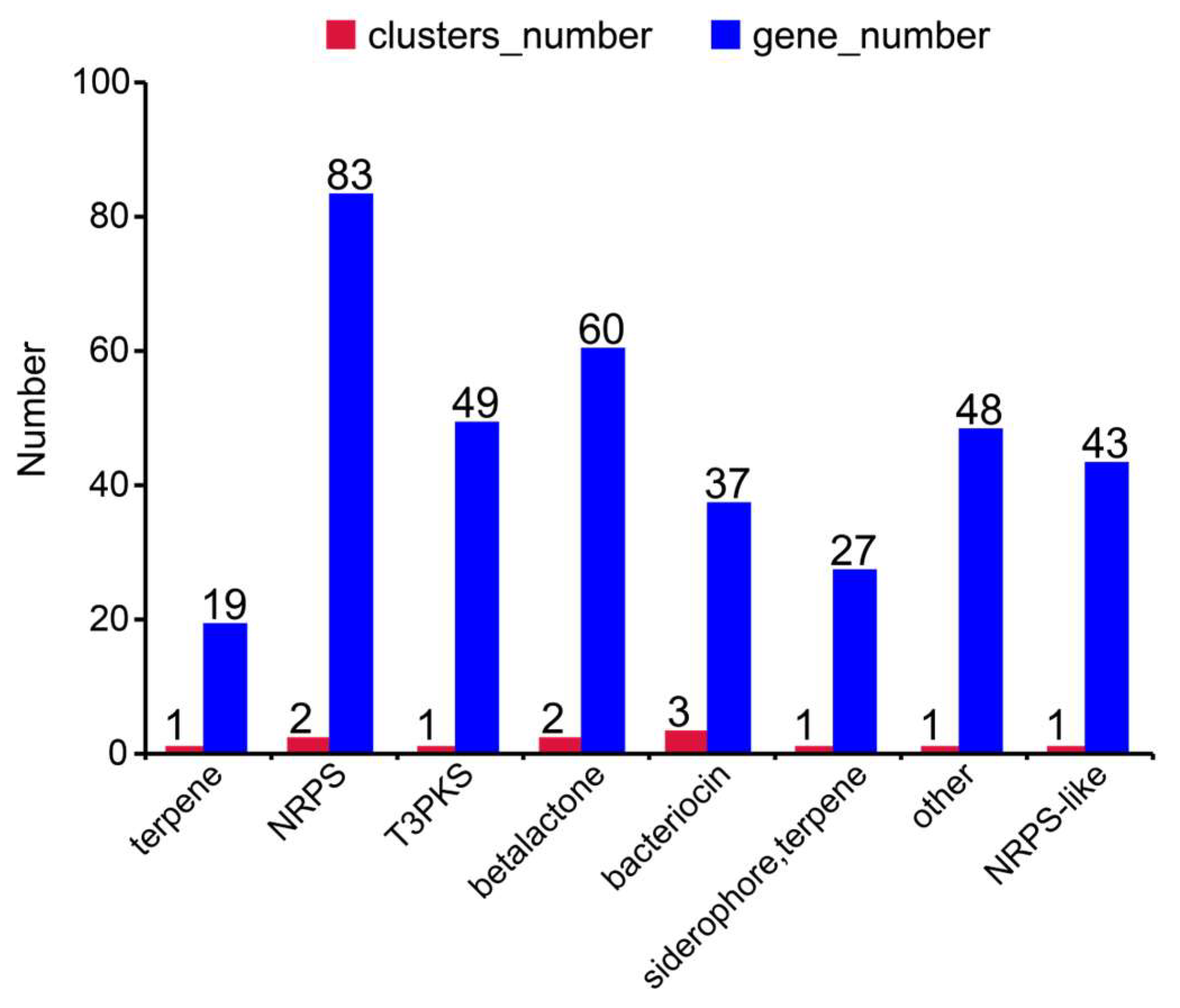
3.3. Genomic Characteristics of Bacillus altitudinis Strain L7
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barker, N.P.; Clark, L.G.; Davis, J.I.; Duvall, M.R.; Guala, G.F.; Hsiao, C.; Kellogg, E.A.; Linder, H.P.; Mason-Gamer, R.J.; Mathews, S.Y.; et al. Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 2001, 88, 373–457. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, T.; Ma, L. Rapidly evolving genetic features for desert adaptations in Stipagrostis pennata. BMC Genom. 2021, 22, 846. [Google Scholar] [CrossRef]
- Mao, W.; Wu, Y.; Li, F.; Tang, W.; Gong, W.; Han, X.; White, J.F.; Ji, X.; Li, H. Seed Endophytes and Their Roles in Host Plant Stress Resistance. J. Soil Sci. Plant Nutr. 2023, 23, 2927–2937. [Google Scholar] [CrossRef]
- Choi, B.; Jeong, S.; Kim, E. Variation of the seed endophytic bacteria among plant populations and their plant growth-promoting activities in a wild mustard plant species, Capsella bursa-pastoris. Ecol. Evol. 2022, 12, e8683. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, S.; Liu, N. Diversity of Endophytic Bacteria in Cardamine hupingshanensis and Potential of Culturable Selenium-Resistant Endophytes to Enhance Seed Germination Under Selenate Stress. Curr. Microbiol. 2021, 78, 2091–2103. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Khan, M.S.; Aziz, S.; Ali, H.; Pecoraro, L. Molecular and Biochemical Characterization, Antimicrobial Activity, Stress Tolerance, and Plant Growth-Promoting Effect of Endophytic Bacteria Isolated from Wheat Varieties. Microorganisms 2022, 10, 21. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.; Irizarry, I.; Bergen, M.; Kharwar, R.N.; White, J.F., Jr. Seed-vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 2017, 122, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Joshi, M. Microbial strategies for enhancing wheat and rice resilience to drought, salinity, and heat stress. Rhizosphere 2025, 34, 101108. [Google Scholar] [CrossRef]
- Zahra, S.T.; Tariq, M.; Abdullah, M.; Zafar, M.; Yasmeen, T.; Shahid, M.S.; Zaki, H.E.M.; Ali, A. Probing the potential of salinity-tolerant endophytic bacteria to improve the growth of mungbean Vigna radiata (L.) Wilczek. Front. Microbiol. 2023, 14, 1149004. [Google Scholar] [CrossRef]
- Abideen, Z.; Cardinale, M.; Zulfiqar, F.; Koyro, H.-W.; Rasool, S.G.; Hessini, K.; Darbali, W.; Zhao, F.; Siddique, K.H.M. Seed Endophyte bacteria enhance drought stress tolerance in Hordeum vulgare by regulating, physiological characteristics, antioxidants and minerals uptake. Front. Plant Sci. 2022, 13, 980046. [Google Scholar] [CrossRef]
- Tang, X.; Liu, Q.; Luo, L.; Yin, C. The Endophyte Bacillus amyloliquefaciens from Picea asperata Seeds Promotes Seed Germination and Its Physiological Mechanism. J. Soil Sci. Plant Nutr. 2024, 24, 421–434. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, T.-M.; Choi, B.; Kim, Y.; Kim, E. Invasive Lactuca serriola seeds contain endophytic bacteria that contribute to drought tolerance. Sci. Rep. 2021, 11, 13307. [Google Scholar] [CrossRef]
- Gabriele, M.; Vitali, F.; Chelucci, E.; Chiellini, C. Characterization of the Cultivable Endophytic Bacterial Community of Seeds and Sprouts of Cannabis sativa L. and Perspectives for the Application as Biostimulants. Microorganisms 2022, 10, 1742. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Xiang, M.; Liang, Z.; Zhang, Y.; Wu, J.; Ma, T.; Duo, L.; Zhang, X.; Fu, G. Grazing Intensity Modifies Soil Microbial Diversity and Their Co-Occurrence Networks in an Alpine Steppe, Central Tibet. Microorganisms 2025, 13, 138. [Google Scholar] [CrossRef]
- Khan, A.R.; Wicaksono, W.A.; Ott, N.J.; Poret-Peterson, A.T.; Browne, G.T. Characterization of soils conducive and non-conducive to Prunus replant disease. PLoS ONE 2021, 16, e0260394. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Gao, M.; Zhou, J.-J.; Wang, E.-T.; Chen, Q.; Xu, J.; Sun, J.-G. Multiphasic characterization of a plant growth promoting bacterial strain, Burkholderia sp. 7016 and its effect on tomato growth in the field. J. Integr. Agric. 2015, 14, 1855–1863. [Google Scholar] [CrossRef]
- Ramzi, Y.; Alsalim, H.A.A. Isolation and identification of phosphate solubilizing bacteria and evaluate its effect on of Mung bean (Vigna radita [L.] R.Wilczek) growth. Iraqi J. Sci. 2019, 60, 985–995. [Google Scholar] [CrossRef]
- Slimani, A.; Raklami, A.; Oufdou, K.; Meddich, A. Isolation and Characterization of PGPR and Their Potenzial for Drought Alleviation in Barley Plants. Gesunde Pflanz. 2022, 75, 377–391. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Ghavami, N.; Alikhani, H.A.; Pourbabaei, A.A.; Besharati, H. Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. J. Plant Nutr. 2017, 40, 736–746. [Google Scholar] [CrossRef]
- Singh, R.K.; Singh, P.; Li, H.-B.; Guo, D.-J.; Song, Q.-Q.; Yang, L.-T.; Malviya, M.K.; Song, X.-P.; Li, Y.-R. Plant-PGPR interaction study of plant growth-promoting diazotrophs Kosakonia radicincitans BA1 and Stenotrophomonas maltophilia COA2 to enhance growth and stress-related gene expression in Saccharum spp. J. Plant Interact. 2020, 15, 427–445. [Google Scholar] [CrossRef]
- Devi, S.; Sharma, S.; Tiwari, A.; Bhatt, A.K.; Singh, N.K.; Singh, M.; Kaushalendra; Kumar, A. Screening for Multifarious Plant Growth Promoting and Biocontrol Attributes in Bacillus Strains Isolated from Indo Gangetic Soil for Enhancing Growth of Rice Crops. Microorganisms 2023, 11, 1085. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xie, Y.; Liao, Y.; Li, X.; Li, Y.; Li, S.; Ma, X.; Lei, S.; Lin, F.; Jiang, W.; et al. Characterization of a Bacillus velezensis strain isolated from Bolbostemmatis Rhizoma displaying strong antagonistic activities against a variety of rice pathogens. Front. Microbiol. 2022, 13, 983781. [Google Scholar] [CrossRef]
- Weng, W.; Yao, X.; Zhao, M.; Fang, Z.; Yang, S.; Ruan, J. Enhanced reactive oxygen species and metabolism are involved in the reduction of tribenuron-methyl residues in Tartary buckwheat mutants. Ind. Crops Prod. 2024, 223, 120120. [Google Scholar] [CrossRef]
- Wang, H.; Jia, Y.; Bai, X.; Gong, W.; Liu, G.; Wang, H.; Xin, J.; Wu, Y.; Zheng, H.; Liu, H.; et al. Whole-Transcriptome Profiling and Functional Prediction of Long Non-Coding RNAs Associated with Cold Tolerance in Japonica Rice Varieties. Int. J. Mol. Sci. 2024, 25, 2310. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Zeeshan Ul Haq, M.; Zhang, L.; Wu, S.; Mushtaq, N.; Tahir, H.; Wang, Z. Transcriptomic Insights into Salt Stress Response in Two Pepper Species: The Role of MAPK and Plant Hormone Signaling Pathways. Int. J. Mol. Sci. 2024, 25, 9355. [Google Scholar] [CrossRef]
- Mondal, H.K.; Gera, R. Screening for drought-tolerant mungbean root nodule bacteria with multiple plant growth promoting traits in Aridisol. Appl. Soil Ecol. 2024, 201, 105510. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Z.; Zhao, Q.; Yang, X.; Li, Y.; Zhou, H.; Zhao, M.; Zheng, H. Whole-genome analysis revealed the growth-promoting and biological control mechanism of the endophytic bacterial strain Bacillus halotolerans Q2H2, with strong antagonistic activity in potato plants. Front. Microbiol. 2024, 14, 1287921. [Google Scholar] [CrossRef]
- Mount, D.W. Using the Basic Local Alignment Search Tool (BLAST). Cold Spring Harb. Protocols. 2007, 2007, pdb.top17. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-H.; Liao, Y.-C. CISA: Contig Integrator for Sequence Assembly of Bacterial Genomes. PLoS ONE 2013, 8, e60843. [Google Scholar] [CrossRef]
- Sohaib, H.; Fays, M.; Khatib, A.; Riviere, J.; El Aouad, N.; Desoignies, N. Contribution to the characterization of the seed endophyte microbiome of Argania spinosa across geographical locations in Central Morocco using metagenomic approaches. Front. Microbiol. 2024, 15, 1310395. [Google Scholar] [CrossRef] [PubMed]
- Roswell, M.; Dushoff, J.; Winfree, R. A conceptual guide to measuring species diversity. Oikos 2021, 130, 321–338. [Google Scholar] [CrossRef]
- Yang, F.; Huang, M.; Li, C.; Wu, X.; Fang, L. Vegetation restoration increases the diversity of bacterial communities in deep soils. Appl. Soil Ecol. 2022, 180, 104631. [Google Scholar] [CrossRef]
- Simonin, M.; Briand, M.; Chesneau, G.; Rochefort, A.; Marais, C.; Sarniguet, A.; Barret, M. Seed microbiota revealed by a large-scale meta-analysis including 50 plant species. New Phytol. 2022, 234, 1448–1463. [Google Scholar] [CrossRef]
- War, A.F.; Bashir, I.; Reshi, Z.A.; Kardol, P.; Rashid, I. Insights into the seed microbiome and its ecological significance in plant life. Microbiol. Res. 2023, 269, 127318. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, X.; Zhu, R.; Liu, Z.; Gu, M.; Zhang, J.; Li, Y.; Xu, Y.; Zhu, D. Analysis of the Endophytic Bacteria Community Structure and Function of Panax notoginseng Based on High-Throughput Sequencing. Curr. Microbiol. 2020, 77, 2745–2750. [Google Scholar] [CrossRef]
- Cankar, K.; Kraigher, H.; Ravnikar, M.; Rupnik, M. Bacterial endophytes from seeds of Norway spruce (Picea abies L. Karst). FEMS Microbiol. Lett. 2005, 244, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Hawkins, S.; Winters, A.; Farrar, K. Bacterial endophytic community composition varies by hemp cultivar in commercially sourced seed. Environ. Microbiol. Rep. 2024, 16, e13259. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda, M.d.C.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.-W.; Li, X.-W.; Wang, T.-T.; Gong, Y.; Zhang, C.-M.; Xing, K.; Qin, S. Root exudates-driven rhizosphere recruitment of the plant growth-promoting rhizobacterium Bacillus flexus KLBMP 4941 and its growth-promoting effect on the coastal halophyte Limonium sinense under salt stress. Ecotoxicol. Environ. Saf. 2020, 194, 110374. [Google Scholar] [CrossRef]
- Xu, J.-C.; Xie, X.-G.; Bi, X.-W.; Zhang, J.-H.; Zhao, Z.-H.; Rahman, K.; Zhu, B.; Qin, L.-P.; Han, T. Isolation of Rhizobacteria from Crocus Sativus L. Rhizosphere and Their Effects on Host-Growth Promotion. J. Plant Growth Regul. 2024, 43, 1536–1547. [Google Scholar] [CrossRef]
- Lamarche, M.G.; Wanner, B.L.; Crepin, S.; Harel, J. The phosphate regulon and bacterial virulence: A regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 2008, 32, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, D.; Stringer, A.; Smith, C.; Lapierre, P.; Wade, J.T. Genome-wide mapping of the Escherichia coli PhoB regulon reveals many transcriptionally inert, intragenic binding sites. bioRxiv 2023. [Google Scholar] [CrossRef]
- Qiu, G.-W.; Zheng, W.-C.; Yang, H.-M.; Wang, Y.-Y.; Qi, X.; Huang, D.; Dai, G.-Z.; Shi, H.; Price, N.M.; Qiu, B.-S. Phosphorus deficiency alleviates iron limitation in Synechocystis cyanobacteria through direct PhoB-mediated gene regulation. Nat. Commun. 2024, 15, 4426. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Saavedra, D.E.M.; Thomson, B.; Garcia, J.A.L.; Zhao, Z.; Patrick, W.M.; Herndl, G.J.; Baltar, F. Enzyme promiscuity in natural environments: Alkaline phosphatase in the ocean. ISME J. 2021, 15, 3375–3383. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Q.; Sun, Z.; Li, Y.; He, H.; Zhang, Y.; Yang, X.; Wang, D.; Dong, B.; Zhou, H.; et al. Whole-genome analysis revealed the growth-promoting mechanism of endophytic bacterial strain Q2H1 in potato plants. Front. Microbiol. 2022, 13, 1035901. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Al-Harrasi, A.; Lee, I.-J. Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. 3 Biotech 2018, 8, 389. [Google Scholar] [CrossRef]
- Ramos, J.V.; Kulka-Peschke, C.J.; Bechtel, D.F.; Zebger, I.; Pierik, A.J.; Layer, G. Characterization of the iron-sulfur clusters in the nitrogenase-like reductase CfbC/D required for coenzyme F430 biosynthesis. FEBS J. 2024, 291, 3233–3248. [Google Scholar] [CrossRef]
- Guo, D.-J.; Singh, R.K.; Singh, P.; Li, D.-P.; Sharma, A.; Xing, Y.-X.; Song, X.-P.; Yang, L.-T.; Li, Y.-R. Complete Genome Sequence of Enterobacter roggenkampii ED5, a Nitrogen Fixing Plant Growth Promoting Endophytic Bacterium with Biocontrol and Stress Tolerance Properties, Isolated from Sugarcane Root. Front. Microbiol. 2020, 11, 580081. [Google Scholar] [CrossRef]
- Wang, N.; Wang, T.; Chen, Y.; Wang, M.; Lu, Q.; Wang, K.; Dou, Z.; Chi, Z.; Qiu, W.; Dai, J.; et al. Microbiome convergence enables siderophore-secreting-rhizobacteria to improve iron nutrition and yield of peanut intercropped with maize. Nat. Commun. 2024, 15, 839. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, Q.; Hou, J.; Tu, C.; Luo, Y.; Christie, P. Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 2016, 6, 26710. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, R.N.; Song, X.-P.; Singh, R.K.; Guo, D.-J.; Singh, P.; Verma, K.K.; Li, Y.-R. Genome analysis of a halophilic Virgibacillus halodenitrificans ASH15 revealed salt adaptation, plant growth promotion, and isoprenoid biosynthetic machinery. Front. Microbiol. 2023, 14, 1229955. [Google Scholar] [CrossRef]
- Petrezselyova, S.; Zahradka, J.; Sychrova, H. Saccharomyces cerevisiae BY4741 and W303-1A laboratory strains differ in salt tolerance. Fungal Biol. 2010, 114, 144–150. [Google Scholar] [CrossRef]
- Vaish, M.; Price-Whelan, A.; Reyes-Robles, T.; Liu, J.; Jereen, A.; Christie, S.; Alonzo, F., III; Benson, M.A.; Torres, V.J.; Krulwich, T.A. Roles of Staphylococcus aureus Mnh1 and Mnh2 Antiporters in Salt Tolerance, Alkali Tolerance, and Pathogenesis. J. Bacteriol. 2018, 200, e00611-17. [Google Scholar] [CrossRef] [PubMed]
- du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Castiglione, A.M.; Mannino, G.; Contartese, V.; Bertea, C.M.; Ertani, A. Microbial Biostimulants as Response to Modern Agriculture Needs: Composition, Role and Application of These Innovative Products. Plants 2021, 10, 1533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Z.; Pu, Y.; Zhang, B.; Wang, B.; Xing, L.; Li, Y.; Zhang, Y.; Gu, R.; Jia, F.; et al. Antagonistic Strain Bacillus velezensis JZ Mediates the Biocontrol of Bacillus altitudinis m-1, a Cause of Leaf Spot Disease in Strawberry. Int. J. Mol. Sci. 2024, 25, 8872. [Google Scholar] [CrossRef]
- Lemjiber, N.; Naamani, K.; Merieau, A.; Dihazi, A.; Zhar, N.; Jediyi, H.; Boukerb, A.M. Identification and Genomic Characterization of Pathogenic Bacillus altitudinis from Common Pear Trees in Morocco. Agronomy 2021, 11, 1344. [Google Scholar] [CrossRef]
- Hu, J.; Qian, W.; He, C. The Xanthomonas oryzae pv. oryzae eglXoB endoglucanase gene is required for virulence to rice. FEMS Microbiol. Lett. 2007, 269, 273–279. [Google Scholar] [CrossRef]
- Boccara, M.; Aymeric, J.L.; Camus, C. Role of endoglucanases in Erwinia chrysanthemi 3937 virulence on Saintpaulia ionantha. J. Bacteriol. 1994, 176, 1524–1526. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.M.; Kim, Y.K.; Guo, W.; González-Candelas, L.; Li, D.; Kolattukudy, P.E. Requirement for either a host- or pectin-induced pectate lyase for infection of Pisum sativum by Nectria hematococca. Proc. Natl. Acad. Sci. USA 2000, 97, 9813–9818. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liao, Y.Y.; Huang, R.X.; Li, A.Z.; An, S.Q.; Tang, J.L.; Tang, D.J. Extracellular Amylase Is Required for Full Virulence and Regulated by the Global Posttranscriptional Regulator RsmA in Xanthomonas campestris Pathovar campestris. Phytopathology 2021, 111, 1104–1113. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Y.; Pang, S.; Niu, W.; Zhang, T.; Ma, L. Community Structure, Growth-Promoting Potential, and Genomic Analysis of Seed-Endophytic Bacteria in Stipagrostis pennata. Microorganisms 2025, 13, 1754. https://doi.org/10.3390/microorganisms13081754
Yuan Y, Pang S, Niu W, Zhang T, Ma L. Community Structure, Growth-Promoting Potential, and Genomic Analysis of Seed-Endophytic Bacteria in Stipagrostis pennata. Microorganisms. 2025; 13(8):1754. https://doi.org/10.3390/microorganisms13081754
Chicago/Turabian StyleYuan, Yuanyuan, Shuyue Pang, Wenkang Niu, Tingting Zhang, and Lei Ma. 2025. "Community Structure, Growth-Promoting Potential, and Genomic Analysis of Seed-Endophytic Bacteria in Stipagrostis pennata" Microorganisms 13, no. 8: 1754. https://doi.org/10.3390/microorganisms13081754
APA StyleYuan, Y., Pang, S., Niu, W., Zhang, T., & Ma, L. (2025). Community Structure, Growth-Promoting Potential, and Genomic Analysis of Seed-Endophytic Bacteria in Stipagrostis pennata. Microorganisms, 13(8), 1754. https://doi.org/10.3390/microorganisms13081754





