Improved Growth Media for Isolation and Identification of Fish Pathogenic Tenacibaculum spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Nutrient Agar Media Preparation
2.2. Comparison of Bacterial Growth on Five Different Agar Media
2.3. Phenotypic Characterization
3. Results
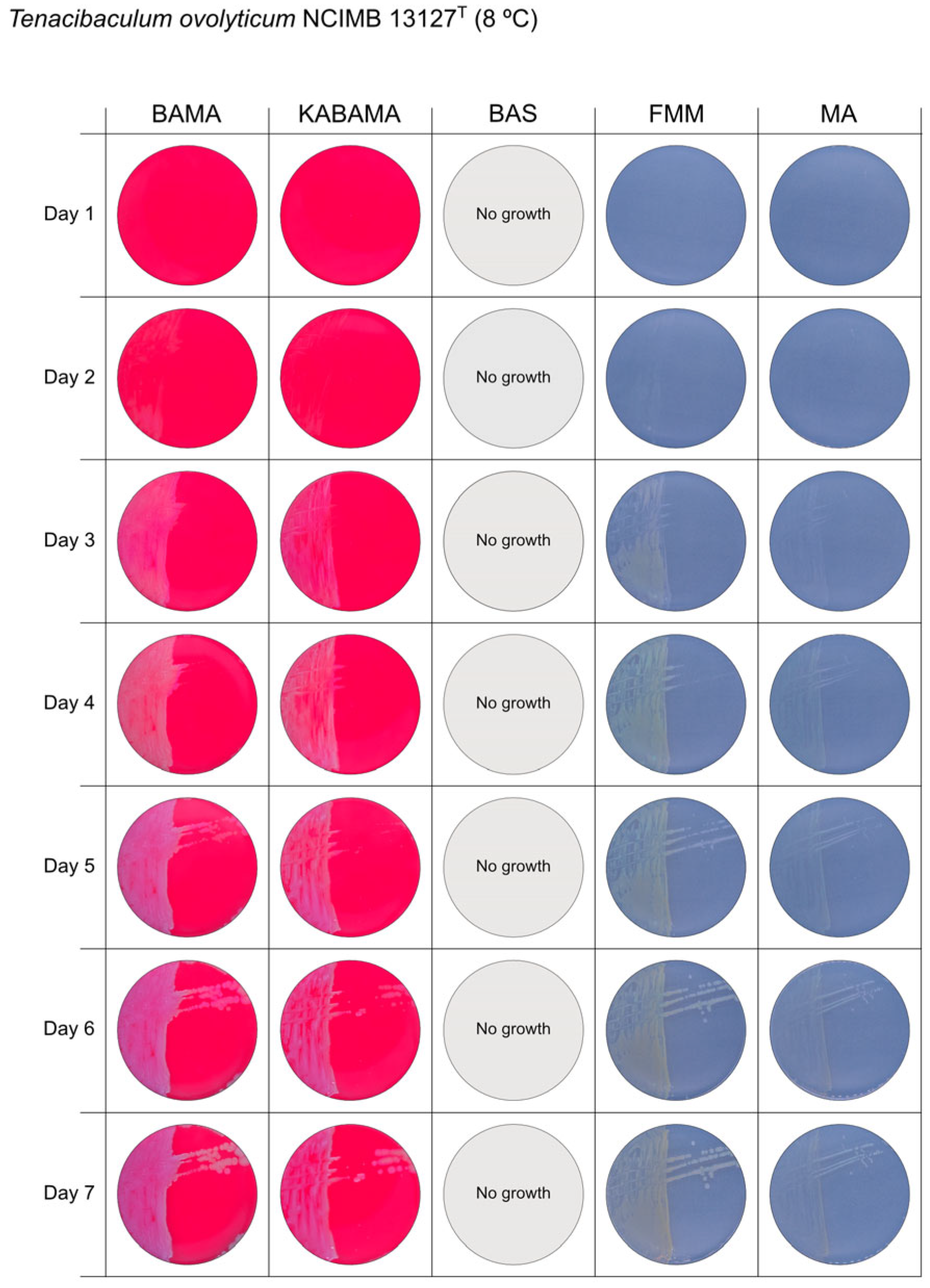
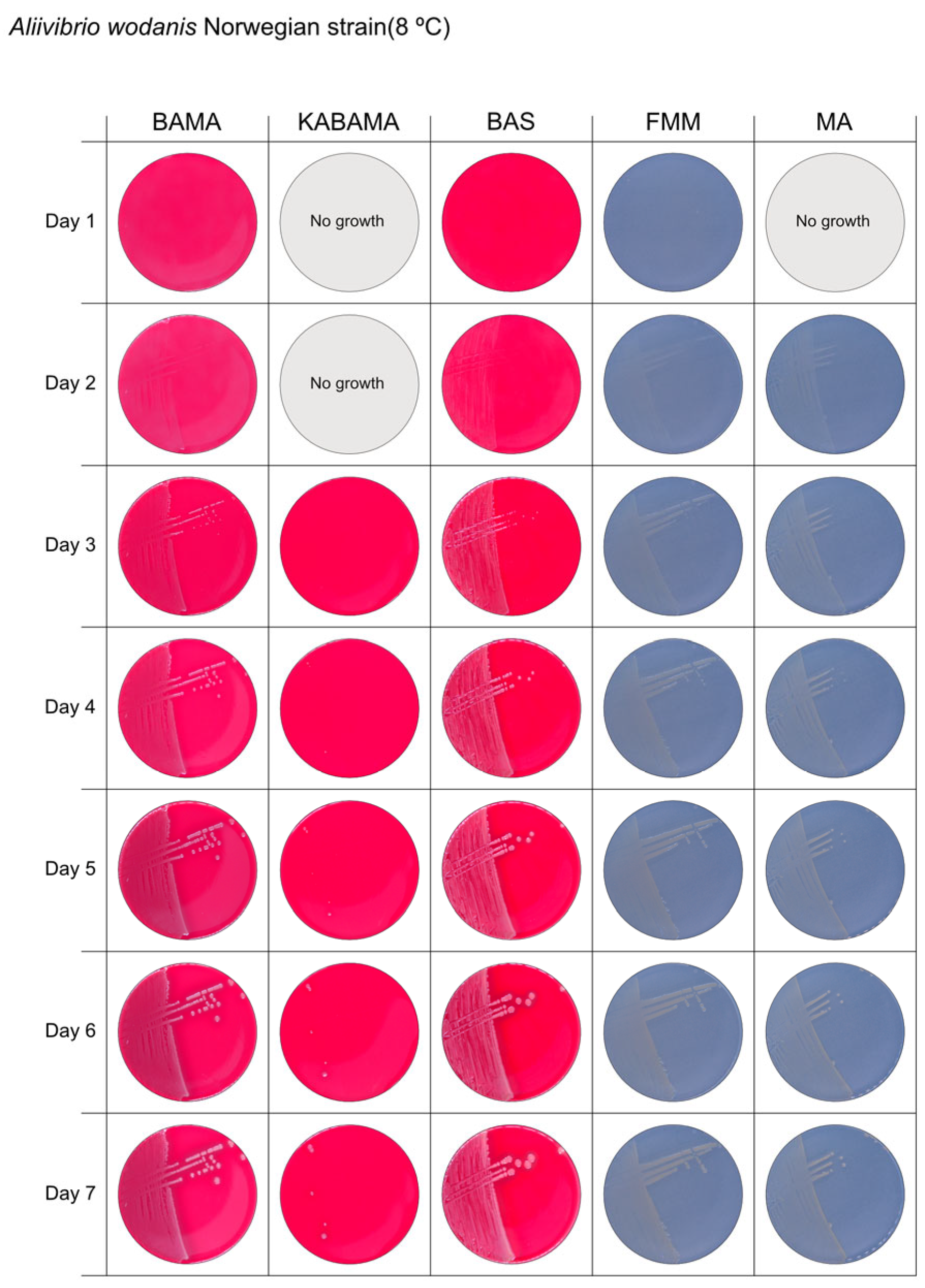
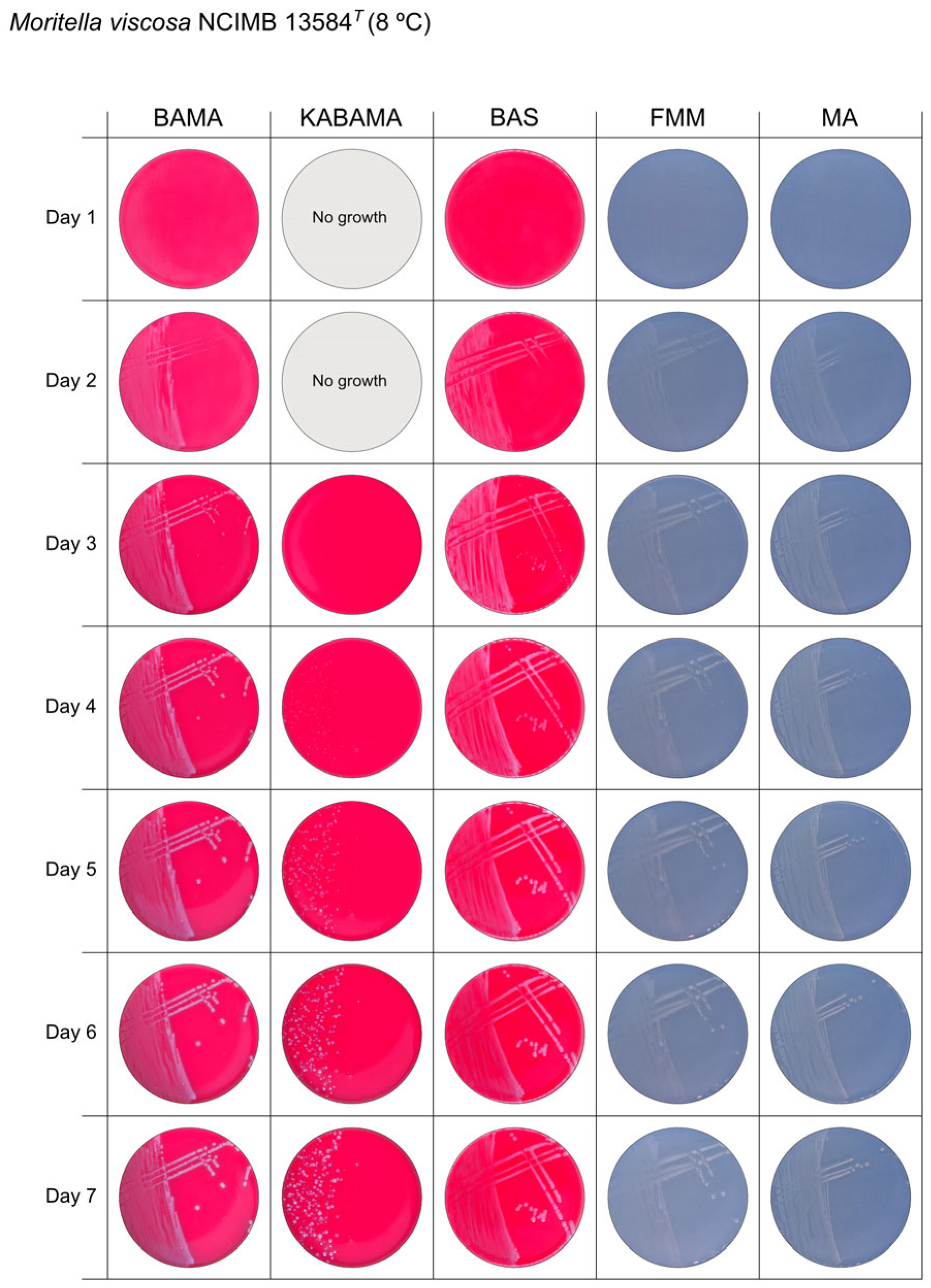
3.1. Growth Comparison
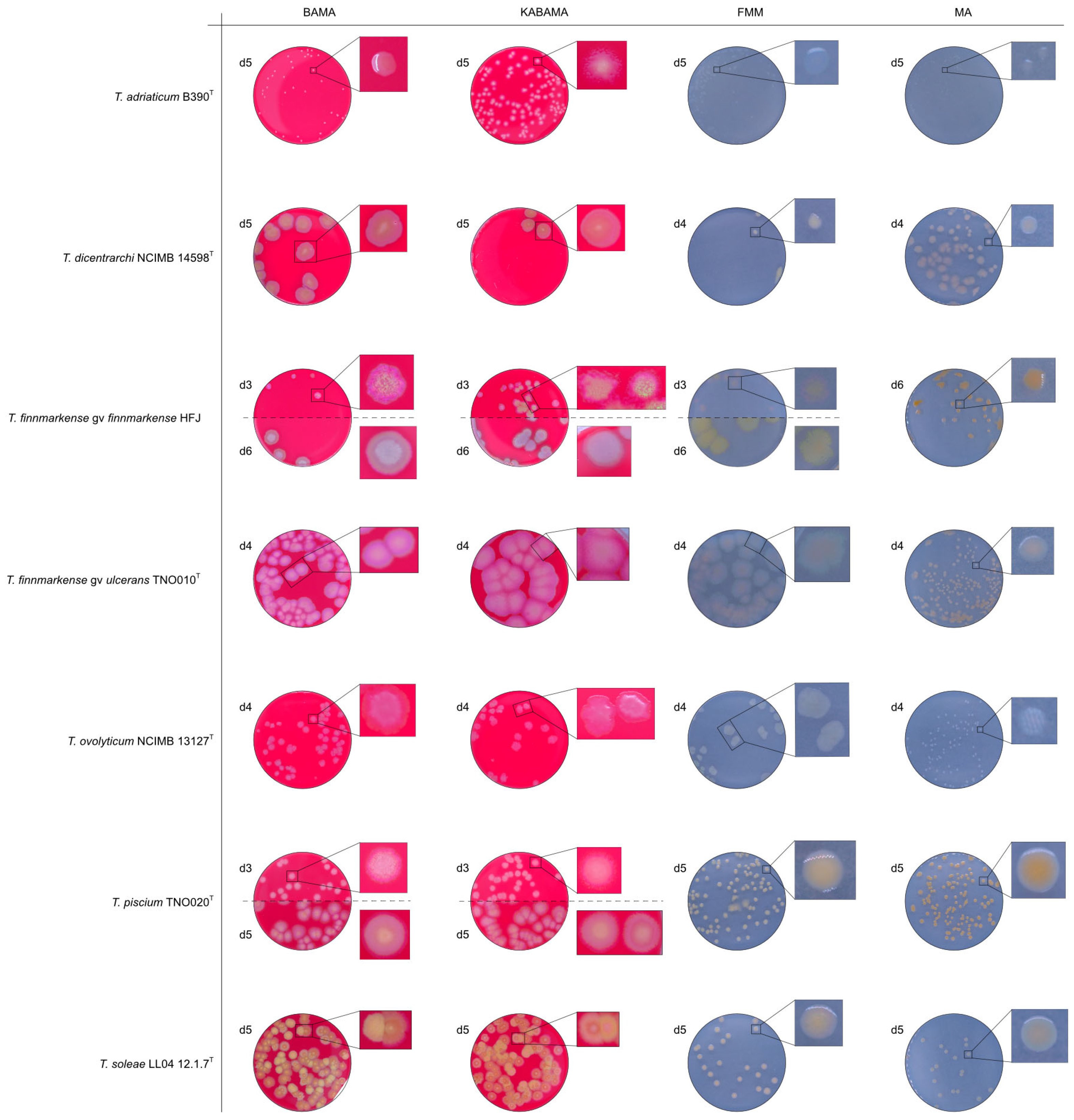
3.2. Phenotypical Characteristics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAS | Blood agar medium supplemented with 2% NaCl |
| FMM | Flexibacter maritimus medium |
| MA | Marine Agar |
References
- Toranzo, A.E.; Magariños, B.; Romalde, J.L. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 2005, 246, 37–61. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Toranzo, A.E.; Magariños, B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Dis. Aquat. Org. 2006, 71, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Herrera, R.; Irgang, R.; Magariños, B.; Romalde, J.L.; Toranzo, A.E. Use of microcosms to determine the survival of the fish pathogen Tenacibaculum maritimum in seawater. Environ. Microbiol. 2006, 8, 921–928. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Irgang, R.; Sandoval, C.; Moreno-Lira, P.; Houel, A.; Duchaud, E.; Poblete-Morales, M.; Nicolas, P.; Ilardi, P. Isolation, Characterization and Virulence Potential of Tenacibaculum dicentrarchi in Salmonid Cultures in Chile. Transbound. Emerg. Dis. 2016, 63, 121–126. [Google Scholar] [CrossRef]
- Klakegg, Ø.; Abayneh, T.; Fauske, A.K.; Fülberth, M.; Sørum, H. An outbreak of acute disease and mortality in Atlantic salmon (Salmo salar) post-smolts in Norway caused by Tenacibaculum dicentrarchi. J. Fish Dis. 2019, 42, 789–807. [Google Scholar] [CrossRef]
- Olsen, A.B.; Spilsberg, B.; Nilsen, H.K.; Lagesen, K.; Gulla, S.; Avendaño-Herrera, R.; Irgang, R.; Duchaud, E.; Colquhoun, D.J. Tenacibaculum piscium sp. nov., isolated from skin ulcers of sea-farmed fish, and description of Tenacibaculum finnmarkense sp. nov. with subdivision into genomovars finnmarkense and ulcerans. Int. J. Syst. Evol. Microbiol. 2020, 70, 6079–6090. [Google Scholar] [CrossRef]
- Småge, S.B.; Brevik, O.J.; Duesund, H.; Ottem, K.F.; Watanabe, K.; Nylund, A. Tenacibaculum finnmarkense sp. nov., a fish pathogenic bacterium of the family Flavobacteriaceae isolated from Atlantic salmon. Antonie Van Leeuwenhoek 2016, 109, 273–285. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Olsen, A.B.; Saldarriaga-Cordoba, M.; Colquhoun, D.J.; Reyes, V.; Rivera-Bohle, J.; Duchaud, E.; Irgang, R. Isolation, identification, virulence potential and genomic features of Tenacibaculum piscium isolates recovered from Chilean salmonids. Transbound. Emerg. Dis. 2022, 69, e3305–e3315. [Google Scholar] [CrossRef]
- Lagadec, E.; Småge, S.B.; Trösse, C.; Nylund, A. Phylogenetic analyses of Norwegian Tenacibaculum strains confirm high bacterial diversity and suggest circulation of ubiquitous virulent strains. PLoS ONE 2021, 16, e0259215. [Google Scholar] [CrossRef]
- Mabrok, M.; Algammal, A.M.; Sivaramasamy, E.; Hetta, H.F.; Atwah, B.; Alghamdi, S.; Fawzy, A.; Avendaño-Herrera, R.; Rodkhum, C. Tenacibaculosis caused by Tenacibaculum maritimum: Updated knowledge of this marine bacterial fish pathogen. Front. Cell. Infect. Microbiol. 2022, 12, 1068000. [Google Scholar] [CrossRef]
- Frisch, K.; Småge, S.B.; Johansen, R.; Duesund, H.; Brevik, Ø.J.; Nylund, A. Pathology of experimentally induced mouthrot caused by Tenacibaculum maritimum in Atlantic salmon smolts. PLoS ONE 2018, 13, e0206951. [Google Scholar] [CrossRef] [PubMed]
- Handlinger, J.; Soltani, M.; Percival, S. The pathology of Flexibacter maritimus in aquaculture species in Tasmania, Australia. J. Fish Dis. 1997, 20, 159–168. [Google Scholar] [CrossRef]
- Escribano, M.P.; Balado, M.; Toranzo, A.E.; Lemos, M.L.; Magariños, B. The secretome of the fish pathogen Tenacibaculum maritimum includes soluble virulence-related proteins and outer membrane vesicles. Front. Cell. Infect. Microbiol. 2023, 13, 1197290. [Google Scholar] [CrossRef] [PubMed]
- Løvoll, M.; Wiik-Nielsen, C.R.; Tunsjø, H.S.; Colquhoun, D.; Lunder, T.; Sørum, H.; Grove, S. Atlantic salmon bath challenged with Moritella viscosa--pathogen invasion and host response. Fish Shellfish. Immunol. 2009, 26, 877–884. [Google Scholar] [CrossRef]
- Tingbø, M.G.; Haugen Tunheim, S.; Klevan, A.; Kamisinska, A.; Behzaad, H.; Sandtrø, A.; Furevik, A. Antigenic similarities and clinical cross-protection between variant and classic non-viscous strains of Moritella viscosa in Atlantic salmon in Norway. Fish Shellfish. Immunol. 2024, 145, 109306. [Google Scholar] [CrossRef] [PubMed]
- Lillehaug, A.; Lunestad, B.T.; Grave, K. Epidemiology of bacterial diseases in Norwegian aquaculture--a description based on antibiotic prescription data for the ten-year period 1991 to 2000. Dis. Aquat. Org. 2003, 53, 115–125. [Google Scholar] [CrossRef]
- Lunder, T.; Evensen, O.; Holstad, G.; Hastein, T.; Evensen, O. ‘Winter ulcer’ in the Atlantic salmon Salmo salar. Pathological and bacteriological investigations and transmission experiments. Dis. Aquat. Org. 1995, 23, 39–49. [Google Scholar] [CrossRef]
- Olsen, A.B.; Nilsen, H.; Sandlund, N.; Mikkelsen, H.; Sorum, H.; Colquhoun, D.J. Tenacibaculum sp. associated with winter ulcers in sea-reared Atlantic salmon Salmo salar. Dis. Aquat. Organ. 2011, 94, 189–199. [Google Scholar] [CrossRef]
- Benediktsdottir, E.; Verdonck, L.; Sproer, C.; Helgason, S.; Swings, J. Characterization of Vibrio viscosus and Vibrio wodanis isolated at different geographical locations: A proposal for reclassification of Vibrio viscosus as Moritella viscosa comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 479–488. [Google Scholar] [CrossRef]
- Småge, S.B.; Brevik, O.J.; Frisch, K.; Watanabe, K.; Duesund, H.; Nylund, A. Concurrent jellyfish blooms and tenacibaculosis outbreaks in Northern Norwegian Atlantic salmon (Salmo salar) farms. PLoS ONE 2017, 12, e0187476. [Google Scholar] [CrossRef]
- Suzuki, M.; Nakagawa, Y.; Harayama, S.; Yamamoto, S. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: Proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int. J. Syst. Evol. Microbiol. 2001, 51 Pt 5, 1639–1652. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.E.; Henry-Ford, D.; Groff, J.M. Isolation and Characterization of Flexibacter maritimus from Marine Fishes of California. J. Aquat. Anim. Health 1995, 7, 318–326. [Google Scholar] [CrossRef]
- Kent, M.L.; Dungan, C.; Elston, R.A.; Holt, R.A. Cytophaga sp. (Cytophagales) infection in seawater pen-reared Atlantic salmon Salmo salar. Dis. Aquat. Org. 1988, 4, 173–179. [Google Scholar] [CrossRef]
- Hahnke, R.L.; Harder, J. Phylogenetic diversity of Flavobacteria isolated from the North Sea on solid media. Syst. Appl. Microbiol. 2013, 36, 497–504. [Google Scholar] [CrossRef]
- Toranzo, A.E. Tenacibaculosis of Farmed Fish in Southern Europe; Tenacibaculum Maritimum Workshop Maritimum Heritage Center: Campbell River, BC, Canada, 2015. [Google Scholar]
- Frisch, K.; Småge, S.B.; Brevik, O.J.; Duesund, H.; Nylund, A. Genotyping of Tenacibaculum maritimum isolates from farmed Atlantic salmon in Western Canada. J. Fish Dis. 2018, 41, 131–137. [Google Scholar] [CrossRef]
- Kumanan, K.; von Ammon, U.; Fidler, A.; Symonds, J.E.; Walker, S.P.; Carson, J.; Hutson, K.S. Advantages of selective medium for surveillance of Tenacibaculum species in marine fish aquaculture. Aquaculture 2022, 558, 738365. [Google Scholar] [CrossRef]
- Pazos, F.; Santos, Y.; Macías, A.R.; Núñez, S.; Toranzo, A.E. Evaluation of media for the successful culture of Flexibacter maritimus. J. Fish Dis. 1996, 19, 193–197. [Google Scholar] [CrossRef]
- Shieh, H. Studies on the nutrition of a fish pathogen, Flexibacter columnaris. Microbios Lett. 1980, 13, 129–133. [Google Scholar]
- Wilson, T.K.; Douglas, M.; Dunn, V. First identification in Tasmania of fish pathogens Tenacibaculum dicentrarchi and T. soleae and multiplex PCR for these organisms and T. maritimum. Dis. Aquat. Org. 2019, 136, 219–226. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kawai, K.; Oshima, S.-i. Evaluation of an Experimental Immersion Infection Method with Tenacibaculum maritimum in Japanese Flounder Paralichthys olivaceus. Aquac. Sci. 2010, 58, 481–489. [Google Scholar] [CrossRef]
- Mabrok, M.; Machado, M.; Serra, C.R.; Afonso, A.; Valente, L.M.; Costas, B. Tenacibaculosis induction in the Senegalese sole (Solea senegalensis) and studies of Tenacibaculum maritimum survival against host mucus and plasma. J. Fish Dis. 2016, 39, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Suga, K.; Kanai, K.; Sugihara, Y. Biological and Serological Characterization of a Non-gliding Strain of Tenacibaculum maritimum Isolated from a Diseased Puffer Fish Takifugu rubripes. Fish Pathol. 2014, 49, 121–129. [Google Scholar] [CrossRef]
- Apablaza, P.; Frisch, K.; Brevik Ø, J.; Småge, S.B.; Vallestad, C.; Duesund, H.; Mendoza, J.; Nylund, A. Primary Isolation and Characterization of Tenacibaculum maritimum from Chilean Atlantic Salmon Mortalities Associated with a Pseudochattonella spp. Algal Bloom. J. Aquat. Anim. Health 2017, 29, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Småge, S.B.; Frisch, K.; Brevik, Ø.J.; Watanabe, K.; Nylund, A. First isolation, identification and characterisation of Tenacibaculum maritimum in Norway, isolated from diseased farmed sea lice cleaner fish Cyclopterus lumpus L. Aquaculture 2016, 464, 178–184. [Google Scholar] [CrossRef]
- Blodgett, R. BAM Appendix 2: Most Probable Number from Serial Dilutions. In Bacteriological Analytical Manual, 8th ed.; Revision A; FDA: Silver Spring, MD, USA, 1998. [Google Scholar]
- Heindl, H.; Wiese, J.; Imhoff, J.F. Tenacibaculum adriaticum sp. nov., from a bryozoan in the Adriatic Sea. Int. J. Syst. Evol. Microbiol. 2008, 58, 542–547. [Google Scholar] [CrossRef]
- Piñeiro-Vidal, M.; Gijón, D.; Zarza, C.; Santos, Y. Tenacibaculum dicentrarchi sp. nov., a marine bacterium of the family Flavobacteriaceae isolated from European sea bass. Int. J. Syst. Evol. Microbiol. 2012, 62, 425–429. [Google Scholar] [CrossRef]
- Wakabayashi, H.; Hikida, M.; Masumura, K. Flexibacter maritimus sp. nov., a pathogen of marine fishes. Int. J. Syst. Evol. Microbiol. 1986, 36, 396–398. [Google Scholar] [CrossRef]
- Hansen, G.H.; Bergh, O.; Michaelsen, J.; Knappskog, D. Flexibacter ovolyticus sp. nov., a pathogen of eggs and larvae of Atlantic halibut, Hippoglossus hippoglossus L. Int. J. Syst. Evol. Microbiol. 1992, 42, 451–458. [Google Scholar] [CrossRef]
- Piñeiro-Vidal, M.; Carballas, C.G.; Gómez-Barreiro, O.; Riaza, A.; Santos, Y. Tenacibaculum soleae sp. nov., isolated from diseased sole (Solea senegalensis Kaup). Int. J. Syst. Evol. Microbiol. 2008, 58, 881–885. [Google Scholar] [CrossRef]
- Lunder, T.; Sørum, H.; Holstad, G.; Steigerwalt, A.G.; Mowinckel, P.; Brenner, D.J. Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from Atlantic salmon (Salmo salar) with ‘winter ulcer’. Int. J. Syst. Evol. Microbiol. 2000, 50, 427–450. [Google Scholar] [CrossRef]
- Frelier, F.P.; Elton, A.R.; Loy, K.J.; Mincher, C. Macroscopic and microscopic features of ulcerative stomatitis in farmed Atlantic salmon Salmo salar. Dis. Aquat. Org. 1994, 18, 227–231. [Google Scholar] [CrossRef]
- Flint, K.P. A note on a selective agar medium for the enumeration of Flavobacterium species in water. J. Appl. Bacteriol. 1985, 59, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Bridel, S.; Olsen, A.B.; Nilsen, H.; Bernardet, J.F.; Achaz, G.; Avendaño-Herrera, R.; Duchaud, E. Comparative genomics of Tenacibaculum dicentrarchi and “Tenacibaculum finnmarkense” highlights intricate evolution of fish-pathogenic species. Genome Biol. Evol. 2018, 10, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pascual, D.; Lunazzi, A.; Magdelenat, G.; Rouy, Z.; Roulet, A.; Lopez-Roques, C.; Larocque, R.; Barbeyron, T.; Gobet, A.; Michel, G.; et al. The Complete Genome Sequence of the Fish Pathogen Tenacibaculum maritimum Provides Insights into Virulence Mechanisms. Front. Microbiol. 2017, 8, 1542. [Google Scholar] [CrossRef]
- Teramoto, M.; Zhai, Z.; Komatsu, A.; Shibayama, K.; Suzuki, M. Genome Sequence of the Psychrophilic Bacterium Tenacibaculum ovolyticum Strain da5A-8 Isolated from Deep Seawater. Genome Announc. 2016, 4, 10–1128. [Google Scholar] [CrossRef]
- Lujan, K.M.; Eisen, J.A.; Coil, D.A. Draft Genome Sequence of Tenacibaculum soleae UCD-KL19. Genome Announc. 2016, 4, e01120-16. [Google Scholar] [CrossRef]
- Sum, R.; Swaminathan, M.; Rastogi, S.K.; Piloto, O.; Cheong, I. Beta-Hemolytic bacteria selectively trigger liposome lysis, enabling rapid and accurate pathogen detection. ACS Sens. 2017, 2, 1441–1451. [Google Scholar] [CrossRef]
- Penttinen, R.; Hoikkala, V.; Sundberg, L.-R. Gliding Motility and Expression of Motility-Related Genes in Spreading and Non-spreading Colonies of Flavobacterium columnare. Front. Microbiol. 2018, 9, 525. [Google Scholar] [CrossRef]
- Litwin, C.M.; Calderwood, S.B. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 1993, 6, 137–149. [Google Scholar] [CrossRef]
- Saldarriaga-Córdoba, M.; Irgang, R.; Avendaño-Herrera, R. Comparison between genome sequences of Chilean Tenacibaculum dicentrarchi isolated from red conger eel (Genypterus chilensis) and Atlantic salmon (Salmo salar) focusing on bacterial virulence determinants. J. Fish Dis. 2021, 44, 1843–1860. [Google Scholar] [CrossRef]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef]
- Johansen, V.E.; Catón, L.; Hamidjaja, R.; Oosterink, E.; Wilts, B.D.; Rasmussen, T.S.; Sherlock, M.M.; Ingham, C.J.; Vignolini, S. Genetic manipulation of structural color in bacterial colonies. Proc. Natl. Acad. Sci. USA 2018, 115, 2652–2657. [Google Scholar] [CrossRef] [PubMed]
- Brosnahan, C.L.; Munday, J.S.; Ha, H.J.; Preece, M.; Jones, J.B. New Zealand rickettsia-like organism (NZ-RLO) and Tenacibaculum maritimum: Distribution and phylogeny in farmed Chinook salmon (Oncorhynchus tshawytscha). J. Fish Dis. 2019, 42, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, S.J.; Rappé, M.S.; Vergin, K.L.; Adair, N.L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the Green Non-Sulfur bacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 7979–7984. [Google Scholar] [CrossRef] [PubMed]
- Habib, C.; Houel, A.; Lunazzi, A.; Bernardet, J.F.; Olsen, A.B.; Nilsen, H.; Toranzo, A.E.; Castro, N.; Nicolas, P.; Duchaud, E. Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl. Environ. Microbiol. 2014, 80, 5503–5514. [Google Scholar] [CrossRef]







| Component | Quantity |
|---|---|
| Peptone from animal tissue (P7750, Merck, Darmstadt, Germany) | 5.0 g |
| Yeast extract (Y1625, Merck) | 1.0 g |
| Coral pro salt (Red Sea Fish Farm Ltd., Herzliya, Israel) | 37.0 g |
| Bacteriological agar (A5306, Merck) | 15.0 g |
| Milli-Q water (Sigma-Aldrich, Burlington, MA, USA) | 950 mL |
| Sterile defibrinated sheep blood (SR0051E, Thermo Fisher, Waltham, MA, USA) | 50 mL |
| * Kanamycin 50 mg ml−1 (Merck) | 1 mL |
| Bacterial Strain | Cell Number per Agar Plate | Incubation (°C) | Reference |
|---|---|---|---|
| T. adriaticum B390T | >1.8 × 108 | 8, 16 | [37] |
| T. dicentrarchi NCIMB 14598T | 1.21 × 107 | 8, 16 | [38] |
| T. finnmarkense genomovar finnmarkense HFJ | 2.9 × 106 | 4, 8, 16 | [7] |
| T. finnmarkense genomovar ulcerans TNO010T | 2.52 × 107 | 4, 8, 16 | [6] |
| T. maritimum NCIMB 2154T | 2.31 × 106 | 12, 16, 25 | [39] |
| T. maritimum CAN 15-1 | 1.14 × 108 | 12, 16, 25 | [26] |
| T. maritimum NLF-15 | 4.8 × 107 | 12, 16, 25 | [35] |
| T. maritimum Ch-2402 | 1.21 × 108 | 12, 16, 25 | [34] |
| T. ovolyticum NCIMB 13127T | 9.3 × 107 | 4, 8, 16 | [40] |
| T. piscium TNO020T | 1.96 × 107 | 4, 8, 16 | [6] |
| T. soleae LL04 12.1.7T | 1.8 × 108 | 8, 16 | [41] |
| Aliivibrio wodanis Norwegian strain | 1.41 × 106 | 8, 16 | This study |
| Moritella viscosa NCIMB 13584T | 5.7 × 107 | 8, 16 | [42] |
| Bacterial Strain | Agar Medium | Hemolytic Activity | Morphological Characteristics |
|---|---|---|---|
| T. adriaticum B390T | BAMA | - | Small grey-purple, round colonies, entire margins |
| KABAMA | - | Medium-size, yellow round colonies with purple irregular margins | |
| BAS | ND | No growth | |
| FMM | ND | Small, round, grey (faint yellow) colonies with entire margins | |
| MA | ND | Very small, round, and grey colonies with entire margins | |
| T. dicentrarchi NCIMB 14598T | BAMA | ++ | Large umbonate colonies with yellow centers and purple undulating margins |
| KABAMA | ++ | Purple iridescent colonies (2 first days), then large umbonate colonies with yellow center and purple undulating margins | |
| BAS | ND | No growth | |
| FMM | ND | Small yellow-grey entire colonies | |
| MA | ND | Medium-sized faint yellow, entire colonies | |
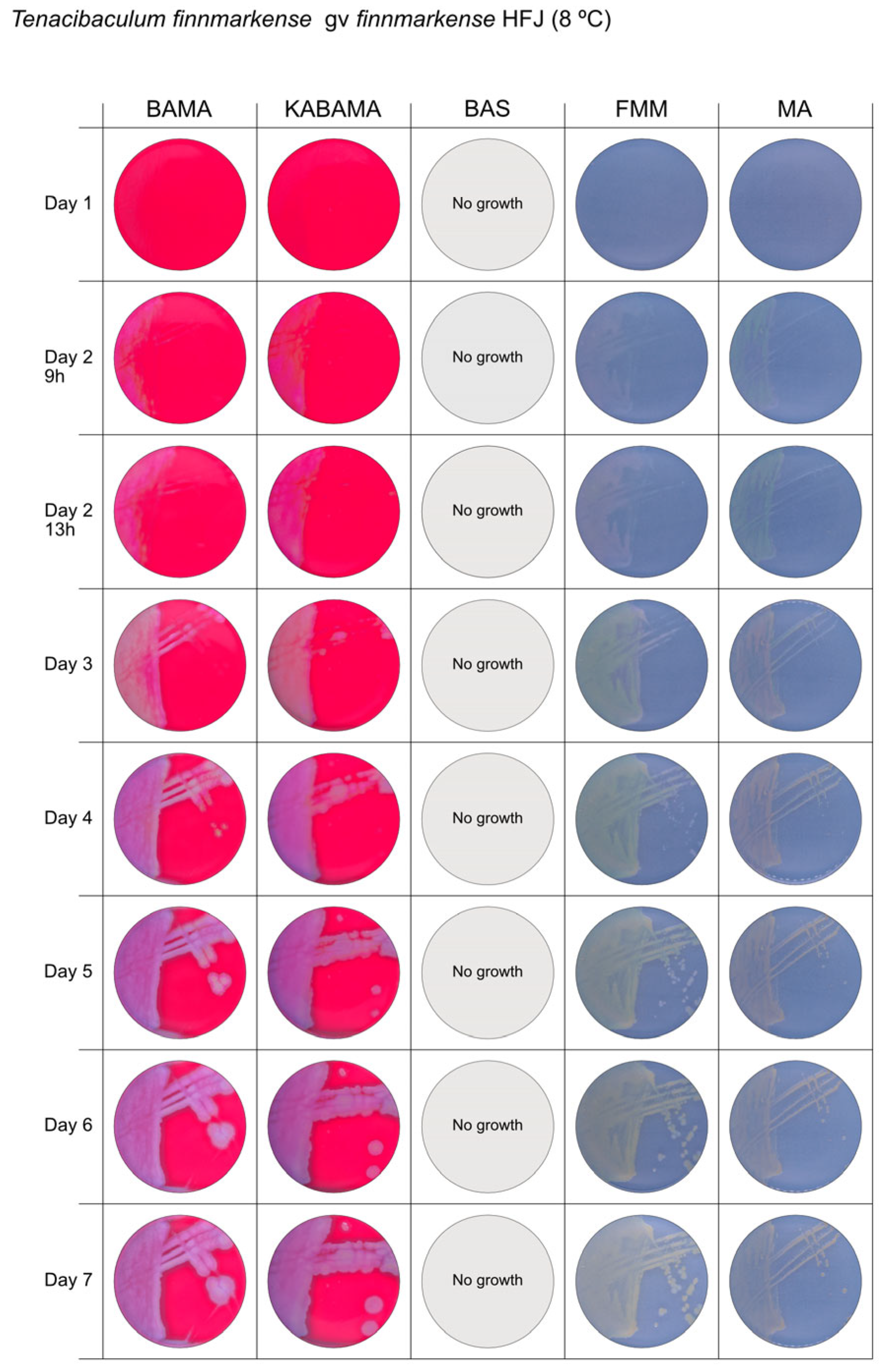
| T. finnmarkense genomovar finnmarkense HFJ | BAMA | +++ | Large, flat, very iridescent colonies, with green centers and purple irregular margins. Very large blue-green non-iridescent entire colonies after 96 h. |
| KABAMA | +++ | Large, flat, very iridescent colonies, with green centers and purple irregular margins. Very large blue-green non-iridescent entire colonies after 96 h. | |
| BAS | ND | No growth | |
| FMM | ND | Weak yellow-green color, slightly iridescent, large colonies with purple margins | |
| MA | ND | Medium-size green then yellow (after 72 h) entire colonies | |
| T. finnmarkense genomovar ulcerans TNO010T | BAMA | - | Large purple iridescent undulating colonies. Purple-white without iridescence after 72 h |
| KABAMA | - | Large purple iridescent undulating colonies. Purple-white without iridescence after 72 h | |
| BAS | ND | No growth | |
| FMM | ND | Large faint yellow-green color colonies with slight iridescent margins | |
| MA | ND | Small yellow entire colonies | |
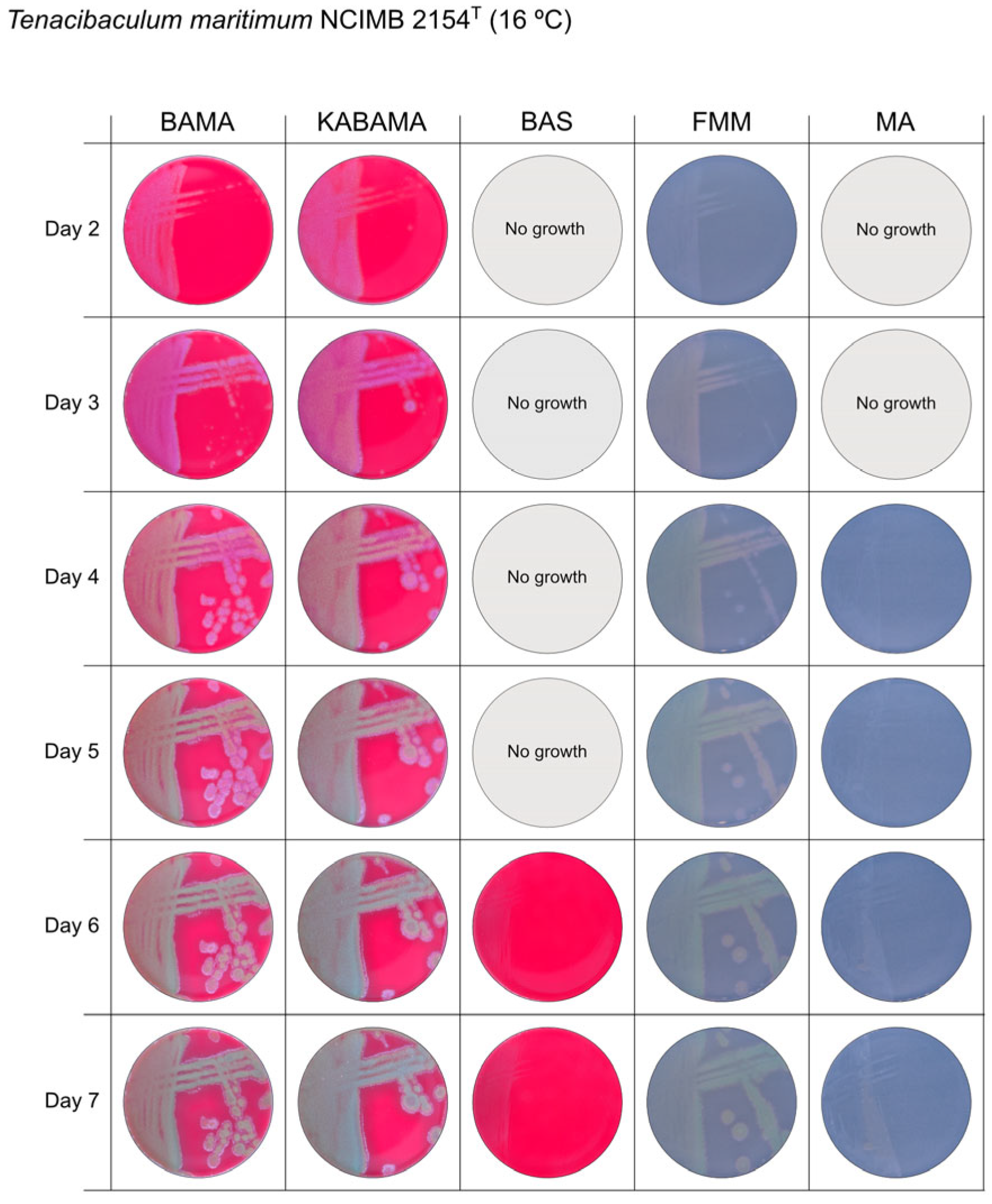
| T. maritimum NCIMB 2154T | BAMA | ++ | Medium-size purple flat colonies with irregular margins displaying important ruggedness. Large dark green with irregular margins and important ruggedness after 96 h |
| KABAMA | ++ | Medium-size purple flat colonies with irregular margins displaying important ruggedness. Large dark green with irregular margins and important ruggedness after 96 h | |
| BAS | + | Slight growth after 120 h. | |
| FMM | ND | Small white-grey colonies with ruggedness. Medium-size yellow-grey colonies after 120 h | |
| MA | ND | Very small, faint, grey-yellow colonies after 96 h | |
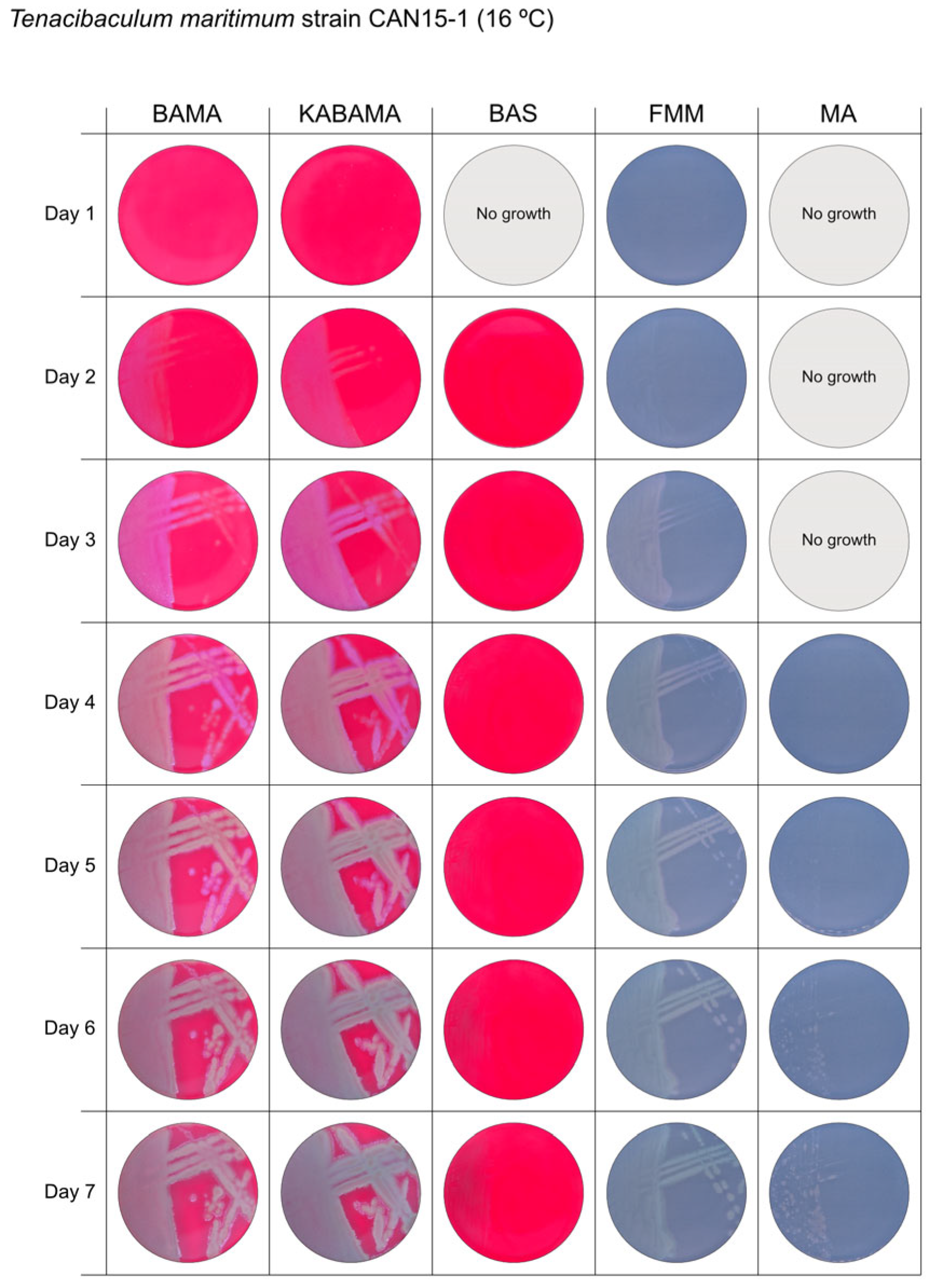
| T. maritimum CAN 15-1 | BAMA | ++ | Purple medium-size colonies with important ruggedness and irregular margins. |
| KABAMA | ++ | Purple medium-size colonies with important ruggedness and irregular margins. | |
| BAS | +++ | Very slow growth of small, faint purple color colonies starting at 48 h | |
| FMM | ND | Grey medium-size colonies | |
| MA | ND | Very slow growth of small white-grey colonies displaying ruggedness | |
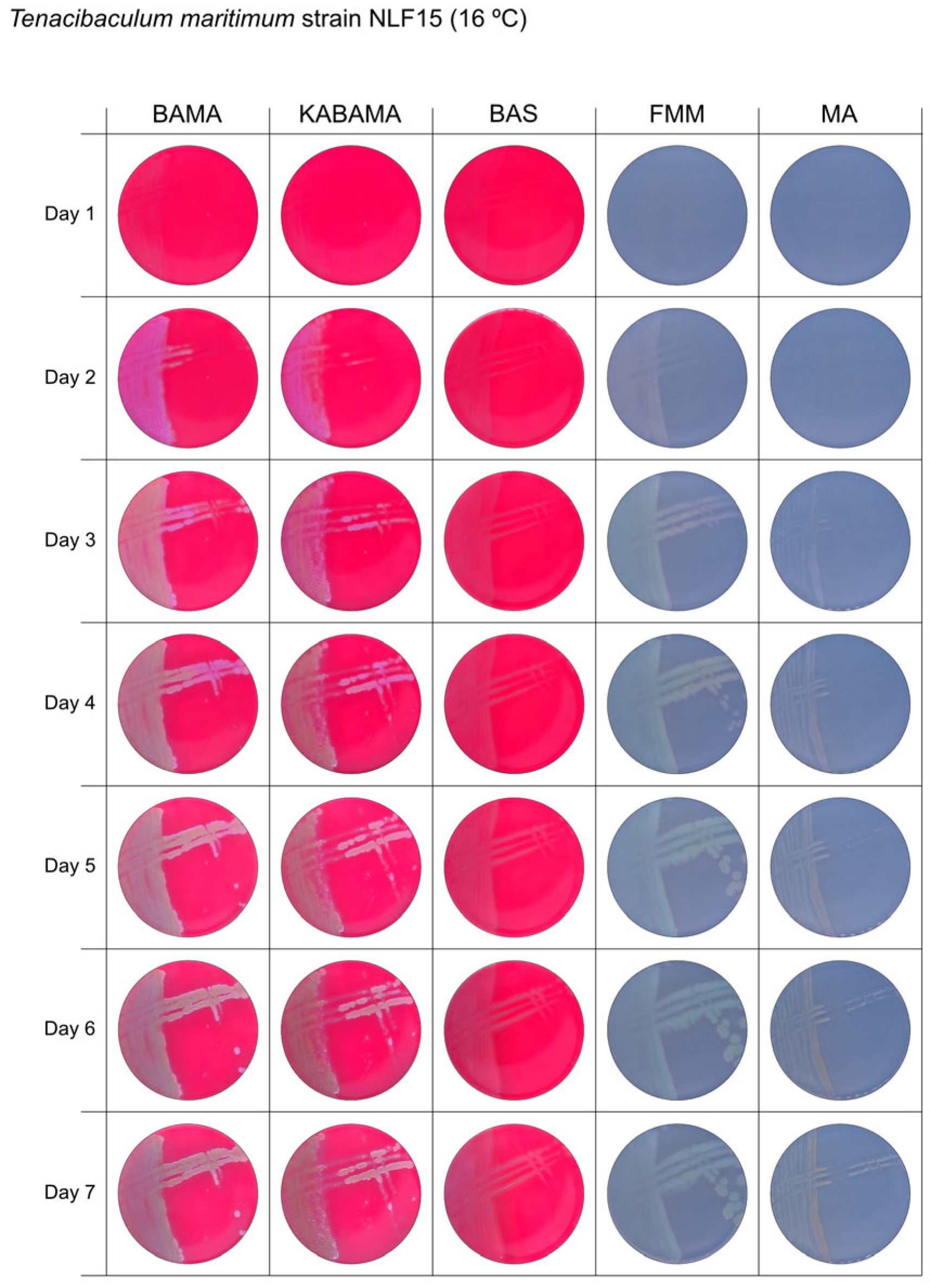
| T. maritimum NLF-15 | BAMA | ++ | Purple medium-size colonies with important ruggedness and irregular margins. Turn large green-yellow colonies with ruggedness after 96 h |
| KABAMA | ++ | Purple medium-size colonies with important ruggedness and irregular margins. Turn large green-yellow colonies with ruggedness after 96 h | |
| BAS | +++ | Slight growth of beige entire colonies | |
| FMM | ND | Medium-size colonies are grey−yellow colonies. Faint yellow colonies with light margin after 120 h | |
| MA | ND | Very small light colonies | |
| T. maritimum Ch-2402 | BAMA | + | Small purple colonies with irregular margins and strong ruggedness. Turn dark green after prolonged growth |
| KABAMA | + | Medium-size purple colonies with irregular margins and strong ruggedness. Turn dark green after prolonged growth | |
| BAS | ++ | Very small white colonies | |
| FMM | ND | Medium-size faint yellow colonies with light margins | |
| MA | ND | Very small light colonies | |
| T. ovolyticum NCIMB 13127T | BAMA | + (weak) | Medium-size purple undulating colonies |
| KABAMA | + (weak) | Medium-size purple undulating colonies | |
| BAS | ND | No growth | |
| FMM | ND | Medium-large yellow−grey colonies | |
| MA | ND | Small white colonies | |
| T. piscium TNO020T | BAMA | - | Medium-large yellow−white colonies with purple iridescent margins. Undulating entire colonies |
| KABAMA | - | Medium-large yellow−white colonies with purple iridescent margins. Undulating entire colonies | |
| BAS | - | White small colonies | |
| FMM | ND | Small white-yellow colonies | |
| MA | ND | Small yellow colonies | |
| T. soleae LL04 12.1.7T | BAMA | - | Medium-size yellow colonies with purple and green iridescent margins |
| KABAMA | - | Medium-size yellow colonies with purple and green iridescent margins | |
| BAS | ND | No growth | |
| FMM | ND | Small yellow colonies | |
| MA | ND | Small colonies with a faint yellow-green color | |
| Aliivibrio wodanis Norwegian strain | BAMA | ++ | Small white entire colonies |
| KABAMA | ++ | Very slow growth of small white colonies | |
| BAS | ++ | Medium size white colonies | |
| FMM | ND | Small white entire colonies | |
| MA | ND | Small white entire colonies | |
| Moritella viscosa NCIMB 13584T | BAMA | ++ | Small grey entire colonies with classic viscous characteristics |
| KABAMA | ++ | Very slow growth of small white colonies with classic viscous characteristics | |
| BAS | ++ | Small white colonies with classic viscous characteristics | |
| FMM | ND | Small white entire colonies with classic viscous characteristics | |
| MA | ND | Small white entire colonies with classic viscous characteristics |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagadec, E.; Kahrs, I.E.B.; Frisch, K.; Duesund, H.; Nylund, A.; Småge, S.B. Improved Growth Media for Isolation and Identification of Fish Pathogenic Tenacibaculum spp. Microorganisms 2025, 13, 1567. https://doi.org/10.3390/microorganisms13071567
Lagadec E, Kahrs IEB, Frisch K, Duesund H, Nylund A, Småge SB. Improved Growth Media for Isolation and Identification of Fish Pathogenic Tenacibaculum spp. Microorganisms. 2025; 13(7):1567. https://doi.org/10.3390/microorganisms13071567
Chicago/Turabian StyleLagadec, Erwan, Ingeborg Emilie Berg Kahrs, Kathleen Frisch, Henrik Duesund, Are Nylund, and Sverre Bang Småge. 2025. "Improved Growth Media for Isolation and Identification of Fish Pathogenic Tenacibaculum spp." Microorganisms 13, no. 7: 1567. https://doi.org/10.3390/microorganisms13071567
APA StyleLagadec, E., Kahrs, I. E. B., Frisch, K., Duesund, H., Nylund, A., & Småge, S. B. (2025). Improved Growth Media for Isolation and Identification of Fish Pathogenic Tenacibaculum spp. Microorganisms, 13(7), 1567. https://doi.org/10.3390/microorganisms13071567






