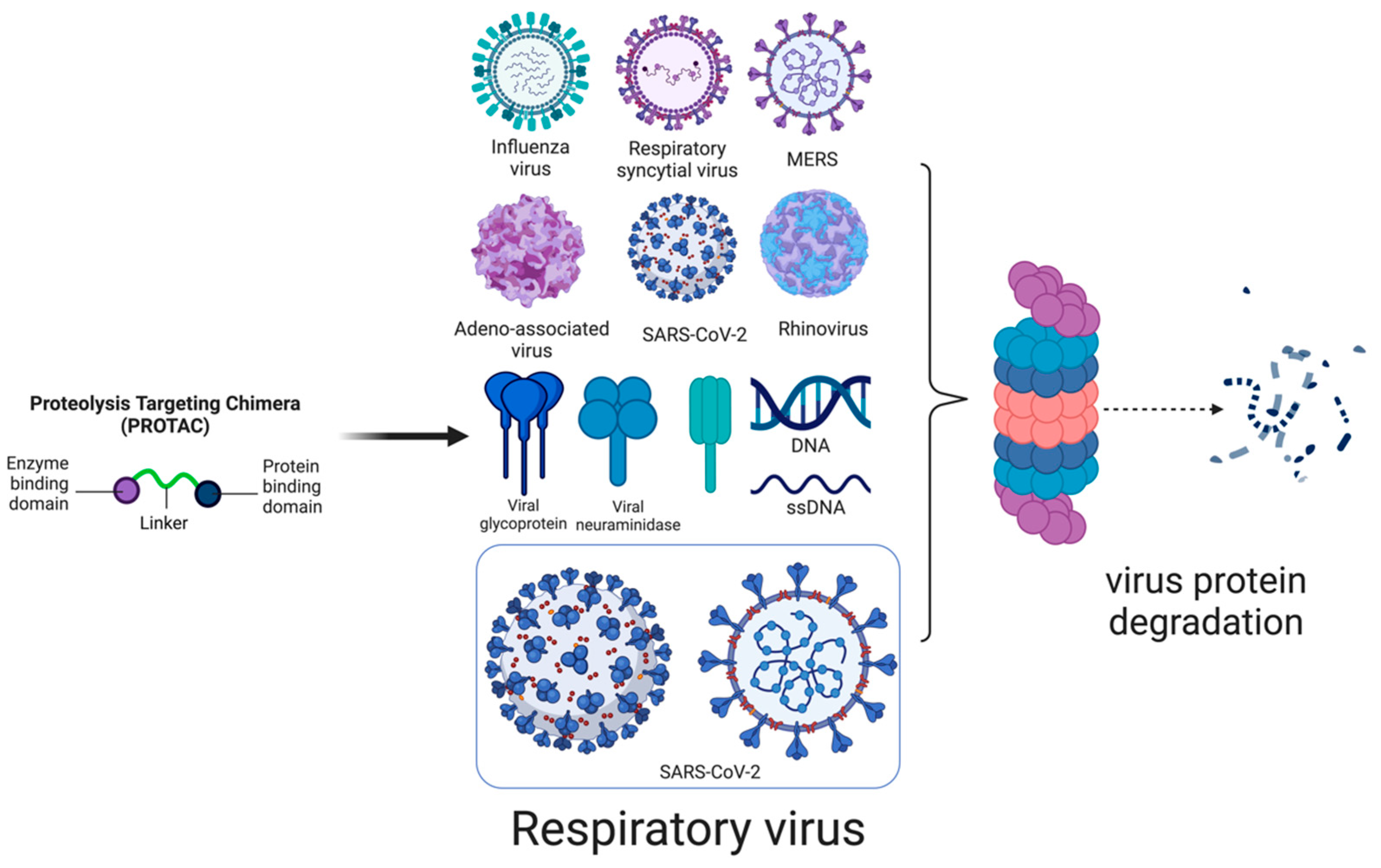
PROTAC-Based Antivirals for Respiratory Viruses: A Novel Approach for Targeted Therapy and Vaccine Development
Abstract
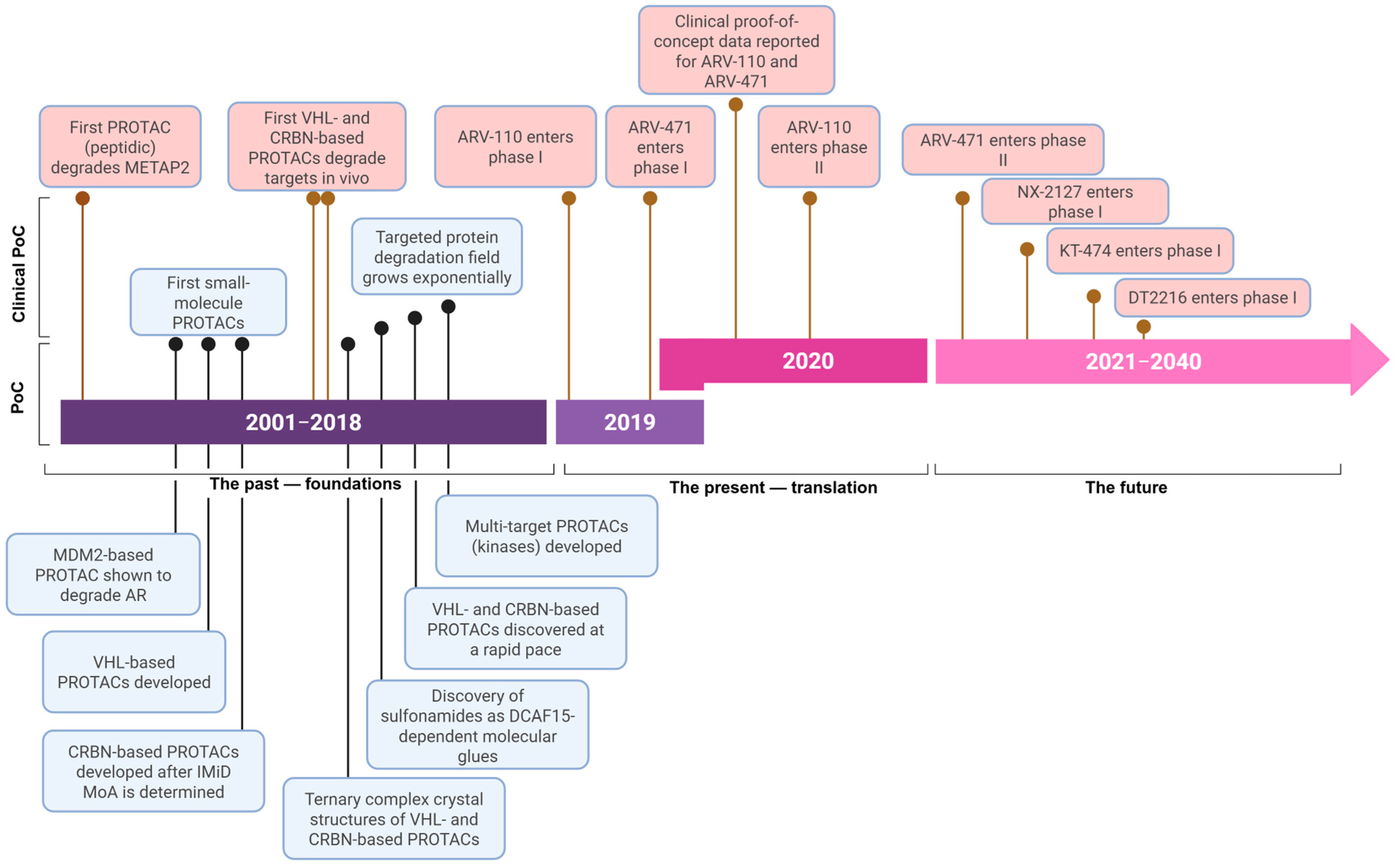
1. Introduction
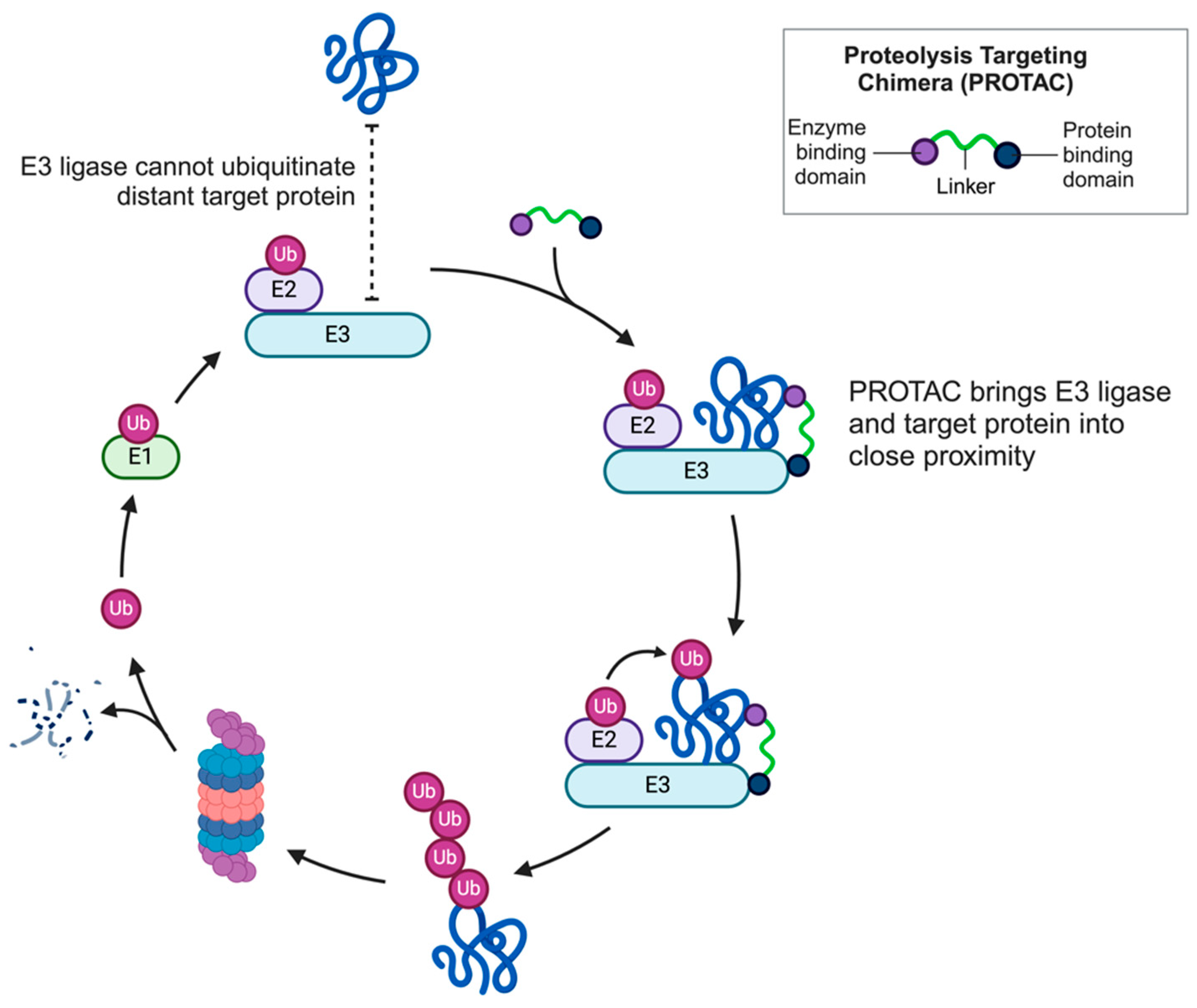
2. PROTAC
2.1. PROTAC Design and Synthesis
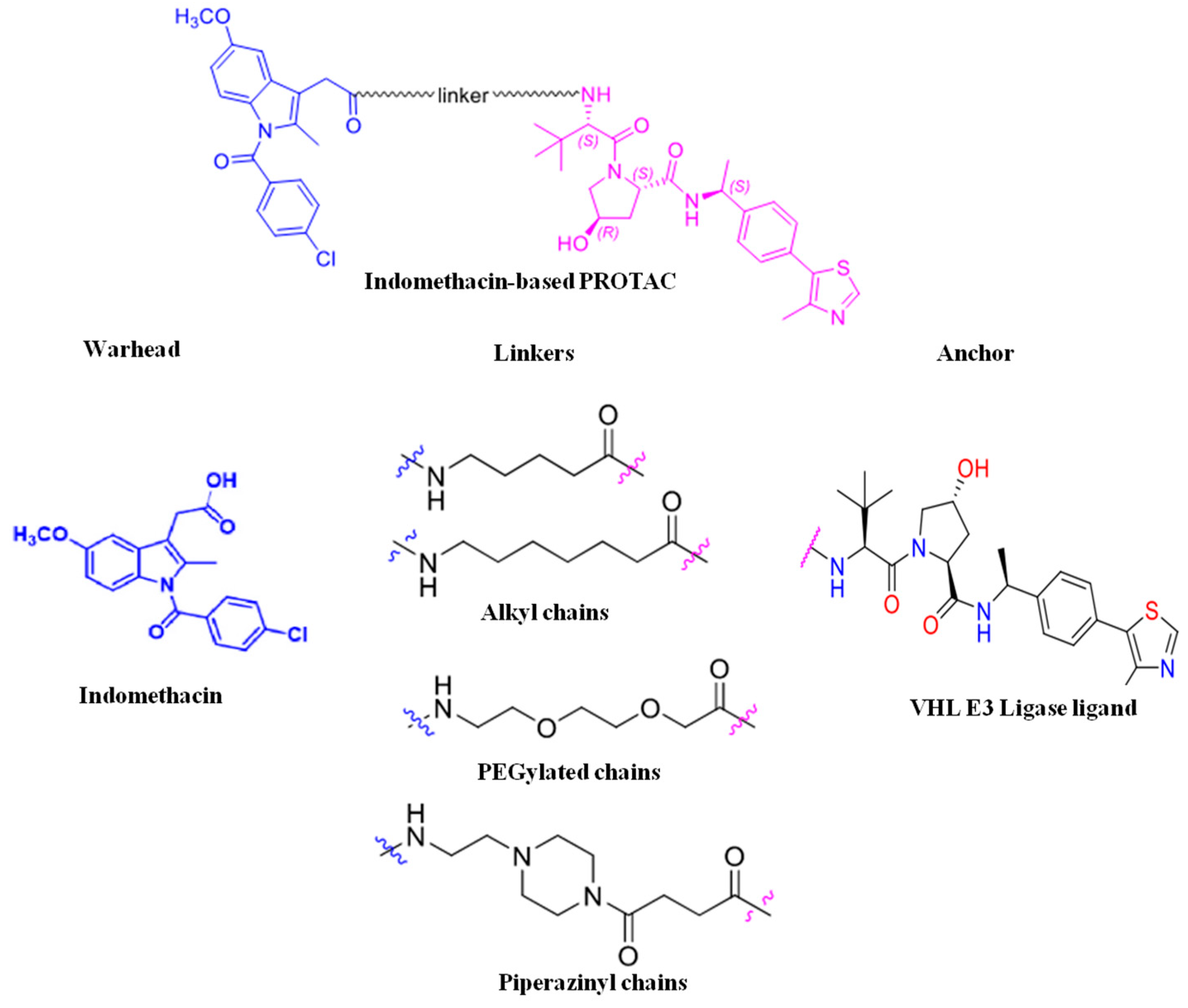
2.1.1. Warhead
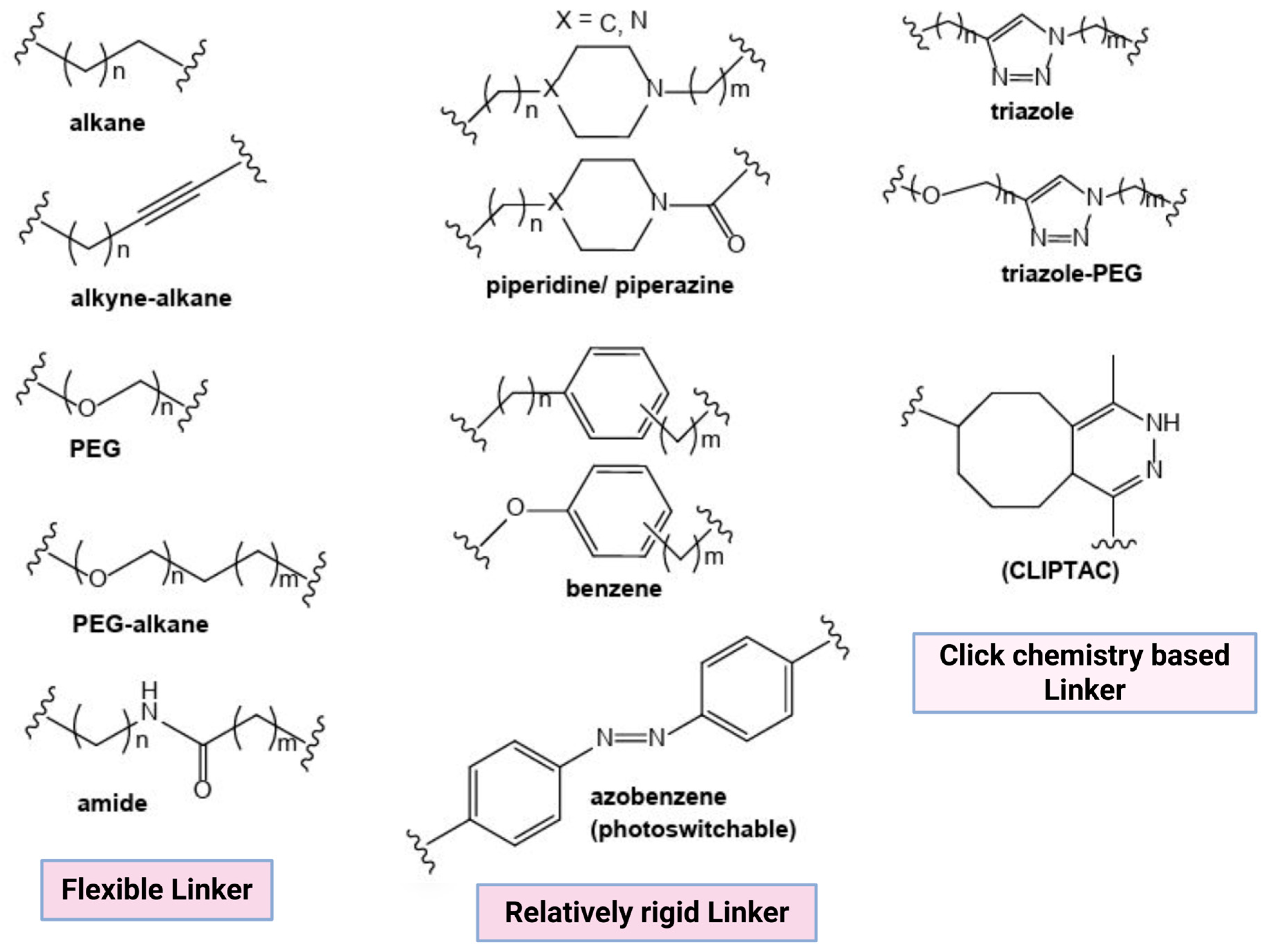
2.1.2. Linker
- Linker length: Linker length determines the PROTAC target protein degradation efficacy; long linkers may hinder POI ubiquitination, whereas short linkers minimize the chances of ternary complex (TC) formation and result in a hook effect, i.e., binary complex formation (PROTAC-E3 ligase complex) [15].
- Flexibility: Flexible linkers, such as alkyl chains and PEG, increase PROTAC flexibility, whereas linkers like alkynes, piperazine, and triazole increase PROTAC rigidity. Alkyl, PEG, and extended PEG are the most commonly used linkers because of their easily controlled flexibility [15].
- Chemical composition: PROTACs with a high molecular weight result in poor pharmacokinetic–pharmacodynamic (PK/PD) properties. Multiple warhead–linker–anchor combinations can be screened to select a PROTAC with the desired PK/PD properties. The hydrophilic linkers (PEG) increase the bioavailability and solubility of PROTAC molecules. On the contrary, hydrophobic linkers improve the PROTAC’s cellular permeability [14,15].
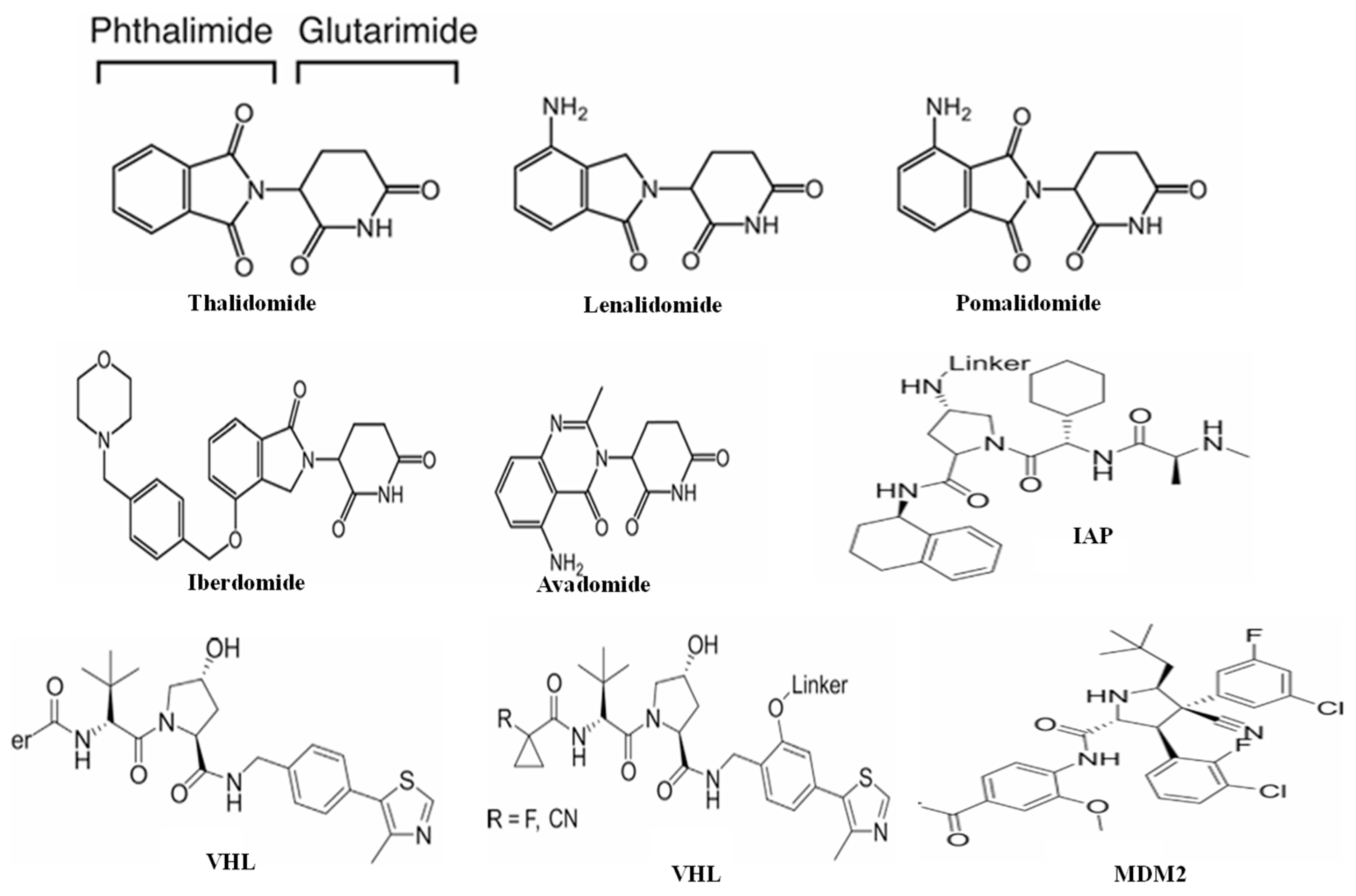
2.1.3. Anchor
2.1.4. PROTAC Synthesis
3. PROTAC-Based Antivirals
3.1. Protac-Based Antivirals for Influenza Virus
3.1.1. PROTAC-Based Antivirals Targeting Hemagglutinin
3.1.2. PROTAC-Based Antiviral Targeting Neuraminidase
3.1.3. Others
3.2. Protac-Based Antiviral for SARS-CoV-2
4. PROTAC Targeting Host Proteins
4.1. Human Prostaglandin E Synthase Type 2 (PGES-2)-Based PROTAC
4.2. Cyclin Dependent Kinase (CDK)-Based PROTAC
4.3. PROTAC-Based Antiviral for Virus-Induced Cytokine Storm
5. Protac Vaccine
6. Challenges
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Weston, S.; Frieman, M.B. Respiratory Viruses. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2018; p. B9780128012383661615. ISBN 978-0-12-801238-3. [Google Scholar]
- Cherry, J.D.; Krogstad, P. SARS: The First Pandemic of the 21st Century. Pediatr. Res. 2004, 56, 1–5. [Google Scholar] [CrossRef]
- Azhar, E.I.; Hui, D.S.C.; Memish, Z.A.; Drosten, C.; Zumla, A. The Middle East Respiratory Syndrome (MERS). Infect. Dis. Clin. North. Am. 2019, 33, 891–905. [Google Scholar] [CrossRef] [PubMed]
- Acuti Martellucci, C.; Flacco, M.E.; Cappadona, R.; Bravi, F.; Mantovani, L.; Manzoli, L. SARS-CoV-2 pandemic: An overview. Adv. Biol. Regul. 2020, 77, 100736. [Google Scholar] [CrossRef] [PubMed]
- Charostad, J.; Rezaei Zadeh Rukerd, M.; Mahmoudvand, S.; Bashash, D.; Hashemi, S.M.A.; Nakhaie, M.; Zandi, K. A comprehensive review of highly pathogenic avian influenza (HPAI) H5N1: An imminent threat at doorstep. Travel. Med. Infect. Dis. 2023, 55, 102638. [Google Scholar] [CrossRef] [PubMed]
- Lampejo, T. Influenza and antiviral resistance: An overview. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1201–1208. [Google Scholar] [CrossRef]
- Gao, H.; Sun, X.; Rao, Y. PROTAC Technology: Opportunities and Challenges. ACS Med. Chem. Lett. 2020, 11, 237–240. [Google Scholar] [CrossRef]
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. PROTACs: Great opportunities for academia and industry. Sig Transduct. Target. Ther. 2019, 4, 64. [Google Scholar] [CrossRef]
- Garber, K. The PROTAC gold rush. Nat. Biotechnol. 2022, 40, 12–16. [Google Scholar] [CrossRef]
- Samarasinghe, K.T.G.; Crews, C.M. Targeted Protein Degradation: A Promise for Undruggable Proteins. Cell Chem. Biol. 2021, 28, 934–951. [Google Scholar] [CrossRef]
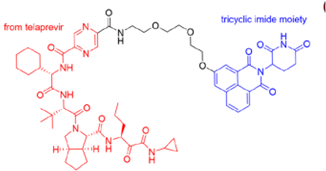
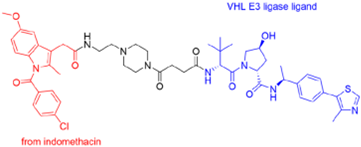
- Desantis, J.; Mercorelli, B.; Celegato, M.; Croci, F.; Bazzacco, A.; Baroni, M.; Siragusa, L.; Cruciani, G.; Loregian, A.; Goracci, L. Indomethacin-based PROTACs as pan-coronavirus antiviral agents. Eur. J. Med. Chem. 2021, 226, 113814. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, X.; Ma, X.; Zou, W.; Chen, Q.; Chen, F.; Deng, X.; Liang, J.; Dong, C.; Lan, K.; et al. Discovery of oseltamivir-based novel PROTACs as degraders targeting neuraminidase to combat H1N1 influenza virus. Cell Insight 2022, 1, 100030. [Google Scholar] [CrossRef]
- de Wispelaere, M.; Du, G.; Donovan, K.A.; Zhang, T.; Eleuteri, N.A.; Yuan, J.C.; Kalabathula, J.; Nowak, R.P.; Fischer, E.S.; Gray, N.S.; et al. Small molecule degraders of the hepatitis C virus protease reduce susceptibility to resistance mutations. Nat. Commun. 2019, 10, 3468. [Google Scholar] [CrossRef] [PubMed]
- Pravin, N.; Jóźwiak, K. PROTAC unleashed: Unveiling the synthetic approaches and potential therapeutic applications. Eur. J. Med. Chem. 2024, 279, 116837. [Google Scholar] [CrossRef] [PubMed]
- Troup, R.I.; Fallan, C.; Baud, M.G.J. Current strategies for the design of PROTAC linkers: A critical review. Explor. Target. Antitumor Ther. 2020, 1, 273–312. [Google Scholar] [CrossRef] [PubMed]
- Wurz, R.P.; Dellamaggiore, K.; Dou, H.; Javier, N.; Lo, M.-C.; McCarter, J.D.; Mohl, D.; Sastri, C.; Lipford, J.R.; Cee, V.J. A “Click Chemistry Platform” for the Rapid Synthesis of Bispecific Molecules for Inducing Protein Degradation. J. Med. Chem. 2018, 61, 453–461. [Google Scholar] [CrossRef]
- Xu, H.; Kurohara, T.; Takano, R.; Yokoo, H.; Shibata, N.; Ohoka, N.; Inoue, T.; Naito, M.; Demizu, Y. Development of Rapid and Facile Solid-Phase Synthesis of PROTACs via a Variety of Binding Styles. ChemistryOpen 2022, 11, e202200131. [Google Scholar] [CrossRef]
- Rapid PROTAC Discovery Platform: Nanomole-Scale Array Synthesis and Direct Screening of Reaction Mixtures|ACS Medicinal Chemistry Letters. Available online: https://pubs.acs.org/doi/10.1021/acsmedchemlett.3c00314 (accessed on 20 June 2025).
- Bemis, T.A.; Clair, J.J.L.; Burkart, M.D. Traceless Staudinger ligation enabled parallel synthesis of proteolysis targeting chimera linker variants. Chem. Commun. 2021, 57, 1026–1029. [Google Scholar] [CrossRef]
- Imrie, F.; Bradley, A.R.; van der Schaar, M.; Deane, C.M. Protein Family-Specific Models Using Deep Neural Networks and Transfer Learning Improve Virtual Screening and Highlight the Need for More Data. J. Chem. Inf. Model. 2018, 58, 2319–2330. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, S.; Su, S.; Zhao, C.; Xu, J.; Chen, H. SyntaLinker: Automatic fragment linking with deep conditional transformer neural networks. Chem. Sci. 2020, 11, 8312–8322. [Google Scholar] [CrossRef]
- Rovers, E.; Schapira, M. Chapter Ten—Methods for computer-assisted PROTAC design. In Methods in Enzymology; Lloyd, M., Ed.; Modern Methods of Drug Design and Development; Academic Press: Cambridge, MA, USA, 2023; Volume 690, pp. 311–340. [Google Scholar]
- Fermin, G. Virion Structure, Genome Organization, and Taxonomy of Viruses. In Viruses; Academic Press: Cambridge, MA, USA, 2018; pp. 17–54. [Google Scholar]
- Louten, J. Virus Replication. Essent. Human. Virol. 2016, 49–70. [Google Scholar]
- Millet, J.K.; Jaimes, J.A.; Whittaker, G.R. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol. Rev. 2020, 45, fuaa057. [Google Scholar] [CrossRef]
- Byrd-Leotis, L.; Cummings, R.D.; Steinhauer, D.A. The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase. Int. J. Mol. Sci. 2017, 18, 1541. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Chen, W.; Wu, Y.; Xing, D. New-generation advanced PROTACs as potential therapeutic agents in cancer therapy. Mol. Cancer 2024, 23, 110. [Google Scholar] [CrossRef] [PubMed]
- Petrylak, D.P.; Gao, X.; Vogelzang, N.J.; Garfield, M.H.; Taylor, I.; Moore, M.D.; Peck, R.A.; Burris, H.A. First-in-Human Phase I Study of ARV-110, an Androgen Receptor (AR) PROTAC Degrader in Patients (pts) with Metastatic Castrate-Resistant Prostate Cancer (mCRPC) Following Enzalutamide (ENZ) and/or Abiraterone (ABI). J. Clin. Oncol. 2020, 38, 3500. [Google Scholar] [CrossRef]
- Nelson, P.P.; Papadopoulos, N.G.; Skevaki, C. Respiratory Viral Pathogens. In Encyclopedia of Respiratory Medicine; Elsevier: Amsterdam, The Netherlands, 2022; pp. 129–137. ISBN 978-0-08-102724-0. [Google Scholar]
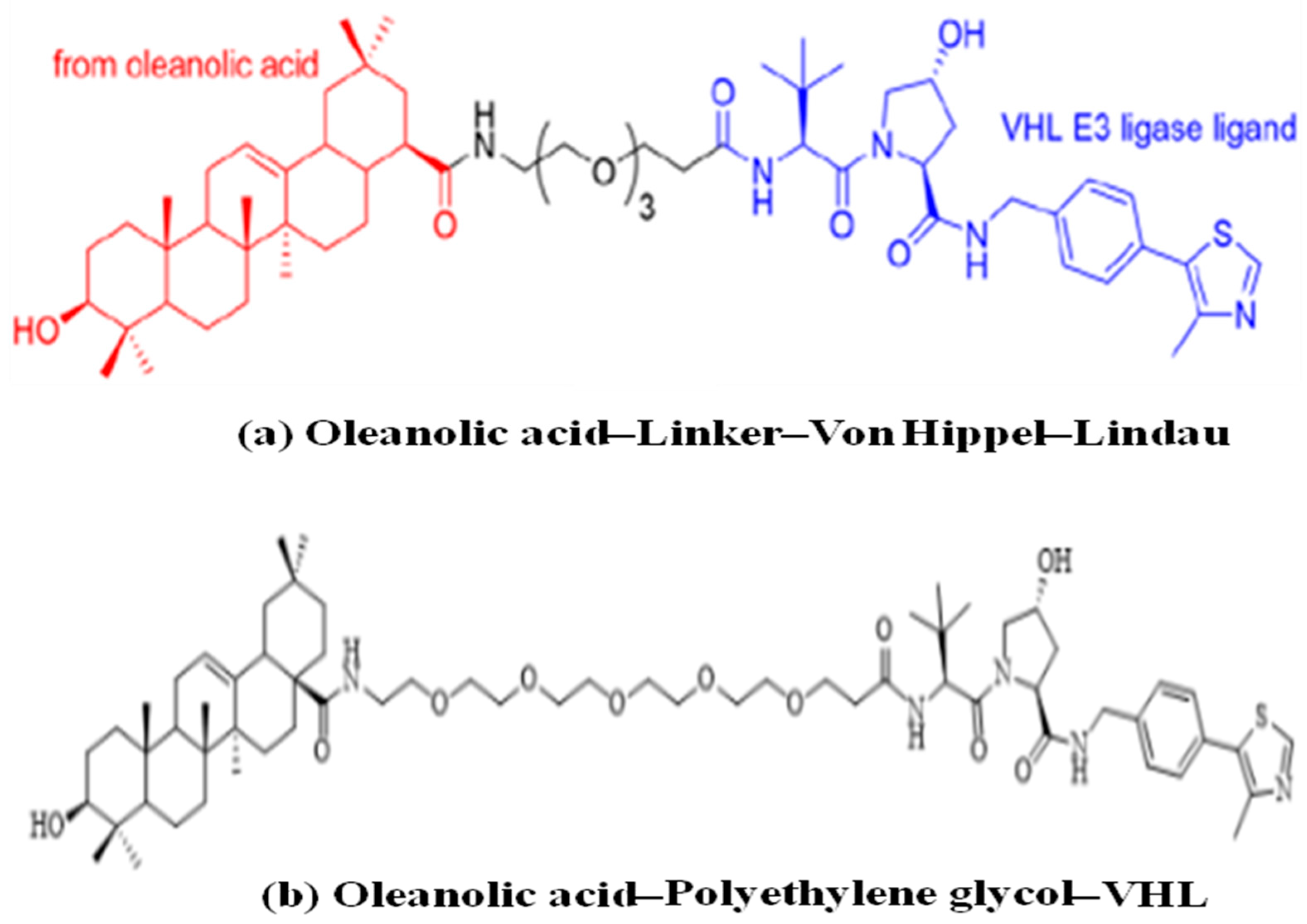
- Li, H.; Wang, S.; Ma, W.; Cheng, B.; Yi, Y.; Ma, X.; Xiao, S.; Zhang, L.; Zhou, D. Discovery of Pentacyclic Triterpenoid PROTACs as a Class of Effective Hemagglutinin Protein Degraders. J. Med. Chem. 2022, 65, 7154–7169. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, J.; Pang, X.; Liu, Z.; Li, Q.; Yi, D.; Zhang, Y.; Fang, X.; Zhang, T.; Zhou, R.; et al. An anti-influenza A virus microbial metabolite acts by degrading viral endonuclease PA. Nat. Commun. 2022, 13, 2079. [Google Scholar] [CrossRef] [PubMed]
- McAuley, J.L.; Gilbertson, B.P.; Trifkovic, S.; Brown, L.E.; McKimm-Breschkin, J.L. Influenza Virus Neuraminidase Structure and Functions. Front. Microbiol. 2019, 10, 39. [Google Scholar] [CrossRef]
- Farrukee, R.; Hurt, A.C. Antiviral Drugs for the Treatment and Prevention of Influenza. Curr. Treat. Options Infect. Dis. 2017, 9, 318–332. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Bonilla, H.; Jagannathan, P.; Shafer, R.W. SARS-CoV-2 Antiviral Therapy. Clin. Microbiol. Rev. 2021, 34, e00109-21. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Ponnapati, M.; Kramme, C.; Plesa, A.M.; Church, G.M.; Jacobson, J.M. Targeted intracellular degradation of SARS-CoV-2 via computationally optimized peptide fusions. Commun. Biol. 2020, 3, 715. [Google Scholar] [CrossRef] [PubMed]
- Shaheer, M.; Singh, R.; Sobhia, M.E. Protein degradation: A novel computational approach to design protein degrader probes for main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2022, 40, 10905–10917. [Google Scholar] [CrossRef]
- De Jesús-González, L.A.; León-Juárez, M.; Lira-Hernández, F.I.; Rivas-Santiago, B.; Velázquez-Cervantes, M.A.; Méndez-Delgado, I.M.; Macías-Guerrero, D.I.; Hernández-Castillo, J.; Hernández-Rodríguez, X.; Calderón-Sandate, D.N.; et al. Advances and Challenges in Antiviral Development for Respiratory Viruses. Pathogens 2025, 14, 20. [Google Scholar] [CrossRef]
- Hajjo, R.; Sabbah, D.A.; Abusara, O.H.; Kharmah, R.; Bardaweel, S. Targeting Human Proteins for Antiviral Drug Discovery and Repurposing Efforts: A Focus on Protein Kinases. Viruses 2023, 15, 568. [Google Scholar] [CrossRef]
- Łukasik, P.; Załuski, M.; Gutowska, I. Cyclin-Dependent Kinases (CDK) and Their Role in Diseases Development–Review. Int. J. Mol. Sci. 2021, 22, 2935. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wang, Q.; Chen, S.; Gao, S.; Song, L.; Liu, P.; Huang, W. Influenza A virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein. J. Virol. 2013, 87, 3039–3052. [Google Scholar] [CrossRef]
- Sui, L.; Li, L.; Zhao, Y.; Zhao, Y.; Hao, P.; Guo, X.; Wang, W.; Wang, G.; Li, C.; Liu, Q. Host cell cycle checkpoint as antiviral target for SARS-CoV-2 revealed by integrative transcriptome and proteome analyses. Sig Transduct. Target. Ther. 2023, 8, 21. [Google Scholar] [CrossRef]
- Gutierrez-Chamorro, L.; Felip, E.; Ezeonwumelu, I.J.; Margelí, M.; Ballana, E. Cyclin-dependent Kinases as Emerging Targets for Developing Novel Antiviral Therapeutics. Trends Microbiol. 2021, 29, 836–848. [Google Scholar] [CrossRef]
- Riyaz Tramboo, S.; Elkhalifa, A.M.E.; Quibtiya, S.; Ali, S.I.; Nazir Shah, N.; Taifa, S.; Rakhshan, R.; Hussain Shah, I.; Ahmad Mir, M.; Malik, M.; et al. The critical impacts of cytokine storms in respiratory disorders. Heliyon 2024, 10, e29769. [Google Scholar] [CrossRef]
- Li, H.; Yang, W.; Li, H.; Bai, X.; Zhang, H.; Fan, W.; Liu, W.; Sun, L. PROTAC targeting cyclophilin A controls virus-induced cytokine storm. iScience 2023, 26, 107535. [Google Scholar] [CrossRef]
- Ghattas, M.; Dwivedi, G.; Lavertu, M.; Alameh, M.-G. Vaccine Technologies and Platforms for Infectious Diseases: Current Progress, Challenges, and Opportunities. Vaccines 2021, 9, 1490. [Google Scholar] [CrossRef] [PubMed]
- Groenke, N.; Trimpert, J.; Merz, S.; Conradie, A.M.; Wyler, E.; Zhang, H.; Hazapis, O.-G.; Rausch, S.; Landthaler, M.; Osterrieder, N.; et al. Mechanism of Virus Attenuation by Codon Pair Deoptimization. Cell Rep. 2020, 31, 107586. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Y.; Meng, F.D.; Sang, G.J.; Zhang, H.L.; Tian, Z.J.; Zheng, H.; Cai, X.H.; Tang, Y.D. A novel viral vaccine platform based on engineered transfer RNA. Emerg. Microbes Infect. 2023, 12, 2157339. [Google Scholar] [CrossRef]
- Du, Y.; Salehi-Rad, R.; Zhang, T.-H.; Crosson, W.P.; Abascal, J.; Chen, D.; Shi, Y.; Jiang, H.; Tseng, Y.-W.; Ma, X.; et al. Hyper-Interferon Sensitive Influenza Induces Adaptive Immune Responses and Overcomes Resistance to Anti-PD-1 in Murine Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2024, 12, 1765–1779. [Google Scholar] [CrossRef] [PubMed]
- Subbarao, K. Live Attenuated Cold-Adapted Influenza Vaccines. Cold Spring Harb. Perspect. Med. 2021, 11, a038653. [Google Scholar] [CrossRef]
- Antia, R.; Ahmed, H.; Bull, J.J. Directed attenuation to enhance vaccine immunity. PLoS Comput. Biol. 2021, 17, e1008602. [Google Scholar] [CrossRef]
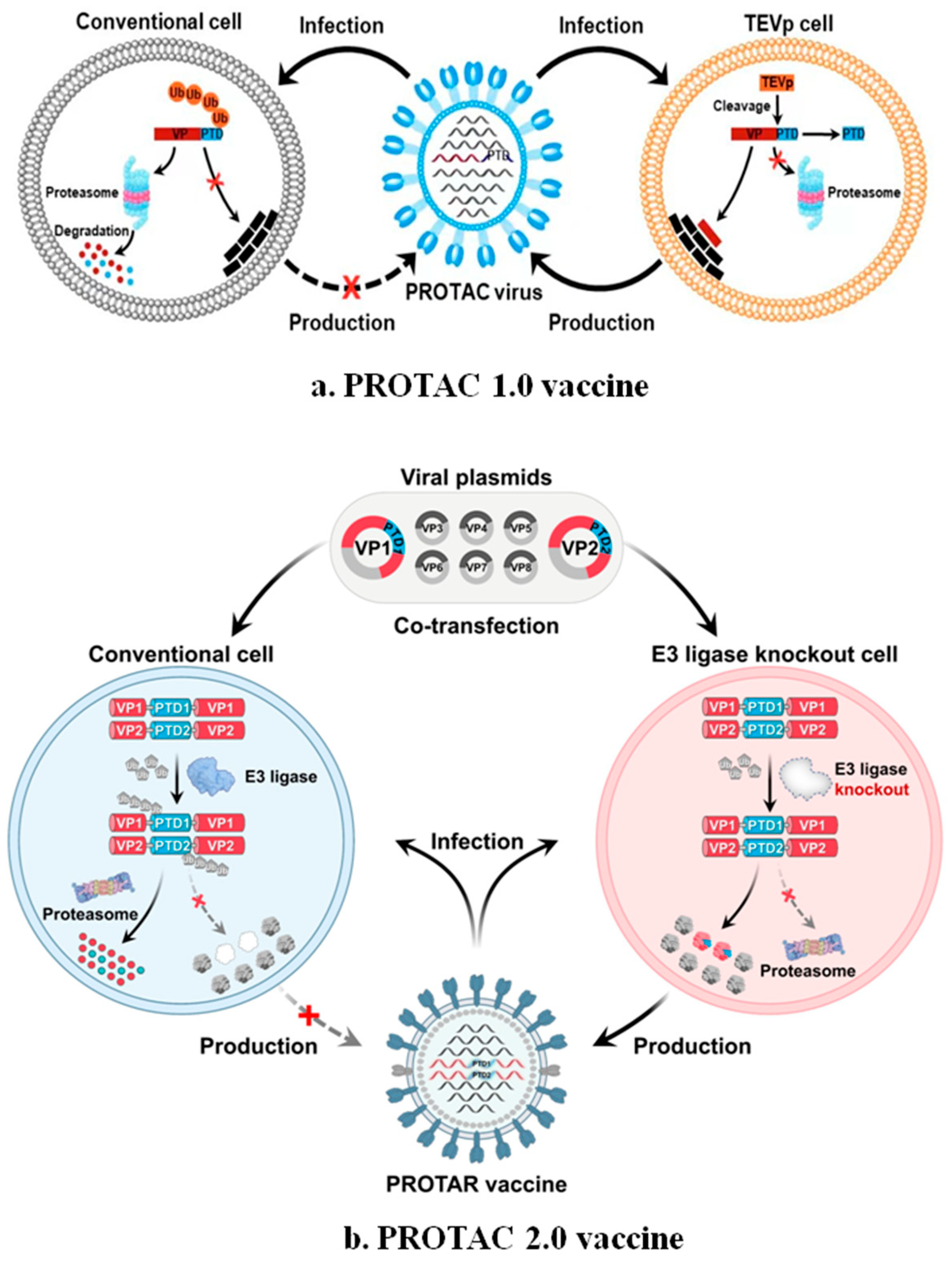
- Li, Z.; Bai, H.; Xi, X.; Tian, W.; Zhang, J.Z.H.; Zhou, D.; Si, L. PROTAC vaccine: A new way to live attenuated vaccines. Clin. Transl. Med. 2022, 12, e1081. [Google Scholar] [CrossRef]
- Si, L.; Shen, Q.; Li, J.; Chen, L.; Shen, J.; Xiao, X.; Bai, H.; Feng, T.; Ye, A.Y.; Li, L.; et al. Generation of a live attenuated influenza A vaccine by proteolysis targeting. Nat. Biotechnol. 2022, 40, 1370–1377. [Google Scholar] [CrossRef]
- Shen, J.; Li, J.; Shen, Q.; Hou, J.; Zhang, C.; Bai, H.; Ai, X.; Su, Y.; Wang, Z.; Zhang, Y.; et al. Proteolysis-targeting influenza vaccine strains induce broad-spectrum immunity and in vivo protection. Nat. Microbiol. 2025, 10, 431–447. [Google Scholar] [CrossRef]
- PROTAR Vaccine 2.0 Generates Influenza Vaccines by Degrading Multiple Viral Proteins|Nature Chemical Biology. Available online: https://www.nature.com/articles/s41589-024-01813-z (accessed on 4 April 2025).
- Ge, J.; Li, S.; Weng, G.; Wang, H.; Fang, M.; Sun, H.; Deng, Y.; Hsieh, C.-Y.; Li, D.; Hou, T. PROTAC-DB 3.0: An updated database of PROTACs with extended pharmacokinetic parameters. Nucleic Acids Res. 2024, 53, D1510–D1515. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hu, Q.; Zhou, Y.; Yang, H.; Bai, F. DiffPROTACs is a deep learning-based generator for proteolysis targeting chimeras. Brief. Bioinform. 2024, 25, bbae358. [Google Scholar] [CrossRef]
- Li, F.; Hu, Q.; Zhang, X.; Sun, R.; Liu, Z.; Wu, S.; Tian, S.; Ma, X.; Dai, Z.; Yang, X.; et al. DeepPROTACs is a deep learning-based targeted degradation predictor for PROTACs. Nat. Commun. 2022, 13, 7133. [Google Scholar] [CrossRef]
- Lebraud, H.; Wright, D.J.; Johnson, C.N.; Heightman, T.D. Protein Degradation by In-Cell Self-Assembly of Proteolysis Targeting Chimeras. ACS Cent. Sci. 2016, 2, 927–934. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, R.; Wang, Y.; Su, Y.; Lan, X. Nano-PROTACs: State of the art and perspectives. Nanoscale 2024, 16, 4378–4391. [Google Scholar] [CrossRef] [PubMed]
- Developing Antibody-Based PROTACs. Available online: https://www.nature.com/articles/d41573-022-00159-2 (accessed on 15 October 2024).
- Dey, S.K.; Jaffrey, S.R. RIBOTACs: Small Molecules Target RNA for Degradation. Cell Chem. Biol. 2019, 26, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Moriyama, J.; Nakamura, T.; Miki, E.; Takahashi, E.; Sato, A.; Akaike, T.; Itto-Nakama, K.; Arimoto, H. AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76, 797–810.e10. [Google Scholar] [CrossRef]
- Ramadas, B.; Pain, P.K.; Manna, D. LYTACs: An Emerging Tool for the Degradation of Non-Cytosolic Proteins. ChemMedChem 2021, 16, 2951–2953. [Google Scholar] [CrossRef]
- De Vita, E.; Lucy, D.; Tate, E.W. Beyond targeted protein degradation: LD·ATTECs clear cellular lipid droplets. Cell Res. 2021, 31, 945–946. [Google Scholar] [CrossRef]


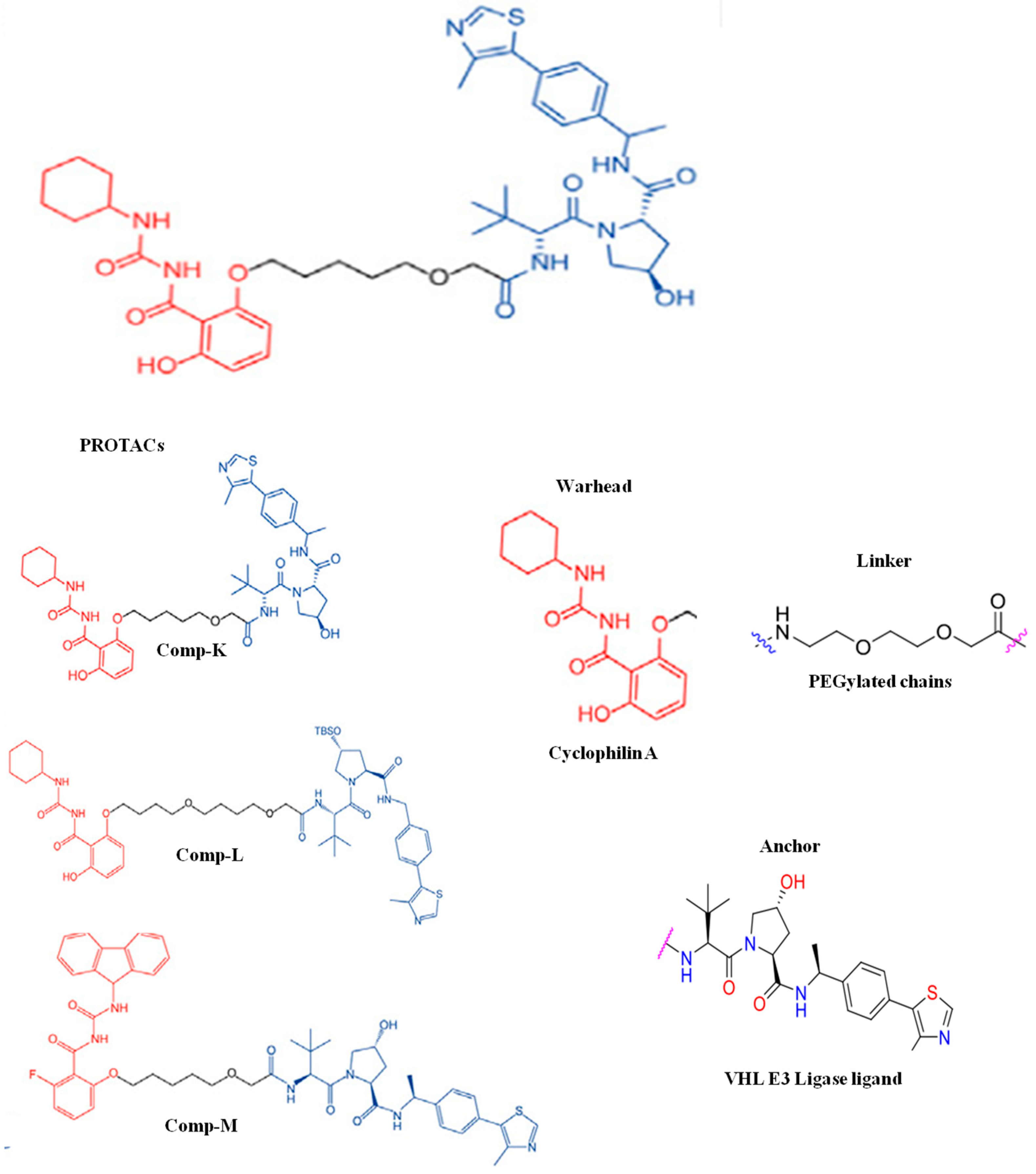
| PROTAC-Based Antivirals | Structure |
|---|---|
|  |
|  |
| 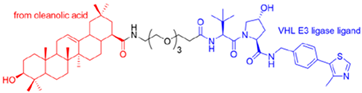 |
|  |
|  |
|  |
|  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anugu, A.; Singh, P.; Kashyap, D.; Joseph, J.; Naik, S.; Sarkar, S.; Zaman, K.; Dhaliwal, M.; Nagar, S.; Gupta, T.; et al. PROTAC-Based Antivirals for Respiratory Viruses: A Novel Approach for Targeted Therapy and Vaccine Development. Microorganisms 2025, 13, 1557. https://doi.org/10.3390/microorganisms13071557
Anugu A, Singh P, Kashyap D, Joseph J, Naik S, Sarkar S, Zaman K, Dhaliwal M, Nagar S, Gupta T, et al. PROTAC-Based Antivirals for Respiratory Viruses: A Novel Approach for Targeted Therapy and Vaccine Development. Microorganisms. 2025; 13(7):1557. https://doi.org/10.3390/microorganisms13071557
Chicago/Turabian StyleAnugu, Amith, Pankaj Singh, Dharambir Kashyap, Jillwin Joseph, Sheetal Naik, Subhabrata Sarkar, Kamran Zaman, Manpreet Dhaliwal, Shubham Nagar, Tanishq Gupta, and et al. 2025. "PROTAC-Based Antivirals for Respiratory Viruses: A Novel Approach for Targeted Therapy and Vaccine Development" Microorganisms 13, no. 7: 1557. https://doi.org/10.3390/microorganisms13071557
APA StyleAnugu, A., Singh, P., Kashyap, D., Joseph, J., Naik, S., Sarkar, S., Zaman, K., Dhaliwal, M., Nagar, S., Gupta, T., & Honnavar, P. (2025). PROTAC-Based Antivirals for Respiratory Viruses: A Novel Approach for Targeted Therapy and Vaccine Development. Microorganisms, 13(7), 1557. https://doi.org/10.3390/microorganisms13071557








