Volatile Metabolome and Transcriptomic Analysis of Kosakonia cowanii Ch1 During Competitive Interaction with Sclerotium rolfsii Reveals New Biocontrol Insights
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial–Fungal Interaction Assays
2.2. Analysis of VOCs Produced by Bacterial Strains via HS-SPME-GC-MS
2.3. Evaluation of Cell-Free Filtrates in In Vitro and In Vivo Competitive Colonization Interaction Essays
2.4. RNA Isolation
2.5. RNA-Seq Library Preparation
2.6. RNA-Seq Data Bioinformatics Analysis
2.7. Statistical Analysis
3. Results
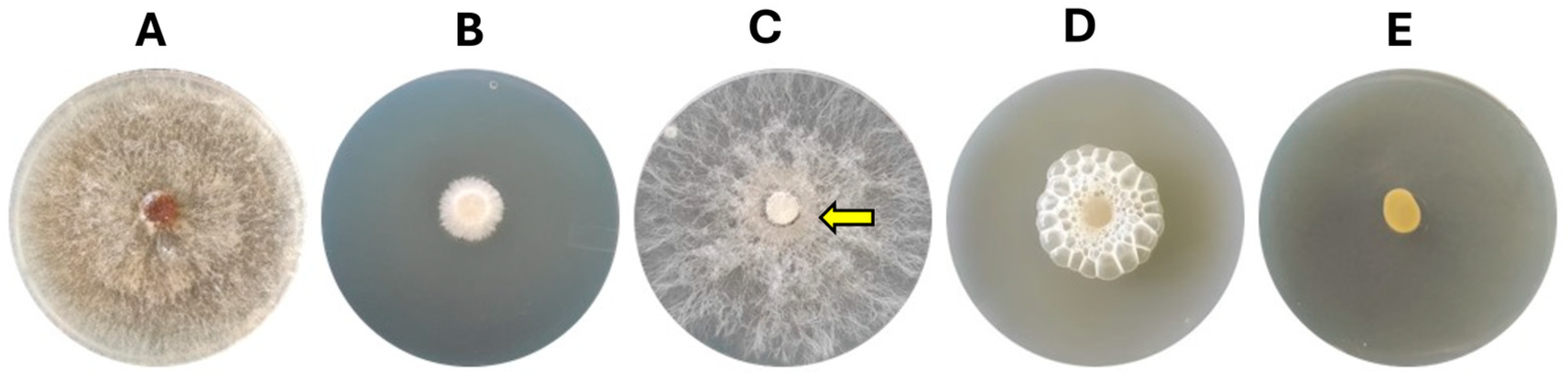
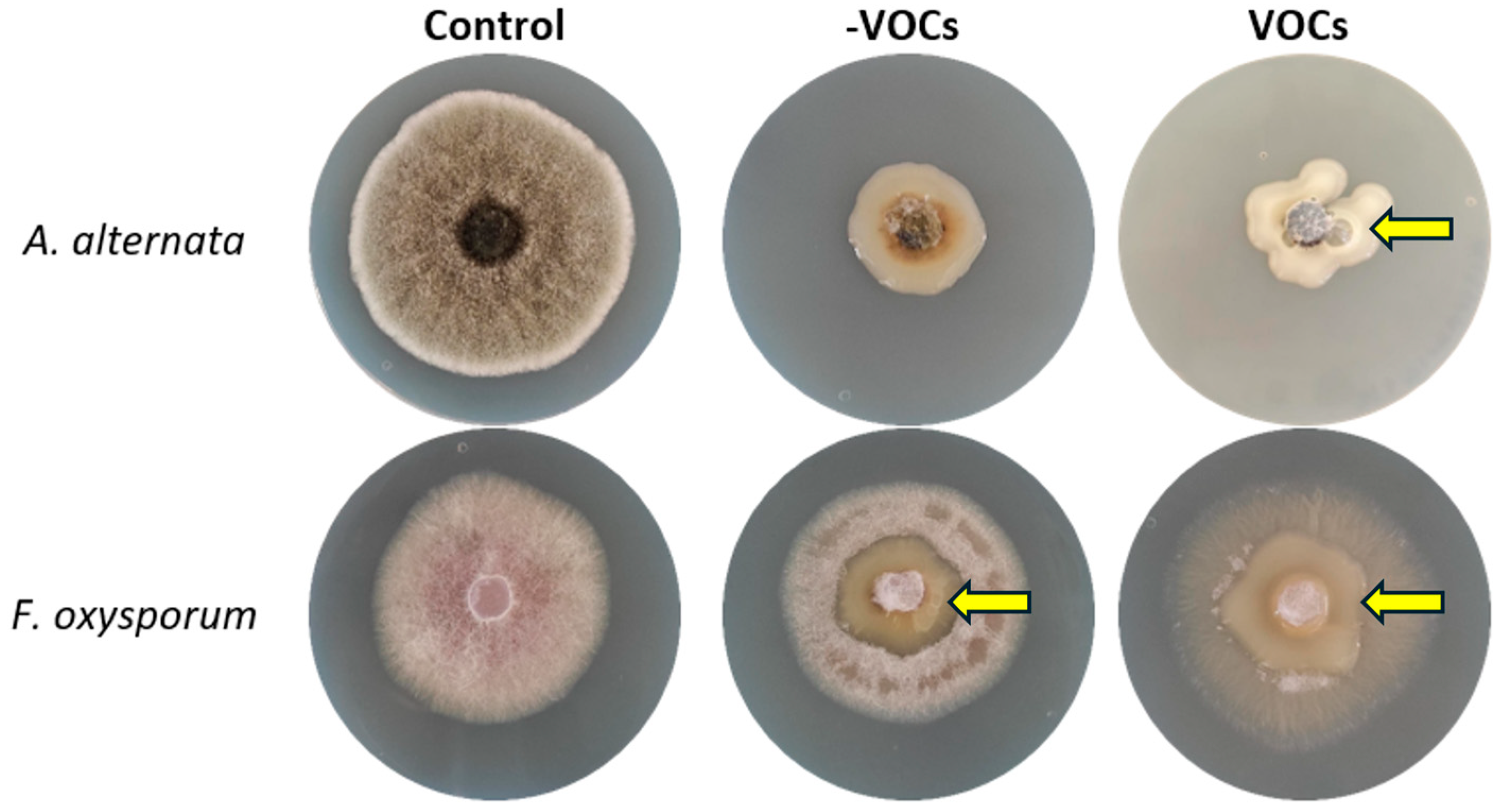
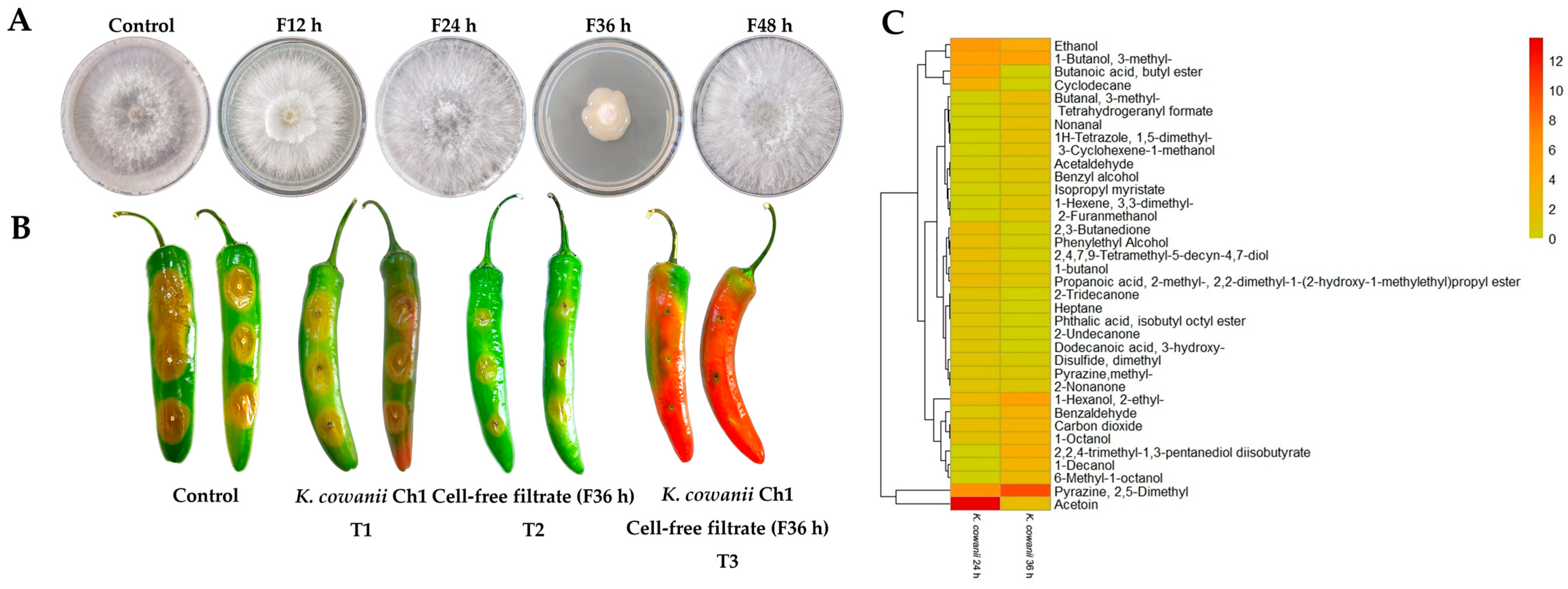
3.1. Bacterial–Fungal Interaction Assays
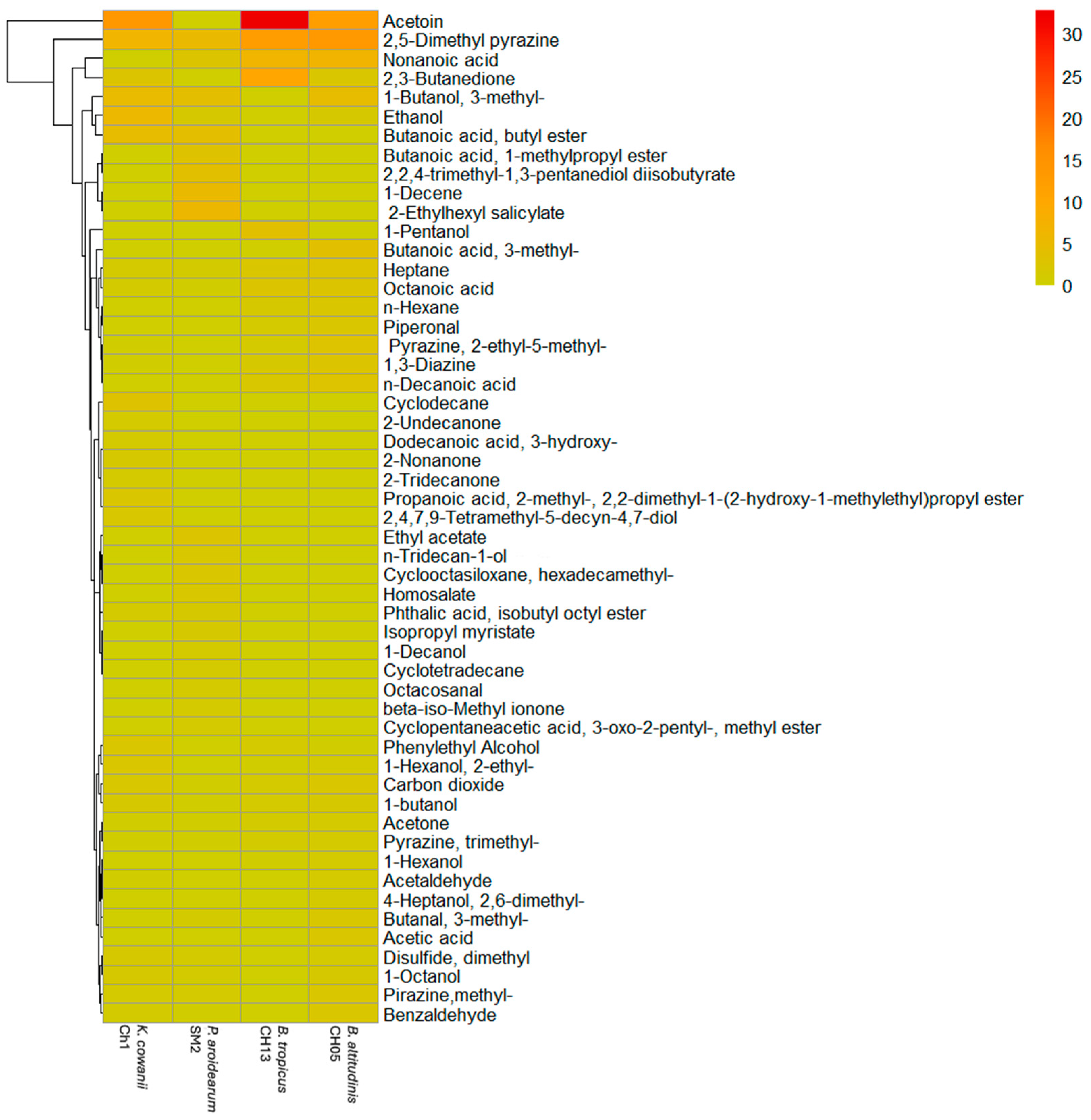
3.2. Identification of VOCs Using HS-SPME-GC-MS
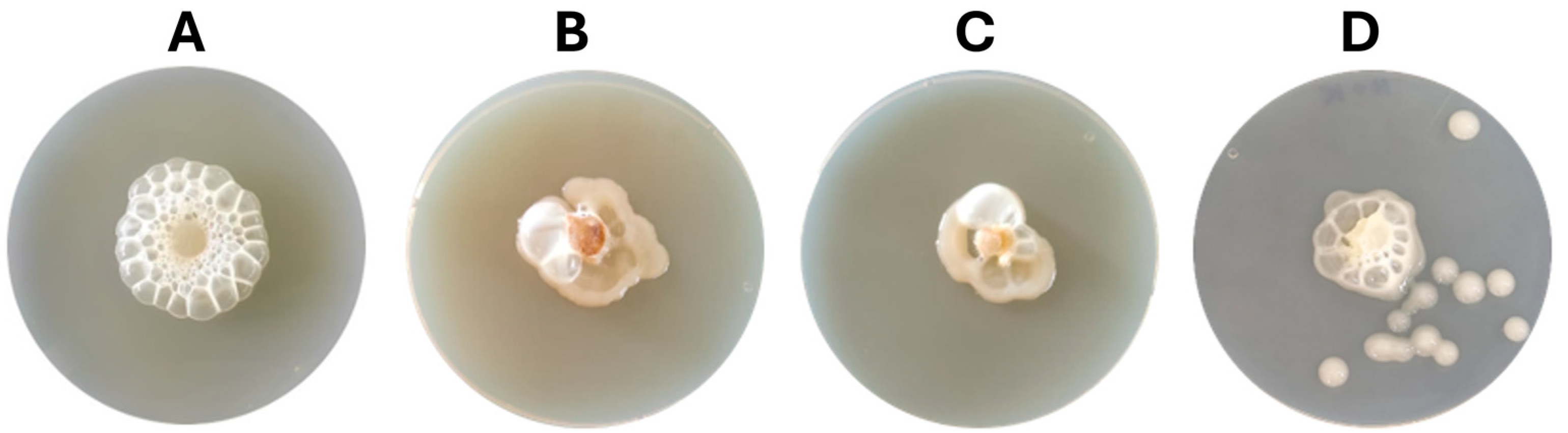
3.3. In Vitro and In Vivo Evaluation of Cell-Free Filtrates
3.4. RNA Sequencing Analysis in K. cowanii Ch1
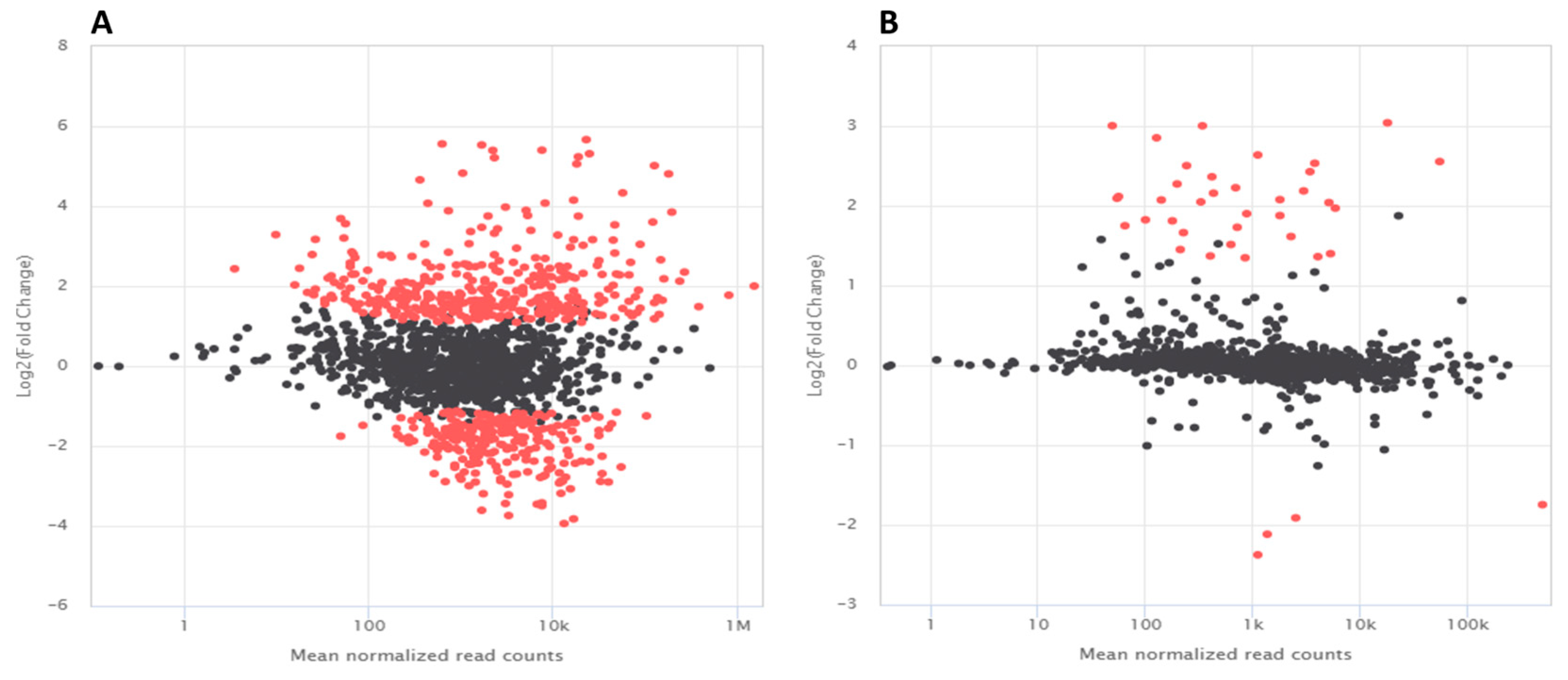
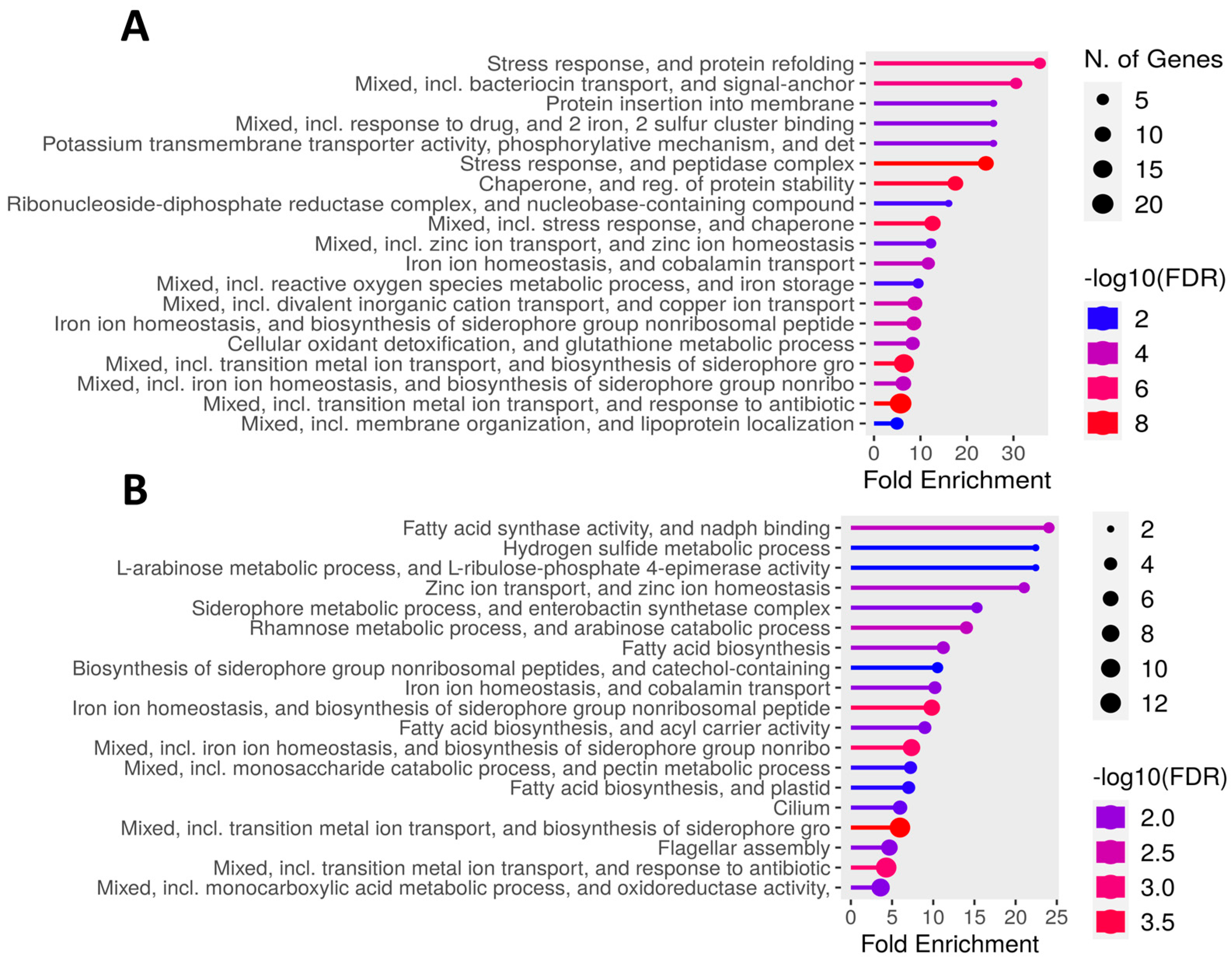
3.5. Differentially Expressed Genes in K. cowanii Ch1 in Absence of VOCs
3.6. Differentially Expressed Genes in K. cowanii Ch1 in Presence of VOCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jan-Roblero, J.; Cruz-Maya, J.A.; Barajas, C.G. Chapter 12—Kosakonia. In Beneficial Microbes in Agro-Ecology; Amaresan, N., Annapurna, K., Sankaranarayanan, A., Senthil Kumar, M., Kumar, K., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 213–231. [Google Scholar]
- Mena Navarro, M.P.; Espinosa Bernal, M.A.; Martinez-Avila, A.E.; Aponte Pineda, L.S.; Montes Flores, L.A.; Chan Ku, C.D.; Hernández Gómez, Y.F.; González Espinosa, J.; Pacheco Aguilar, J.R.; Ramos López, M.Á.; et al. Role of Volatile Organic Compounds Produced by Kosakonia cowanii Cp1 during Competitive Colonization Interaction against Pectobacterium aroidearum SM2. Microorganisms 2024, 12, 930. [Google Scholar] [CrossRef]
- Quintas-Nunes, F.; Rossi, M.J.; Nascimento, F.X. Genomic insights into the plant-associated lifestyle of Kosakonia radicincitans MUSA4, a diazotrophic plant-growth-promoting bacterium. Syst. Appl. Microbiol. 2022, 45, 126303. [Google Scholar] [CrossRef]
- Mertschnigg, T.; Patz, S.; Becker, M.; Feierl, G.; Ruppel, S.; Bunk, B.; Spröer, C.; Overmann, J.; Zarfel, G. First Report of Kosakonia radicincitans Bacteraemia from Europe (Austria)—Identification and Whole-Genome Sequencing of Strain DSM 107547. Sci. Rep. 2020, 10, 1948. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Compant, S.; Franken, P.; Ruppel, S.; Ballhausen, M.-B. It Takes Two to Tango: A Bacterial Biofilm Provides Protection against a Fungus-Feeding Bacterial Predator. Microorganisms 2021, 9, 1566. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B.L.; Maltz, M.A.; Vigneron, A.; Wu, Y.; Walter, K.S.; O’Neill, M.B.; Wang, J.; Aksoy, S. Colonization of the tsetse fly midgut with commensal Kosakonia cowanii Zambiae inhibits trypanosome infection establishment. PLoS Pathog. 2019, 15, e1007470. [Google Scholar] [CrossRef]
- Sherpa, M.T.; Sharma, L.; Bag, N.; Das, S. Isolation, Characterization, and Evaluation of Native Rhizobacterial Consortia Developed From the Rhizosphere of Rice Grown in Organic State Sikkim, India, and Their Effect on Plant Growth. Front. Microbiol. 2021, 12, 713660. [Google Scholar] [CrossRef]
- Remus, R.; Ruppel, S.; Jacob, H.-J.; Hecht-Buchholz, C.; Merbach, W. Colonization behaviour of two enterobacterial strains on cereals. Biol. Fertil. Soils 2000, 30, 550–557. [Google Scholar] [CrossRef]
- Brock, A.K.; Berger, B.; Mewis, I.; Ruppel, S. Impact of the PGPB Enterobacter radicincitans DSM 16656 on growth, glucosinolate profile, and immune responses of Arabidopsis thaliana. Microb. Ecol. 2013, 65, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Thanwisai, L.; Siripornadulsil, W.; Siripornadulsil, S. Kosakonia oryziphila NP19 bacterium acts as a plant growth promoter and biopesticide to suppress blast disease in KDML105 rice. Sci. Rep. 2024, 14, 17944. [Google Scholar] [CrossRef]
- González Espinosa, J.; Hernández Gómez, Y.F.; Javier Martínez, Y.; Flores Gallardo, F.J.; Pacheco Aguilar, J.R.; Ramos López, M.Á.; Arvizu Gómez, J.L.; Saldaña Gutierrez, C.; Rodríguez Morales, J.A.; García Gutiérrez, M.C.; et al. Kosakonia cowanii Ch1 Isolated from Mexican Chili Powder Reveals Growth Inhibition of Phytopathogenic Fungi. Microorganisms 2023, 11, 1758. [Google Scholar] [CrossRef]
- Cirimotich, C.M.; Dong, Y.; Clayton, A.M.; Sandiford, S.L.; Souza-Neto, J.A.; Mulenga, M.; Dimopoulos, G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 2011, 332, 855–858. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Romano, I.; Ventorino, V.; Ambrosino, P.; Testa, A.; Chouyia, F.E.; Pepe, O. Development and Application of Low-Cost and Eco-Sustainable Bio-Stimulant Containing a New Plant Growth-Promoting Strain Kosakonia pseudosacchari TL13. Front. Microbiol. 2020, 11, 2044. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Lu, C.; Wang, H.; Wang, L.; Yang, Y.; Jiang, T.; Li, S.; Xu, D.; Wu, L. Production exopolysaccharide from Kosakonia cowanii LT-1 through solid-state fermentation and its application as a plant growth promoter. Int. J. Biol. Macromol. 2020, 150, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Montejano-Ramírez, V.; Ávila-Oviedo, J.L.; Campos-Mendoza, F.J.; Valencia-Cantero, E. Microbial Volatile Organic Compounds: Insights into Plant Defense. Plants 2024, 13, 2013. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tahir, H.A.S.; Ali, Q.; Rajer, F.U.; Shakeel, Q.; Gillani, W.; Binyamin, R.; Tayyab, H.M.A.; Khan, A.R.; Gu, Q.; Gao, X.; et al. Transcriptomic analysis of Ralstonia solanacearum in response to antibacterial volatiles of Bacillus velezensis FZB42. Arch. Microbiol. 2023, 205, 358. [Google Scholar] [CrossRef]
- Beyari, E.A. Alternatives to chemical pesticides: The role of microbial biocontrol agents in phytopathogen management: A comprehensive review. J. Plant Pathol. 2025, 107, 291–314. [Google Scholar] [CrossRef]
- Patra, G.K.; Acharya, G.K.; Panigrahi, J.; Mukherjee, A.K.; Rout, G.R. The soil-borne fungal pathogen Athelia rolfsii: Past, present, and future concern in legumes. Folia Microbiol. 2023, 68, 677–690. [Google Scholar] [CrossRef]
- Espinosa Bernal, M.A.; Mena Navarro, M.P.; Arvizu Gómez, J.L.; Saldaña, C.; Ramos López, M.Á.; Amaro Reyes, A.; Escamilla García, M.; Pacheco Aguilar, J.R.; Moreno, V.P.; Rodríguez Morales, J.A.; et al. Biocontrol Activity of Bacillus altitudinis CH05 and Bacillus tropicus CH13 Isolated from Capsicum annuum L. Seeds against Fungal Strains. Microorganisms 2024, 12, 1943. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables repro-ducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Patel, H.; Ewels, P.; Peltzer, A.; Hammarén, R.; Botvinnik, O.; Sturm, G.; Davenport, C. nf-core/rnaseq: nf-core/rnaseq v3.0—Silver Shark (Version 3.0). Zenodo: 2020. Available online: https://zenodo.org/records/4323183 (accessed on 11 November 2024).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef]
- Broad Institute. “Picard Toolkit.” Broad Institute, GitHub Repository; Broad Institute: Cambridge, MA, USA, 2019; Available online: http://broadinstitute.github.io/picard/ (accessed on 14 November 2024).
- Daley, T.; Smith, A.D. Predicting the molecular complexity of sequencing libraries. Nat. Methods 2013, 10, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Sayols, S.; Scherzinger, D.; Klein, H. dupRadar: A Bioconductor package for the assessment of PCR artifacts in RNA-Seq data. BMC Bioinform. 2016, 17, 428. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Liu, M.; Blattman, S.B.; Takahashi, M.; Mandayam, N.; Jiang, W.; Oikonomou, P.; Tavazoie, S.F.; Tavazoie, S. Conserved genetic basis for microbial colonization of the gut. Cell 2025, 188, 2823–2824. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Paramasivan, M.; Sahayarayan, J.J. Microbial volatile organic compounds: An alternative for chemical fertilizers in sustainable agriculture development. Microorganisms 2023, 11, 42. [Google Scholar] [CrossRef]
- Weisskopf, L.; Schulz, S.; Garbeva, P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat. Rev. Microbiol. 2021, 19, 391–404. [Google Scholar] [CrossRef]
- Schulz-Bohm, K.; Martín-Sánchez, L.; Garbeva, P. Microbial volatiles: Small molecules with an important role in intra- and inter-kingdom interactions. Front. Microbiol. 2017, 8, 2484. [Google Scholar] [CrossRef] [PubMed]
- Grahovac, J.; Pajčin, I.; Vlajkov, V. Bacillus VOCs in the Context of Biological Control. Antibiotics 2023, 12, 581. [Google Scholar] [CrossRef] [PubMed]
- Koo, M.S.; Lee, J.H.; Rah, S.Y.; Yeo, W.S.; Lee, J.W.; Lee, K.L.; Koh, Y.S.; Kang, S.O.; Roe, J.H. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003, 22, 2614–2622. [Google Scholar] [CrossRef]
- Seo, S.W.; Kim, D.; Szubin, R.; Palsson, B.O. Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Rep. 2015, 12, 1289–1299. [Google Scholar] [CrossRef]
- Chan, E.; Weiss, B. Endonuclease IV of Escherichia coli is induced by paraquat. Proc. Natl. Acad. Sci. USA 1987, 84, 3189–3193. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Prajapati, K.S.; Swamy, M.; Pachauri, V. Heat shock proteins: A therapeutic target worth to consider. Vet World 2015, 8, 46–51. [Google Scholar] [CrossRef]
- Zhang, X.S.; García-Contreras, R.; Wood, T.K. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 2007, 189, 3051–3062. [Google Scholar] [CrossRef]
- Capdevila, D.A.; Wang, J.; Giedroc, D.P. Bacterial Strategies to Maintain Zinc Metallostasis at the Host-Pathogen Interface. J. Biol. Chem. 2016, 291, 20858–20868. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Terán, I.; Goh, K.G.; Ulett, G.C. Resisting death by metal: Metabolism and Cu/Zn homeostasis in bacteria. Emerg. Top. Life Sci. 2024, 8, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Lang, M. Key players in the regulation of iron homeostasis at the host-pathogen interface. Front. Immunol. 2023, 14, 1279826. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Wei, Z.; Shao, Z.; Friman, V.-P.; Cao, K.; Yang, T.; Kramer, J.; Wang, X.; Li, M.; Mei, X.; et al. Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 2020, 5, 1002–1010. [Google Scholar] [CrossRef]
- Lyng, M.; Jørgensen, J.P.B.; Schostag, M.D.; Jarmusch, S.A.; Aguilar, D.K.C.; Lozano-Andrade, C.N.; Kovács, Á.T. Competition for iron shapes metabolic antagonism between Bacillus subtilis and Pseudomonas marginalis. ISME J. 2024, 18, wrad001. [Google Scholar] [CrossRef] [PubMed]
- Seputiene, V.; Motiejūnas, D.; Suziedelis, K.; Tomenius, H.; Normark, S.; Melefors, O.; Suziedeliene, E. Molecular characterization of the acid-inducible asr gene of Escherichia coli and its role in acid stress response. J. Bacteriol. 2003, 185, 2475–2484. [Google Scholar] [CrossRef]
- Nair, A.V.; Singh, A.; Devasurmutt, Y.; Rahman, S.A.; Tatu, U.S.; Chakravortty, D. Spermidine constitutes a key determinant of motility and attachment of Salmonella Typhimurium through a novel regulatory mechanism. Microbiol. Res. 2024, 281, 127605. [Google Scholar] [CrossRef]
- Abergel, R.J.; Warner, J.A.; Shuh, D.K.; Raymond, K.N. Enterobactin protonation and iron release: Structural characterization of the salicylate coordination shift in ferric enterobactin. J. Am. Chem. Soc. 2006, 128, 8920–8931. [Google Scholar] [CrossRef]
- Crain, A.V.; Broderick, J.B. Pyruvate formate-lyase and its activation by pyruvate formate-lyase activating enzyme. J. Biol. Chem. 2014, 289, 5723–5729. [Google Scholar] [CrossRef]
- Dai, Y.; Zhang, J.; Zhang, T.; Chen, J.; Hassanin, H.A.M.; Jiang, B. Characteristics of a fructose 6-phosphate 4-epimerase from Caldilinea aerophila DSM 14535 and its application for biosynthesis of tagatose. Enzym. Microb. Technol. 2020, 139, 109594. [Google Scholar] [CrossRef]
- Zeller, T.; Klug, G. Thioredoxins in bacteria: Functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 2006, 93, 259–266. [Google Scholar] [CrossRef]
- Uhlich, G.A. KatP contributes to OxyR-regulated hydrogen peroxide resistance in Escherichia coli serotype O157: H7. Microbiology 2009, 155 Pt 11, 3589–3598. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, S.; Imlay, J.A. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol. 2010, 75, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Garbeva, P.; Hordijk, C.; Gerards, S.; de Boer, W. Volatile-mediated interactions between phylogenetically different soil bacteria. Front. Microbiol. 2014, 5, 289. [Google Scholar] [CrossRef] [PubMed]
- Sideri, M.; Georgiou, C.D. Differentiation and hydrogen peroxide production in Sclerotium rolfsii are induced by the oxidizing growth factors, light and iron. Mycologia 2000, 92, 1033–1042. [Google Scholar] [CrossRef]
- Massawe, V.C.; Hanif, A.; Farzand, A.; Mburu, D.K.; Ochola, S.O.; Wu, L.; Tahir, H.A.S.; Gu, Q.; Wu, H.; Gao, X. Volatile Compounds of Endophytic Bacillus spp. have Biocontrol Activity Against Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1373–1385. [Google Scholar] [CrossRef]
- Singh, A.; Chaudhary, S.; Shankar, A.; Prasad, V. Chapter 13—Polyketide and Its Derivatives. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, H.B., Gupta, V.K., Jogaiah, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 219–227. [Google Scholar] [CrossRef]
- Tapia-Pastrana, G.; Chavez-Dueñas, L.; Lanz-Mendoza, H.; Teter, K.; Navarro-García, F. VirK is a periplasmic protein required for efficient secretion of plasmid-encoded toxin from enteroaggregative Escherichia coli. Infect. Immun. 2012, 80, 2276–2285. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández Gómez, Y.F.; González Espinosa, J.; Olvera Rivas, G.C.; Arvizu Gómez, J.L.; Valenzuela Soto, J.H.; Ramos López, M.A.; Amaro Reyes, A.; Rodríguez de León, E.; Saldaña, C.; Hernández Flores, J.L.; et al. Volatile Metabolome and Transcriptomic Analysis of Kosakonia cowanii Ch1 During Competitive Interaction with Sclerotium rolfsii Reveals New Biocontrol Insights. Microorganisms 2025, 13, 1483. https://doi.org/10.3390/microorganisms13071483
Hernández Gómez YF, González Espinosa J, Olvera Rivas GC, Arvizu Gómez JL, Valenzuela Soto JH, Ramos López MA, Amaro Reyes A, Rodríguez de León E, Saldaña C, Hernández Flores JL, et al. Volatile Metabolome and Transcriptomic Analysis of Kosakonia cowanii Ch1 During Competitive Interaction with Sclerotium rolfsii Reveals New Biocontrol Insights. Microorganisms. 2025; 13(7):1483. https://doi.org/10.3390/microorganisms13071483
Chicago/Turabian StyleHernández Gómez, Yoali Fernanda, Jacqueline González Espinosa, Griselda Catalina Olvera Rivas, Jackeline Lizzeta Arvizu Gómez, José Humberto Valenzuela Soto, Miguel Angel Ramos López, Aldo Amaro Reyes, Eloy Rodríguez de León, Carlos Saldaña, José Luis Hernández Flores, and et al. 2025. "Volatile Metabolome and Transcriptomic Analysis of Kosakonia cowanii Ch1 During Competitive Interaction with Sclerotium rolfsii Reveals New Biocontrol Insights" Microorganisms 13, no. 7: 1483. https://doi.org/10.3390/microorganisms13071483
APA StyleHernández Gómez, Y. F., González Espinosa, J., Olvera Rivas, G. C., Arvizu Gómez, J. L., Valenzuela Soto, J. H., Ramos López, M. A., Amaro Reyes, A., Rodríguez de León, E., Saldaña, C., Hernández Flores, J. L., & Guillén, J. C. (2025). Volatile Metabolome and Transcriptomic Analysis of Kosakonia cowanii Ch1 During Competitive Interaction with Sclerotium rolfsii Reveals New Biocontrol Insights. Microorganisms, 13(7), 1483. https://doi.org/10.3390/microorganisms13071483








