New Species, New Record, and Antagonistic Potential of Torula (Torulaceae, Pleosporales) from Jilin Province, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Isolation, and Morphological Observation
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analysis
2.4. Preliminary Assessment of Antagonistic Effects on Pathogens
3. Results
3.1. Phylogenetic Analyses
3.2. Taxonomy
3.3. Preliminary Assessment of Antagonistic Activity on Pathogens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sturm, J. Deutschlands Flora Abt III; Die Pilze Deutschlands: Nuremberg, Germany, 1829. [Google Scholar]
- Manawasinghe, I.S.; Calabon, M.S.; Jones, E.B.G.; Zhang, Y.X.; Liao, C.F.; Xiong, Y.R.; Chaiwan, N.; Kularathnage, N.D.; Liu, N.G.; Tang, S.M.; et al. Mycosphere notes 345–386. Mycosphere 2022, 13, 454–557. [Google Scholar] [CrossRef]
- Crane, J.L.; Miller, A.N. Studies in genera Similar to Torula: Bahusaganda, Bahusandhika, Pseudotorula, and Simmonsiella gen. nov. IMA Fungus 2016, 7, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Su, X.J.; Luo, Z.L.; Jeewon, R.; Bhat, D.J.; Bao, D.F.; Li, W.L.; Hao, Y.E.; Su, H.Y.; Hyde, K.D. Morphology and multigene phylogeny reveal new genus and species of Torulaceae from freshwater habitats in northwestern Yunnan, China. Mycol. Prog. 2018, 17, 531–545. [Google Scholar] [CrossRef]
- Crous, P.W.; Carris, L.M.; Giraldo, A.; Groenewald, J.Z.; Hawksworth, D.L.; Hemández-Restrepo, M.; Jaklitsch, W.M.; Lebrun, M.H.; Schumacher, R.K.; Stielow, J.B.; et al. The Genera of Fungi—Fixing the application of the type species of generic names—G2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 2015, 6, 163–198. [Google Scholar] [CrossRef]
- Su, H.Y.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Ariyawansa, H.A.; Luo, Z.L.; Promputtha, I.; Tian, Q.; Lin, C.G.; Shang, Q.J.; Zhao, Y.C.; et al. The Families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 2016, 80, 375–409. [Google Scholar] [CrossRef]
- Li, J.F.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Mapook, A.; Camporesi, E.; Shang, Q.J.; Chukeatirote, E.; Bahkali, A.H.; Hyde, K.D. Molecular taxonomy and morphological characterization reveal new species and new host records of Torula species (Torulaceae, Pleosporales). Mycol. Prog. 2017, 16, 447–461. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Bhat, D.J.; Liu, N.G.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- He, S.; Wei, D.; Bhunjun, C.S.; Jayawardena, R.S.; Thiyagaraja, V.; Zhao, Q.; Fatimah, A.O.; Hyde, K.D. Morphology and multi-gene phylogeny reveal a new species of family Torulaceae from Yunnan Province, China. Diversity 2024, 16, 551. [Google Scholar] [CrossRef]
- Tian, W.; Su, P.; Chen, Y.; Maharachchikumbura, S.S.N. Four new species of Torula (Torulaceae, Pleosporales) from Sichuan, China. J. Fungi 2023, 9, 150. [Google Scholar] [CrossRef]
- Nallathambi, P.; Umamaheswari, C. A new disease of ber (Ziziphus mauritiana Lim) caused by Torula herbarum (Pers) link. J. Mycol. Plant Pathol. 2001, 31, 92. [Google Scholar]
- Grabowski, M. The study of new fungus species causing apple sooty blotch. Folia Hortic. 2007, 19, 89–97. [Google Scholar]
- Li, J.; Bhat, D.J.; Phookamsak, R.; Mapook, A.; Lumyong, S.; Hyde, K. Sporidesmioides thailandica gen. et sp. nov. (Dothideomycetes) from northern Thailand. Mycol. Prog. 2016, 15, 1169–1178. [Google Scholar] [CrossRef]
- Crous, P.W.; Schumacher, R.K.; Wood, A.R.; Groenewald, J.Z. The Genera of Fungi—G5: Arthrinium, Ceratosphaeria, Dimerosporiopsis, Hormodochis, Lecanostictopsis, Lembosina, Neomelanconium, Phragmotrichum, Pseudomelanconium, Rutola, and Trullula. Fungal Syst. Evol. 2020, 5, 77–98. [Google Scholar] [CrossRef]
- Boonmee, S.; Wanasinghe, D.N.; Calabon, M.S.; Huanraluek, N.; Chandrasiri, S.K.U.; Jones, G.E.B.; Rossi, W.; Leonardi, M.; Singh, S.K.; Rana, S.; et al. Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021, 111, 1–335. [Google Scholar]
- Persoon, C.H. Neuer versuch einer systematischen eintheilung der schwaümme. Neues Mag. Für Die Bot. Ihrem Ganzen Umfange 1794, 1, 63–128. [Google Scholar]
- Crane, J.L.; Schoknecht, J.D. Revision of Torula species. Rutola, a new name for Torula graminis. Can. J. Bot. 1977, 55, 3013–3019. [Google Scholar]
- Mason, E.W. Annotated account of fungi received at the Imperial Mycological Institute. List 2. Fascicle 3 (special part). In Mycological Papers; Commonwealth: Kew, Surrey, England, 1941; Volume 5, pp. 1–144. [Google Scholar]
- Hughes, S.J. Conidiophores, conidia, and classification. Can. J. Bot. 1953, 31, 577–659. [Google Scholar] [CrossRef]
- Subramanian, C.V. Hypomycetes: An Account of Indian Species Except Cercospora; Council of Agricultural Research: New Delhi, India, 1971; pp. 180–189. [Google Scholar]
- Ellis, M.B. Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1971; p. 608. [Google Scholar]
- Ellis, M.B. More Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1976; p. 507. [Google Scholar]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Nayak, B.K. Enumeration of phylloplane and endophytic fungi from medicinal plant, Solanum nigrum by two different techniques. Int. J. Chem. Concepts 2018, 4, 1–6. [Google Scholar]
- Karmakar, B.; SenGupta, K.; Kaur, A.; Roy, A.; Bhattacharya, S.G. Fungal bio-aerosol in multiple micro-environments from eastern India: Source, distribution, and health hazards. SN Appl. Sci. 2020, 2, 565. [Google Scholar] [CrossRef]
- Chunyu, W.X.; Zhao, J.Y.; Ding, Z.G.; Wang, Y.X.; Han, X.L.; Li, M.G.; Wen, M.L. A new dichlorinated aromatic lactone from the tin mine tailings-derived fungus Torula sp. YIM DT 10072. Chem. Nat. Compd. 2018, 54, 432–434. [Google Scholar] [CrossRef]
- Kadkol, M.V.; Gopalkrishnan, K.S.; Narasimhachari, N. Isolation and characterization of naphthaquinone pigments from Torula herbarum (Pers.). herbarin and dehydroherbarin. J. Antibiot. 1971, 24, 245–248. [Google Scholar] [CrossRef]
- Narasimhachari, N.; Gopalkrishnan, K.S. Naphthaquinone pigments from Torula herbarum: Structure of O-methylherbarin. J. Antibiot. 1974, 27, 283–287. [Google Scholar] [CrossRef]
- Wang, L.; Dong, J.Y.; Song, H.C.; Shen, K.Z.; Wang, L.M.; Sun, R.; Wang, C.R.; Li, G.H.; Li, L.; Zhang, K.Q. Screening and isolation of antibacterial activities of the fermentative extracts of freshwater fungi from Yunnan Province, China. Ann. Microbiol. 2008, 58, 579–584. [Google Scholar] [CrossRef]
- Osmana, M.E.; El-Beih, A.A.; Khatab, O.-K.H.; Moghannem, S.A.M.; Abdullah, N.H. Production of herbarin and dehydroherbarin by endophytic Chaetosphaeronema sp. (KY321184) isolated from Nepeta septemcrenata and evaluation of their bioactivities. S. Afr. J. Bot. 2018, 117, 174–183. [Google Scholar] [CrossRef]
- Geng, W.L.; Wang, X.Y.; Kurtan, T.; Mandi, A.; Tang, H.; Schulz, B.; Sun, P.; Zhang, W. Herbarone, a rearranged heptaketide derivative from the sea hare associated fungus Torula herbarum. J. Nat. Prod. 2012, 75, 1828–1832. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Crous, P.W.; Gams, W.; Stalpers, J.A.; Robert, V.; Stegehuis, G. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004, 50, 19–22. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Wang, W.P.; Shen, H.W.; Bao, D.F.; Lu, Y.Z.; Yang, Q.X.; Su, X.J.; Luo, Z.L. Two novel species and three new records of Torulaceae from Yunnan Province, China. MycoKeys 2023, 99, 1–24. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest 2.0; Program Distributed by the Author; Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A. FigTree: Tree Figure Drawing Tool, Version 1.2.2; Institute of Evolutionary Biology: Barcelona, Spain; University of Edinburgh: Edinburgh, UK, 2008. [Google Scholar]
- Skidmore, A.M.; Dickinson, C.M. Colony interactions and hyphal interference between Sepatoria nodorum and Phylloplane Fungi. Trans. Br. Mycol. Soc. 1976, 66, 57–64. [Google Scholar] [CrossRef]
- Hamzah, T.N.T.; Lee, S.Y.; Hidayat, A.; Terhem, R.; Faridah-Hanum, I.; Mohamed, R. Diversity and characterization of endophytic fungi isolated from the tropical mangrove species, Rhizophora mucronata, and identification of potential antagonists against the soil-borne fungus, Fusarium solani. Front. Microbiol. 2018, 9, 1707. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.A.; Untereiner, W.A.; Ewaze, J.O.; Wong, B.; Doyle, D. Baudoinia, a new genus to accommodate Torula compniacensis. Mycologia 2007, 99, 592–601. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.; Crane, C.; Barrett, S.; Cano-Lira, J.F.; Roux, J.J.L.; Thangavel, R.; Guarro, J.; et al. Fungal planet description sheets: 469–557. Persoonia 2016, 37, 218–403. [Google Scholar] [CrossRef]
- Li, Y.X.; Doilom, M.; Dong, W.; Liao, C.F.; Manawasinghe, I.S.; Xua, B. Taxonomic and phylogenetic contribution to Torula: T. phytolaccae sp. nov. on Phytolacca acinosa from China. Phytotaxa 2023, 584, 1–17. [Google Scholar] [CrossRef]
- Luan, S.; Shen, H.W.; Bao, D.F.; Luo, Z.L.; Xia, Y. Morphology and multi-gene phylogeny reveal a novel Torula (Pleosporales, Torulaceae) species from the plateau lakes in Yunnan, China. Biodivers. Data J. 2023, 11, e109477. [Google Scholar] [CrossRef]
- Li, X.H.; Phookamsak, R.; Sun, F.Q.; Jiang, H.B.; Xu, J.C.; Li, J.F. Torula aquilariae sp. nov. (Torulaceae, Pleosporales), a new species associated with Aquilaria sinensis from Yunnan, China. Stud. Fungi 2024, 9, e020. [Google Scholar] [CrossRef]
- Li, J.; Jeewon, R.; Mortimer, P.E.; Doilom, M.; Phookamsak, R.; Promputtha, I. Multigene phylogeny and taxonomy of Dendryphion hydei and Torula hydei spp. nov. from herbaceous litter in northern Thailand. PLoS ONE 2020, 15, e0228067. [Google Scholar] [CrossRef]
- Zhao, Z.; Liang, C.J.; Zhang, W.; Yang, Y.Y.; Bi, Q.X.; Yu, H.Y.; Wang, L.B. Genome-wide association analysis identifies a candidate gene controlling seed size and yield in Xanthoceras sorbifolium Bunge. Hortic. Res. 2024, 11, 243. [Google Scholar] [CrossRef]
- Zheng, P. China’s Geography; China Intercontinental Press: Beijing, China, 2006. [Google Scholar]
- Liu, Z.H.; Cong, Y.L.; Sossah, F.L.; Lu, Y.Z.; Kang, J.C.; Li, Y. Characterization and genome analysis of Cladobotryum mycophilum, the causal agent of cobweb disease of Morchella sextelata in China. J. Fungi 2023, 9, 411. [Google Scholar] [CrossRef]
- Xu, R.; Liu, Z.H.; Fu, Y.P.; Li, Y. Identification and biological characteristics of Cladobotryum mycophilum causing cobweb disease on Ganoderma lingzhi. Mycosystema 2019, 38, 669–678. [Google Scholar]
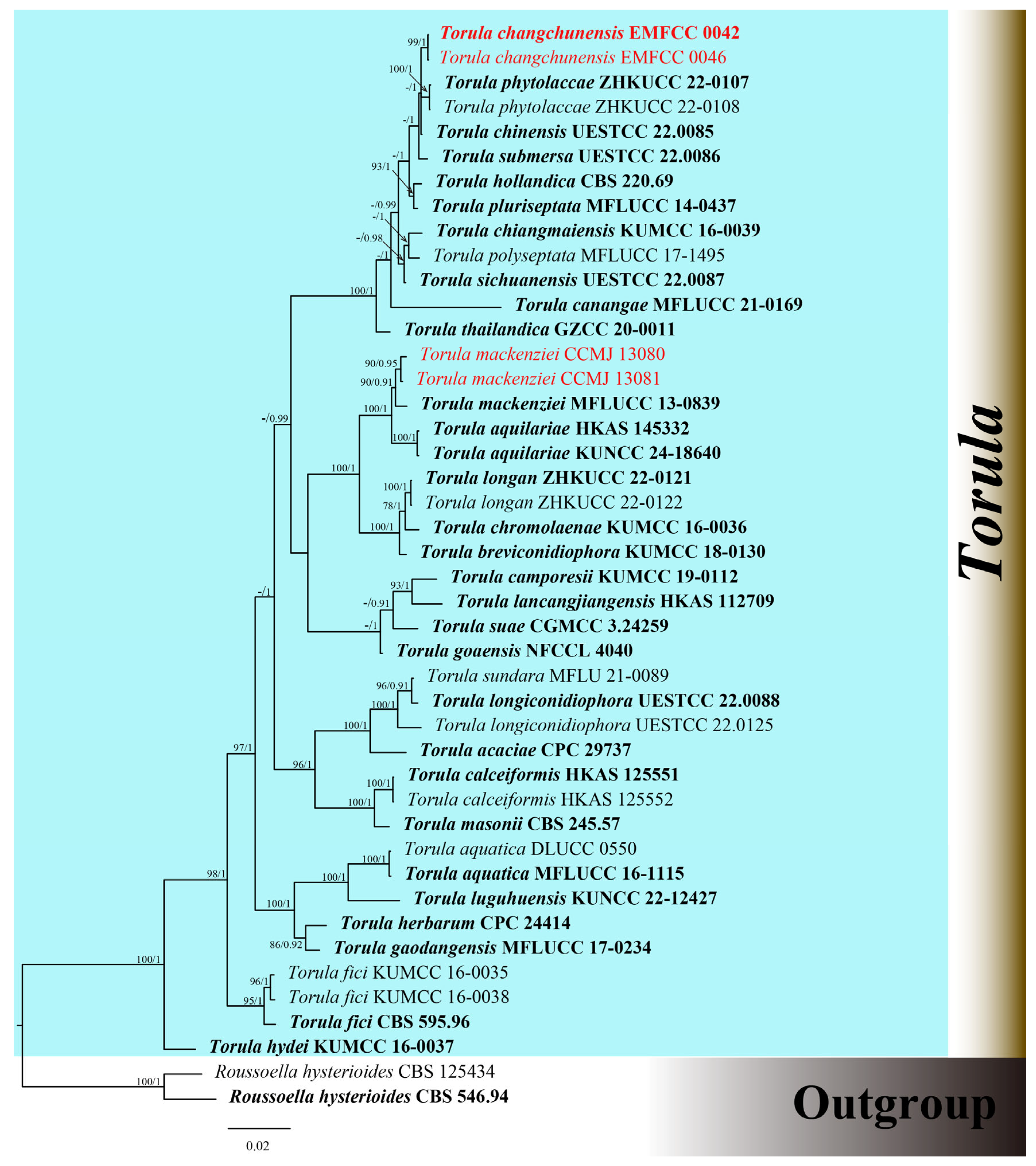

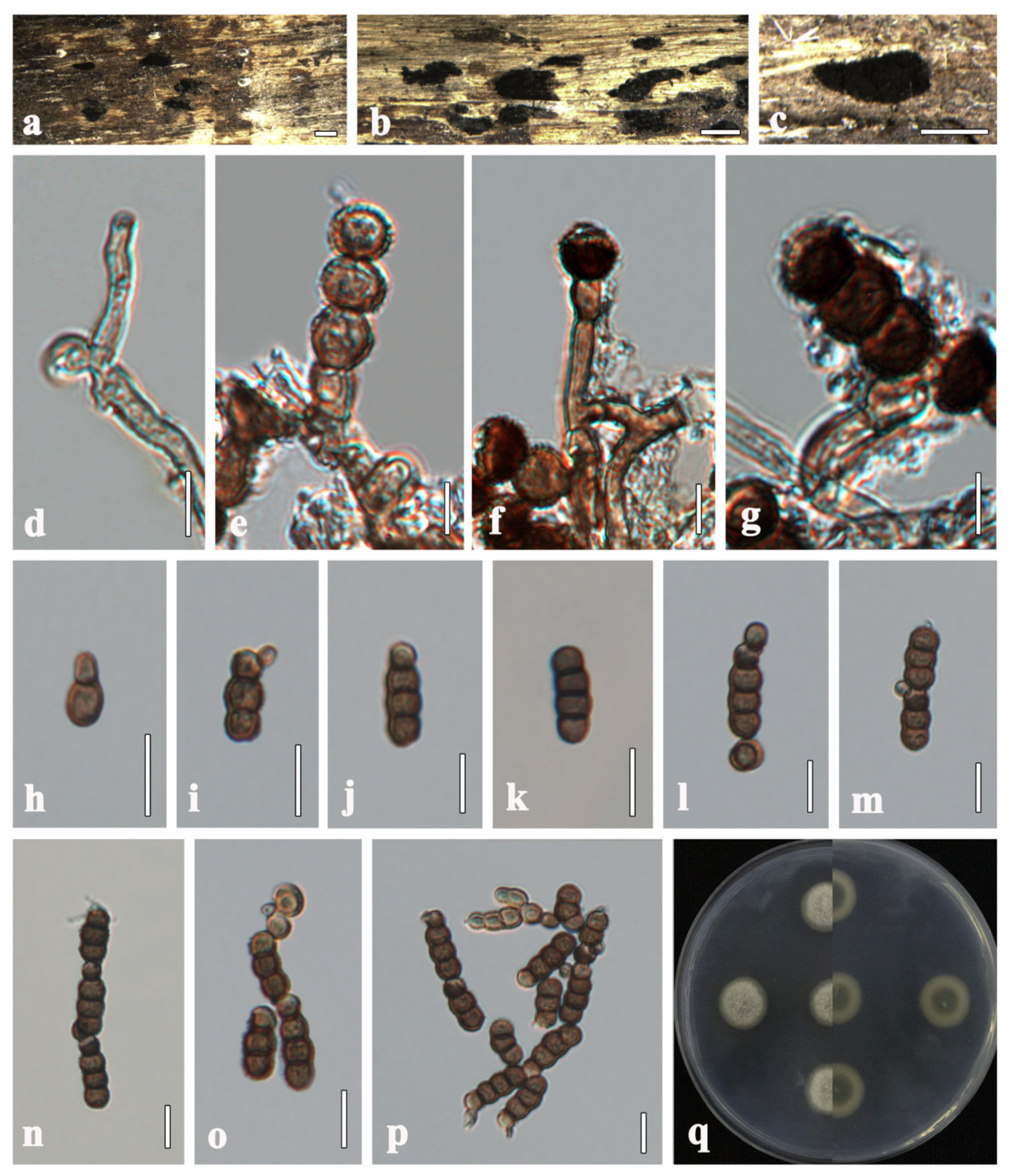


| Species | Strain/Isolate | Host | Country | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1-α | rpb2 | ||||
| Roussoella hysterioides | CBS 546.94 | - | - | KF443405 | KF443381 | AY642528 | KF443399 | KF443392 |
| R. hysterioides | CBS 125434 | - | Japan | MH863689 | MH875155 | - | - | - |
| Torula acaciae | CPC 29737 | Acacia koa | USA | KY173471 | KY173560 | - | - | KY173594 |
| T. aquatica | DLUCC 0550 | submerged wood | China | MG208166 | MG208145 | - | MG207996 | MG207976 |
| T. aquatica | MFLUCC 16-1115 | submerged wood | China | MG208167 | MG208146 | - | - | MG207977 |
| aquilariae | KUNCC 24-18640 | Aquilaria sinensis | China | PQ788522 | PQ788524 | - | PQ810572 | PQ810570 |
| T. aquilariae | HKAS 145332 | Aquilaria sinensis | China | PQ788521 | PQ788523 | - | PQ810571 | PQ810569 |
| T. breviconidiophora | KUMCC 18-0130 | decaying wood | Thailand | MK071670 | MK071672 | MK071697 | MK077673 | - |
| T. calceiformis | HKAS 125551 | dead wood | China | OP751054 | OP751052 | OP751050 | OQ630512 | OQ630510 |
| T. calceiformis | HKAS 125552 | dead wood | China | OP751055 | OP751053 | OP751051 | OQ630513 | OQ630511 |
| T. camporesii | KUMCC 19-0112 | herbaceous litter | China | MN507400 | MN507402 | MN507401 | MN507403 | MN507404 |
| T. canangae | MFLUCC 21-0169 | Cananga odorata | Thailand | OL966950 | OL830816 | - | ON032379 | - |
| T. changchunensis | EMFCC 0042 | submerged wood | China | PP151720 | PP153475 | PQ578289 | PQ601343 | PQ601346 |
| T. changchunensis | EMFCC 0046 | submerged wood | China | PV082156 | PV082158 | PV082159 | PV094898 | PV094899 |
| T. chiangmaiensis | KUMCC 16-0039 | Chromolaena odorata | Thailand | MN061342 | KY197856 | KY197863 | KY197876 | - |
| T. chinensis | UESTCC 22.0085 | submerged wood | China | OQ127986 | OQ128004 | OQ127995 | - | - |
| T. chromolaenae | KUMCC 16-0036 | Chromolaena odorata | Thailand | MN061345 | KY197860 | KY197867 | KY197880 | KY197873 |
| T. fici | CBS 595.96 | Ficus religiosa | Cuba | KF443408 | KF443385 | KF443387 | KF443402 | KF443395 |
| T. fici | KUMCC 16-0038 | Chromolaena odorata | Thailand | MN061341 | KY197859 | KY197866 | KY197879 | KY197872 |
| T. fici | KUMCC 16-0035 | Chromolaena odorata | Thailand | MN061340 | KY197858 | KY197865 | KY197878 | KY197871 |
| T. gaodangensis | MFLUCC 17-0234 | submerged wood | China | MF034135 | MF034133 | MF034134 | - | - |
| T. goaensis | NFCCL 4040 | decaying wood | India | KY440969 | KY440970 | - | - | - |
| T. herbarum | CPC 24414 | Phragmites australis | Netherlands | KR873260 | KR873288 | - | - | - |
| T. hollandica | CBS 220.69 | Delphinium sp. | Netherlands | KF443406 | KF443384 | KF443389 | KF443401 | KF443393 |
| T. hydei | KUMCC 16-0037 | Chromolaena odorata | Thailand | MN061346 | MH253926 | MH253928 | MH253930 | - |
| T. lancangjiangensis | HKAS 112709 | submerged wood | China | MW723059 | MW879526 | MW774582 | MZ567104 | MW729780 |
| T. longan | ZHKUCC 22-0121 | Dimocarpus longan | China | OR194035 | OR194027 | OR194032 | OR228537 | OR228535 |
| T. longan | ZHKUCC 22-0122 | Dimocarpus longan | China | OR194036 | OR194028 | OR194033 | OR228538 | OR228536 |
| T. longiconidiophora | UESTCC 22.0088 | submerged wood | China | OQ127983 | OQ128001 | OQ127992 | OQ158977 | OQ158967 |
| T. longiconidiophora | UESTCC 22.0125 | submerged wood | China | OQ127984 | OQ128002 | OQ127993 | OQ158976 | OQ158972 |
| T. luguhuensis | KUNCC 22-12427 | submerged wood | China | OQ729758 | OQ947766 | - | OQ999004 | OQ999002 |
| T. mackenziei | CCMJ 13080 | Xanthoceras sorbifolium | China | PQ584885 | PQ584887 | PQ578287 | PQ601341 | PQ601344 |
| T. mackenziei | CCMJ 13081 | Xanthoceras sorbifolium | China | PQ584886 | PQ584888 | PQ578288 | PQ601342 | PQ601345 |
| T. mackenziei | MFLUCC 13-0839 | Bidens pilosa | Thailand | MN061344 | KY197861 | KY197868 | KY197881 | KY197874 |
| T. masonii | CBS 245.57 | Brassi sp. | England | KR873261 | KR873289 | - | - | - |
| T. phytolaccae | ZHKUCC 22-0107 | Phytolacca acinosa | China | ON611796 | ON611800 | ON611798 | ON660881 | ON660879 |
| T. phytolaccae | ZHKUCC 22-0108 | Phytolacca acinosa | China | ON611797 | ON611799 | ON611797 | ON660880 | ON660878 |
| T. pluriseptata | MFLUCC 14-0437 | Clematis vitalba | Italy | MN061338 | KY197855 | KY197862 | KY197875 | KY197869 |
| T. polyseptata | MFLUCC 17-1495 | Chromolaena odorata | Thailand | MT214382 | MT214476 | MT214427 | MT235791 | MT235830 |
| T. sichuanensis | UESTCC 22.0087 | submerged wood | China | OQ127981 | OQ127999 | OQ127990 | - | - |
| T. suae | CGMCC 3.24259 | submerged wood | China | OP359406 | OP359415 | OP369300 | OP471618 | OP476730 |
| T. submersa | UESTCC 22.0086 | submerged wood | China | OQ127985 | OQ128003 | OQ127994 | OQ158978 | OQ158968 |
| T. sundara | MFLU 21-0089 | bamboo culms | Thailand | OM276824 | OM287866 | - | - | - |
| T. thailandica | GZCC 20-0011 | decaying wood | Thailand | MN907426 | MN907428 | MN907427 | - | - |
| Species | % PGI | |
|---|---|---|
| Botrytis cinerea | Cladobotryum mycophilum | |
| Torula changchunensis | 19.01 ± 0.0406a | 67.18 ± 0.0169a |
| Torula mackenziei | 10.20 ± 0.0182b | 24.57 ± 0.0259b |
| Species | Host | Distribution |
|---|---|---|
| Torula aquatica | submerged wood | China, Yunnan |
| T. aquilariae | Aquilaria sinensis | China, Yunnan |
| T. calceiformis | dead wood | China, Guizhou |
| T. camporesii | herbaceous litter | China, Yunnan |
| T. canangae | submerged wood | China, Yunnan |
| T. changchunensis | submerged wood | China, Jilin |
| T. chinensis | submerged wood | China, Sichuan |
| T. fici | submerged wood | China, Yunnan |
| T. gaodangensis | submerged wood, Malus sp. | China, Guizhou |
| T. lancangjiangensis | submerged wood | China, Yunnan |
| T. longan | Dimocarpus longan | China, Guangdong |
| T. longiconidiophora | submerged wood | China, Sichuan |
| T. luguhuensis | submerged wood | China, Yunnan |
| T. mackenziei | submerged wood, Xanthoceras sorbifolium | China, Jilin, Yunnan |
| T. masonii | submerged wood, Artemisia carvifolia | China, Yunnan |
| T. phytolaccae | Phytolacca acinosa | China, Yunnan |
| T. sichuanensis | submerged wood | China, Sichuan |
| T. suae | submerged wood | China, Yunnan |
| T. sundara | submerged wood | China, Yunnan |
| T. submersa | submerged wood | China, Sichuan, Yunnan |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, R.; Zhang, Y.; Su, W.; Li, Y. New Species, New Record, and Antagonistic Potential of Torula (Torulaceae, Pleosporales) from Jilin Province, China. Microorganisms 2025, 13, 1459. https://doi.org/10.3390/microorganisms13071459
Xu R, Zhang Y, Su W, Li Y. New Species, New Record, and Antagonistic Potential of Torula (Torulaceae, Pleosporales) from Jilin Province, China. Microorganisms. 2025; 13(7):1459. https://doi.org/10.3390/microorganisms13071459
Chicago/Turabian StyleXu, Rong, Yue Zhang, Wenxin Su, and Yu Li. 2025. "New Species, New Record, and Antagonistic Potential of Torula (Torulaceae, Pleosporales) from Jilin Province, China" Microorganisms 13, no. 7: 1459. https://doi.org/10.3390/microorganisms13071459
APA StyleXu, R., Zhang, Y., Su, W., & Li, Y. (2025). New Species, New Record, and Antagonistic Potential of Torula (Torulaceae, Pleosporales) from Jilin Province, China. Microorganisms, 13(7), 1459. https://doi.org/10.3390/microorganisms13071459





