Limosilactobacillus reuteri M4-100 Mitigates the Pathogenicity of Escherichia coli Strain HMLN-1 in an Intestinal Epithelial Model and Modulates Host Cell Gene Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Cell Culture Experiments
2.3. Adhesion Assay
2.4. Invasion Assay
2.5. Translocation Assay
2.6. Scanning Electron Microscopy (SEM)
2.7. Transmission Electron Microscopy (TEM)
2.8. RNA Extraction
2.9. RNA-Seq Data Analysis
3. Statistical Analysis
4. Results
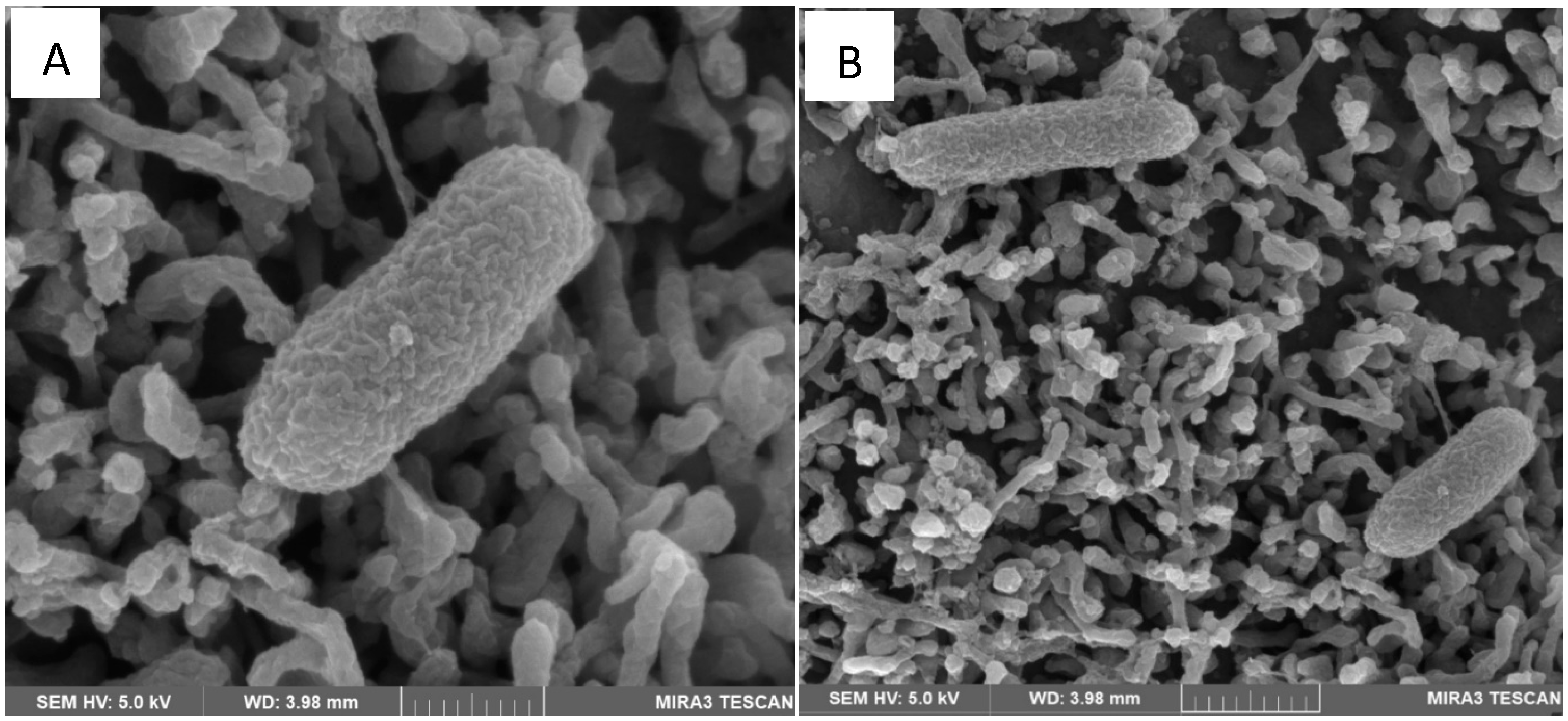
4.1. Scanning Electron Microscopy (SEM)
4.2. Adhesion Assay
4.3. Invasion Assay
4.4. Translocation Assay
4.5. Transmission Electron Microscopy (TEM)
4.6. Differential Gene Expression Results
4.7. Gene Expression Clustering Analysis Results
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, R.D.; Garlington, A.W. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect. Immun. 1979, 23, 403–411. [Google Scholar] [CrossRef] [PubMed]
- MacFie, J.; Reddy, B.S.; Gatt, M.; Jain, P.K.; Sowdi, R.; Mitchell, C.J. Bacterial translocation studied in 927 patients over 13 years. Br. J. Surg. 2006, 93, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Chen, D.; Chen, S.; Ru, X.; Ye, Q. Escherichia coli infection sepsis: An analysis of specifically expressed genes and clinical indicators. Diagnostics 2023, 27, 3542. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Guo, C.; Zeng, Z.; Li, C.; Ding, N. Factors associated with in-hospital mortality in adult sepsis with Escherichia coli infection. BMC Infect. Dis. 2022, 22, 197. [Google Scholar] [CrossRef]
- Bonten, M.; Johnson, J.R.; van den Biggelaar, A.H.J.; Georgalis, L.; Geurtsen, J.; Ibarra de Palacios, P.; Gravenstein, S.; Verstraeten, T.; Hermans, P.; Poolman, J.T. Epidemiology of Escherichia coli bacteremia: A systematic literature review. Clin. Infect. Dis. 2021, 72, 1211–1219. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Katouli, M.; Ramos, N.L.; Nettelbladt, C.G.; Ljungdahl, M.; Robinson, W.; Ison, H.M.; Brauner, A.; Möllby, R. Host species-specific translocation of Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1095–1103. [Google Scholar] [CrossRef]
- Nettelbladt, C.G.; Katouli, M.; Bark, T.; Svenberg, T.; Möllby, R.; Ljunquvist, O. Evidence of bacterial translocation in fatal hemorrhagic pancreatitis: Report of a case. J. Trauma 2000, 48, 314–315. [Google Scholar] [CrossRef]
- Ljungdahl, M.; Lundholm, M.; Katouli, M.; Rasmussen, L.B.; Engstrand, L.; Haglund, U. Bacterial translocation in experimental shock is dependent on the strains in the intestinal flora. Scand. J. gasteroentrol. 2000, 35, 389–397. [Google Scholar] [CrossRef]
- Katouli, M.; Nettelbladt, C.G.; Muratov, V.; Ljunquvist, O.; Bark, T.; Svenberg, T.; Möllby, R. Selective translocation of coliform bacteria adhering to cecal epithelium of rats during catabolic stress. J. Med. Microbiol. 1997, 46, 571–578. [Google Scholar] [CrossRef]
- Ramos, N.L.; Lamprokostopoulou, A.; Chapman, T.A.; Chin, J.C.; Römling, U.; Brauner, A.; Katouli, M. Virulence characteristics of translocating Escherichia coli and the interleukin-8 response to infection. Microb. Pathog. 2011, 50, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.; Zihler Berner, A.; Chervet, N.; Chassard, C.; Lacroix, C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Methods 2013, 94, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhao, J.; Wen, R.; Zhu, X.; Liu, L.; Li, C. 2′-Fucosyllactose promotes Bifidobacterium bifidum DNG6 adhesion to Caco-2 cells. J. Dairy. Sci. 2020, 103, 9825–9834. [Google Scholar] [CrossRef]
- Fonseca, H.C.; de Sousa Melo, D.; Ramos, C.L.; Dias, D.R.; Schwan, R.F. Probiotic properties of lactobacilli and their ability to Iinhibit the adhesion of enteropathogenic bacteria to Caco-2 and HT-29 Cells. Probiotics Antimicrob. Proteins 2021, 13, 102–112. [Google Scholar] [CrossRef]
- Reale, O.; Huguet, A.; Fessard, V. Co-culture model of Caco-2/HT29-MTX cells: A promising tool for investigation of phycotoxins toxicity on the intestinal barrier. Chemosphere 2021, 273, 128497. [Google Scholar] [CrossRef]
- Rout, G.K.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar]
- Vinayamohan, P.; Joseph, D.; Viju, L.S.; Baskaran, S.A.; Venkitanarayanan, K. Efficacy of probiotics in reducing pathogenic potential of infectious agents. Fermentation 2024, 10, 599.19. [Google Scholar] [CrossRef]
- Maftei, N.M.; Raileanu, C.R.; Balta, A.A.; Ambrose, L.; Boev, M.; Marin, D.B.; Lisa, L.E. The potential impact of probiotics on human health: An update on their health-promoting properties. Microorganisms 2024, 12, 234. [Google Scholar] [CrossRef]
- Najafi, S.; Sotoodehnejad Nematalahi, F.; Amiri, M.M.; Pourshafie, M.R.; Rohani, M. Prophylactic vs. therapeutic effect of probiotics on the inflammation mediated by the NF-κB pathway in inflammatory bowel conditions. Biomedicines 2023, 11, 1675. [Google Scholar]
- Aghamohammad, S.; Sepehr, A.; Miri, S.T.; Najafi, S.; Pourshafie, M.R.; Rohani, M. The role of combining probiotics in preventing and controlling inflammation: A focus on the anti-inflammatory and immunomodulatory effects of probiotics in an in vitro model of IBD. Can. J. Gastroenterol. Hepatol. 2022, 2022, 2045572. [Google Scholar] [CrossRef]
- Rohani, M.; Noohi, N.; Talebi, M.; Katouli, M.; Pourshafie, M.R. Highly heterogeneous probiotic Lactobacillus species in healthy Iranians with low functional activities. PLoS ONE 2015, 10, e0144467. [Google Scholar] [CrossRef] [PubMed]
- Bradford, G.; Asgari, B.; Smit, B.; Hatje, E.; Kuballa, A.; Katouli, M. The efficacy of selected probiotic strains and their combination to inhibit the interaction of adherent-iInvasive Escherichia coli (AIEC) with a co-culture of Caco-2: HT29-MTX cells. Microorganisms 2024, 12, 502. [Google Scholar] [CrossRef]
- Asgari, B.; Burke, J.R.; Quigley, B.L.; Bradford, G.; Hatje, E.; Kuballa, A.; Katouli, M. Identification of virulence genes associated with pathogenicity of translocating Escherichia coli with special reference to the type 6 secretion system. Microorganisms 2024, 12, 1851. [Google Scholar] [CrossRef]
- Angelis, I.D.; Turco, L. Caco-2 Cells as a Model for Intestinal Absorption. Curr. Protoc. Toxicol. 2011, 47, 20.6.1–20.6.15. [Google Scholar] [CrossRef]
- Verhoeckx, K.; Cotter, P.; López-Expósito, I.; Kleiveland, C.; Lea, T.; Mackie, A.; Requena, T.; Swiatecka, D.; Wichers, H. Chap 10: Caco-2 cell line. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer: Cham, Switzerland, 2015; pp. 103–111. [Google Scholar]
- Béduneau, A.; Tempesta, C.; Fimbel, S.; Pellequer, Y.; Jannin, V.; Demarne, F.; Lamprecht, A. A tunable Caco-2/HT29-MTX co-culture model mimicking variable permeabilities of the human intestine obtained by an original seeding procedure. Eur. J. Pharm. Biopharm. 2014, 87, 290–298. [Google Scholar] [CrossRef]
- Chen, X.-M.; Elisia, I.; Kitts, D.D. Defining conditions for the co-culture of Caco-2 and HT29-MTX cells using Taguchi design. J. Pharmacol. Toxicol. Methods 2010, 61, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Mondal, S.K.; Sinha, S.K.; Mukhopadhyay, M.; Chakraborty, I. Diagnostic and prognostic significance of different mucin expression, preoperative CEA, and CA-125 in colorectal carcinoma: A clinicopathological study. J. Nat. Sci. Biol. Med. 2014, 5, 404. [Google Scholar]
- Pan, F.; Han, L.; Zhang, Y.; Yu, Y.; Liu, J. Optimization of Caco-2 and HT29 co-culture in vitro cell models for permeability studies. Int. J. Food Sci. Nutr. 2015, 66, 680–685. [Google Scholar] [CrossRef]
- Sato, J.; Kanazawa, A.; Azuma, K.; Ikeda, F.; Goto, H.; Komiya, K.; Kanno, R.; Tamura, Y.; Asahara, T.; Takahashi, T.; et al. Probiotic reduces bacterial translocation in type 2 diabetes mellitus: A randomised controlled study. Sci. Rep. 2017, 7, 12115. [Google Scholar] [CrossRef]
- Lozoya-Agullo, I.; Araújo, F.; González-Álvarez, I.; Merino-Sanjuán, M.; González-Álvarez, M.; Bermejo, M.; Sarmento, B. Usefulness of Caco-2/HT29-MTX and Caco-2/HT29-MTX/Raji B Coculture Models To Predict Intestinal and Colonic Permeability Compared to Caco-2 Monoculture. Mol. Pharm. 2017, 14, 1264–1270. [Google Scholar] [CrossRef]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008, 6, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Hamon, M.A.; Bierne, H.; Cossart, P. Listeria monocytogenes: A multifaceted model. Nat. Rev. Microbiol. 2006, 4, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Ashida, H.; Ogawa, M.; Kim, M.; Mimuro, H.; Sasakawa, C. Bacteria and host interactions in the gut epithelial barrier. Nat. Chem. Biol. 2011, 8, 36–45. [Google Scholar] [CrossRef]
- Derrien, M.; van Passel, M.W.J.; van de Bovenkamp, J.H.B.; Schipper, R.G.; de Vos, W.M.; Dekker, J. Mucin-bacterial interactions in the human oral cavity and di-gestive tract. Gut Microbes 2010, 1, 254–268. [Google Scholar] [CrossRef]
- Aghamohammad, S.; Sepehr, A.; Mir, S.T.; Najaf, S.; Rohani, M.; Pourshafea1, M.R. The efects of the probiotic cocktail on modulation of the NF-kB and JAK/STAT signaling pathways involved in the infammatory response in bowel disease model. BMC Immunol. 2022, 23, 8. [Google Scholar]
- Hasannejad Bibalan, M.; Eshaghi, M.; Rohani, M.; Esghaei, M.; Darban-Sarokhalil, D.; Pourshafie, M.R.; Talebi, M. Isolates of Lactobacillus plantarum and L. reuteri display greater antiproliferative and antipathogenic activity than other Lactobacillus isolates. J. Med. Microbiol. 2017, 66, 1416–1420. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Prete, R.; Battista, N.; Corsetti, A. Adhesion properties of food-associated Lactobacillus plantarum strains on human intestinal epithelial cells and modulation of IL-8 release. Front. Microbiol. 2018, 9, 2392. [Google Scholar] [CrossRef]
- Amiri, S.; Mokarram, R.R.; Khiabani, M.S.; Bari, M.R.; Khaledabad, M.A. Characterization of antimicrobial peptides produced by Lactobacillus acidophilus LA-5 and Bifidobacterium lactis BB-12 and their inhibitory effect against foodborne pathogens. LWT 2022, 153, 112449. [Google Scholar] [CrossRef]
- Capella-Monsonís, H.; Kearns, S.; Kelly, J.; Zeugolis, D.I. Battling adhesions: From understanding to prevention. BMC Biomed. Eng. 2019, 1, 5. [Google Scholar] [CrossRef]
- Campana, R.; van Hemert, S.; Baffone, W.J.G.P. Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Patghog. 2017, 9, 12. [Google Scholar] [CrossRef]
- Hyun Jung Lim, H.J.; Shin, H.S. Antimicrobial and Immunomodulatory Effects of Bifidobacterium Strains: A Review. J. Microbiol. Biotechnol. 2020, 30, 1793–1800. [Google Scholar]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [PubMed]
- Johnson-Henry, K.C.; Donato, K.A.; Shen-Tu, G.; Gordanpour, M.; Sherman, P.M. Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157:H7-induced changes in epithelial barrier function. Infect. Immun. 2008, 76, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Kalischuk, L.D.; Inglis, G.D.; Buret, A.G. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut pathog. 2009, 1, 2. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Garcia-Gil, L.J. Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity. World J. Gastrointest. Pathophysiol. 2014, 5, 213–227. [Google Scholar] [CrossRef]
- Claret, L.; Miquel, S.; Vieille, N.; Ryjenkov, D.A.; Gomelsky, M.; Darfeuille-Michaud, A. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J. Biol. Chem. 2007, 282, 33275–33283. [Google Scholar] [CrossRef]
- Karin, M.; Greten, F.R. NF-κB: Linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005, 5, 749–759. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef]
- Li, M.O.; Flavell, R.A. TGF-β: A master of all T cell trades. Cell. 2008, 134, 392–404. [Google Scholar] [CrossRef]
- Dinarello, C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011, 117, 3720–3732. [Google Scholar] [CrossRef]
- Popov, G.; Petkova, A.R.; Kaneva, R.; Kundurzhiev, G.R.; Popova, T.D. Analysis of bacterial biofilm formation and MUC5AC and MUC5B expression in chronic rhinosinusitis patients. J. Clin. Med. 2023, 23, 1808. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M. Versatile role of Rab27 in membrane trafficking: Focus on the Rab27 effector families. J. Biochem. 2005, 137, 9–16. [Google Scholar] [CrossRef] [PubMed]
 and in the presence of the Limosilactobacillus strain when co-inoculated
and in the presence of the Limosilactobacillus strain when co-inoculated  and pre-inoculated
and pre-inoculated  . E. coli strain JM109 used as the negative control
. E. coli strain JM109 used as the negative control  . (B) The number (mean ± SEM) of adhering HMLN-1
. (B) The number (mean ± SEM) of adhering HMLN-1  alone and in the presence of the M4-100 when co-inoculated
alone and in the presence of the M4-100 when co-inoculated  and pre-inoculated
and pre-inoculated  . E. coli strain JM-109
. E. coli strain JM-109  was used as a negative control. * p < 0.0001 co-inoculation versus pre-inoculation in both (A,B) graphs.
was used as a negative control. * p < 0.0001 co-inoculation versus pre-inoculation in both (A,B) graphs.
 and in the presence of the Limosilactobacillus strain when co-inoculated
and in the presence of the Limosilactobacillus strain when co-inoculated  and pre-inoculated
and pre-inoculated  . E. coli strain JM109 used as the negative control
. E. coli strain JM109 used as the negative control  . (B) The number (mean ± SEM) of adhering HMLN-1
. (B) The number (mean ± SEM) of adhering HMLN-1  alone and in the presence of the M4-100 when co-inoculated
alone and in the presence of the M4-100 when co-inoculated  and pre-inoculated
and pre-inoculated  . E. coli strain JM-109
. E. coli strain JM-109  was used as a negative control. * p < 0.0001 co-inoculation versus pre-inoculation in both (A,B) graphs.
was used as a negative control. * p < 0.0001 co-inoculation versus pre-inoculation in both (A,B) graphs.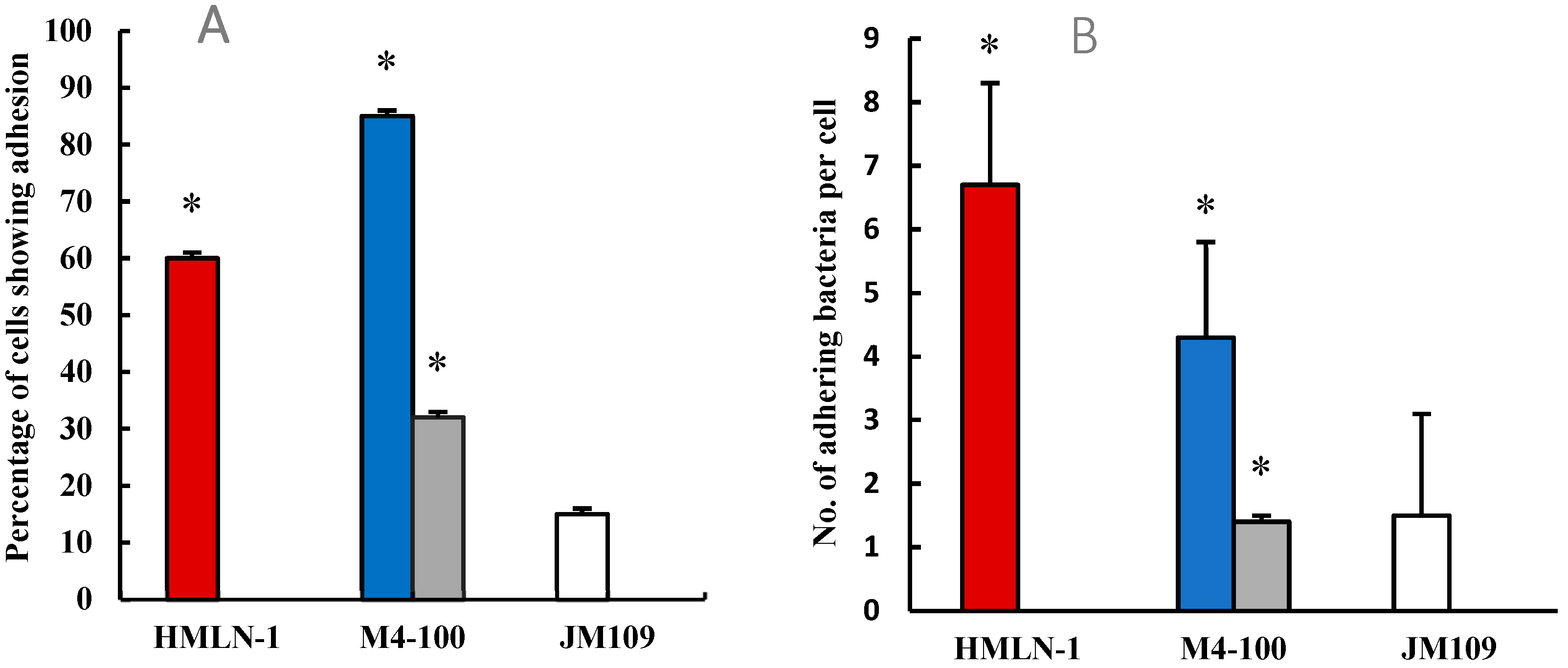
 , in the presence of M4-100 when co-inoculated
, in the presence of M4-100 when co-inoculated  and pre-inoculated.
and pre-inoculated.  E. coli strain JM-109 used as the negative control
E. coli strain JM-109 used as the negative control  . * p < 0.0001 co-inoculation versus pre-inoculation.
. * p < 0.0001 co-inoculation versus pre-inoculation.
 , in the presence of M4-100 when co-inoculated
, in the presence of M4-100 when co-inoculated  and pre-inoculated.
and pre-inoculated.  E. coli strain JM-109 used as the negative control
E. coli strain JM-109 used as the negative control  . * p < 0.0001 co-inoculation versus pre-inoculation.
. * p < 0.0001 co-inoculation versus pre-inoculation.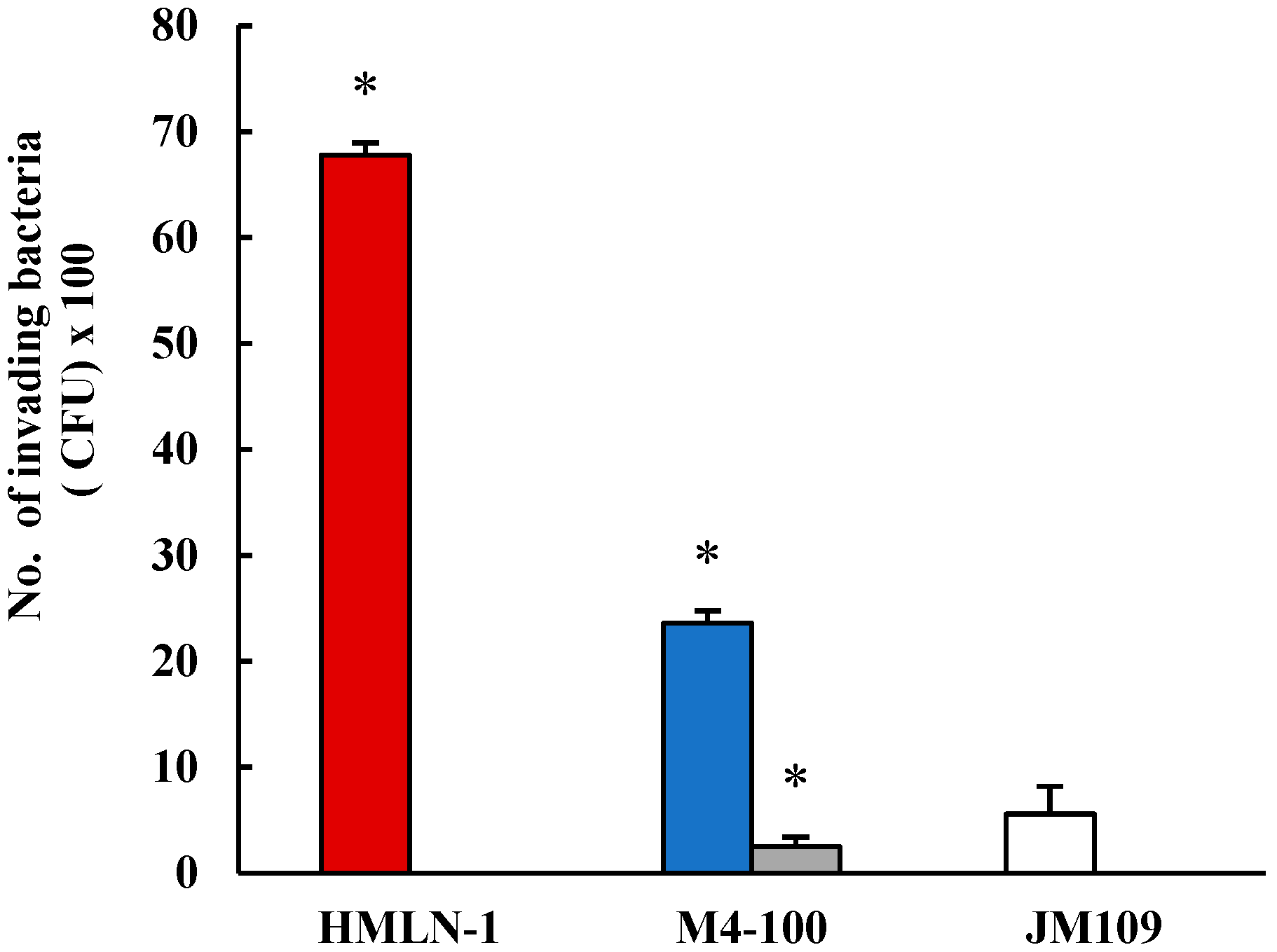
 , and in the presence
of L. reuteri M4-100
, and in the presence
of L. reuteri M4-100  in co-inoculation (a) and in pre-inoculation experiments (b). E. coli strain F44A-1, an AIEC strain, was used as a positive control
in co-inoculation (a) and in pre-inoculation experiments (b). E. coli strain F44A-1, an AIEC strain, was used as a positive control  and E. coli strain JM109 used as a negative control
and E. coli strain JM109 used as a negative control  . Translocation was measured after 30, 60, and 120 min. Results are expressed as mean ± SEM.
. Translocation was measured after 30, 60, and 120 min. Results are expressed as mean ± SEM.
 , and in the presence
of L. reuteri M4-100
, and in the presence
of L. reuteri M4-100  in co-inoculation (a) and in pre-inoculation experiments (b). E. coli strain F44A-1, an AIEC strain, was used as a positive control
in co-inoculation (a) and in pre-inoculation experiments (b). E. coli strain F44A-1, an AIEC strain, was used as a positive control  and E. coli strain JM109 used as a negative control
and E. coli strain JM109 used as a negative control  . Translocation was measured after 30, 60, and 120 min. Results are expressed as mean ± SEM.
. Translocation was measured after 30, 60, and 120 min. Results are expressed as mean ± SEM.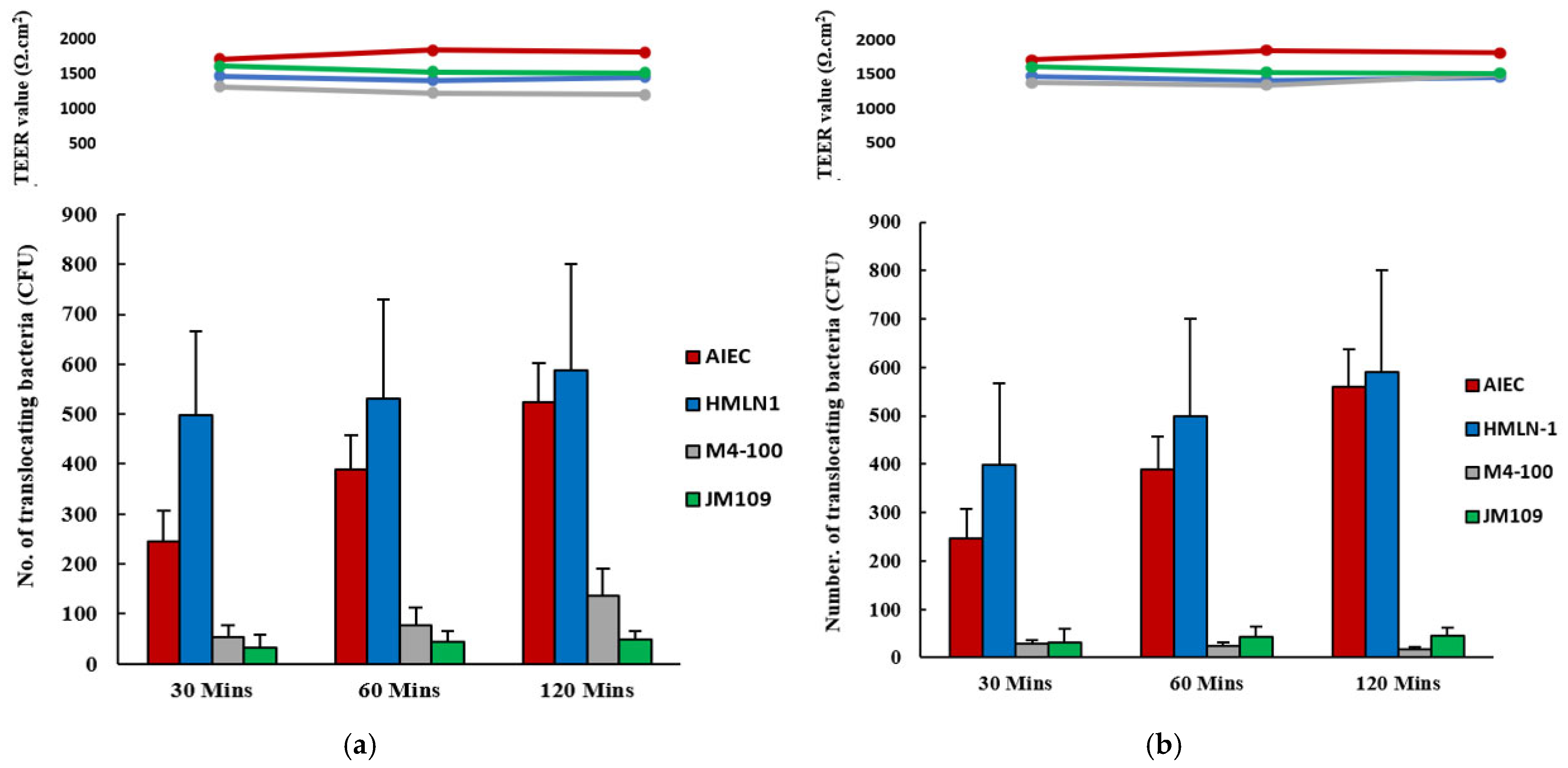

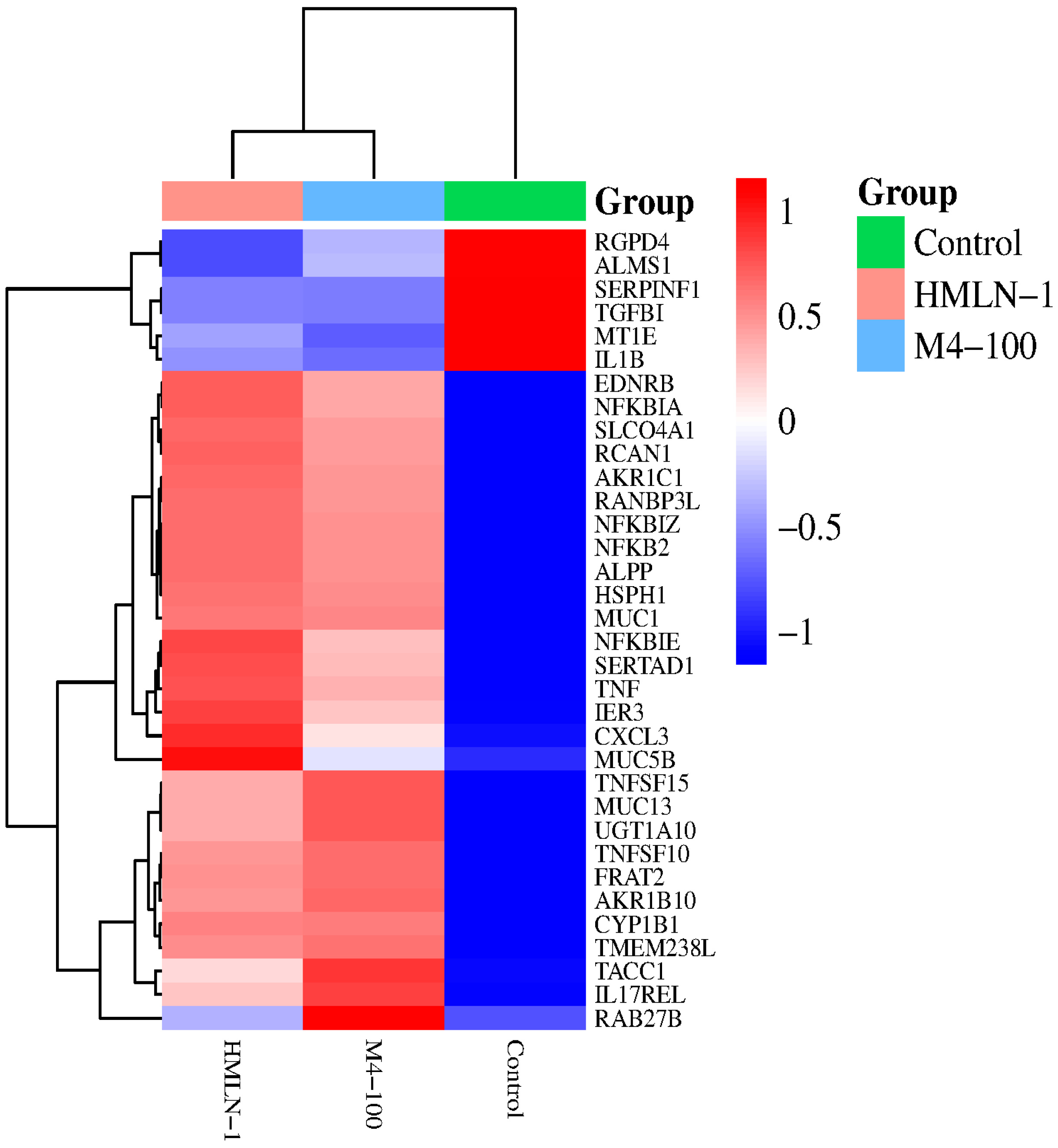
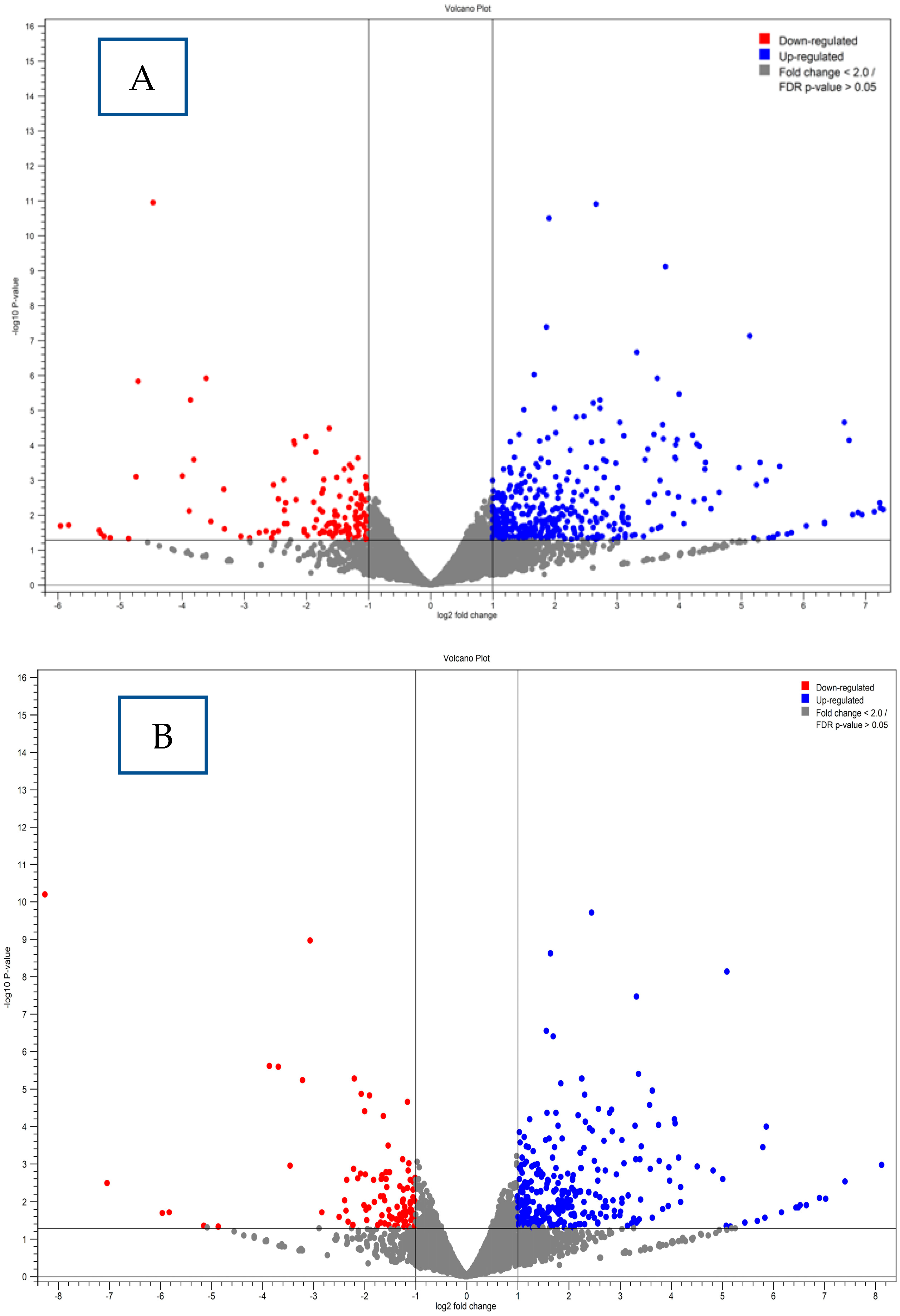
| Gene Category | Gene Name | FDR Value | |
|---|---|---|---|
| HMLN-1 vs. Control | L. reuteri M4-100 vs. Control | ||
| Genes involved in pro-inflammatory response | IL1B | ↓ 0.001895 | |
| IL17REL | ↑ 0.004429 | ||
| NFKBIZ | ↑ 4.25E-08 | ↑ 3.9E-07 | |
| IL6ST | |||
| TNF | ↑ 0.000455 | ↑ 0.001208 | |
| TLR7 | ↑ 0.00033 | ↑ 0.000236 | |
| Genes involved in tight junction and adherence | L1CAM | ||
| MUC13 | ↑ 0.00313096 | ↑ 0.002085 | |
| MUC6 | |||
| MUC5B | |||
| MUC1 | ↑ 0.00313084 | ↑ 0.004085 | |
| MAPK4 | |||
| Genes involved in anti-inflammatory response | TGFB1 | ↓ 0.00546 | |
| CXCL3 | ↑ 0.003130 | ↑ 0.004085 | |
| IL2RA | ↑ 0.009759 | ||
| NFKBIA | ↑ 3.2E-11 | ↑ 2.37E-09 | |
| NFKB2 | ↑ 0.009517 | ||
| TNFRSF19 | ↑ 7.59E-05 | ↓ 0.000132 | |
| CXCL5 | ↑ 0.009013 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asgari, B.; Bradford, G.; Hatje, E.; Kuballa, A.; Katouli, M. Limosilactobacillus reuteri M4-100 Mitigates the Pathogenicity of Escherichia coli Strain HMLN-1 in an Intestinal Epithelial Model and Modulates Host Cell Gene Expression. Microorganisms 2025, 13, 1428. https://doi.org/10.3390/microorganisms13061428
Asgari B, Bradford G, Hatje E, Kuballa A, Katouli M. Limosilactobacillus reuteri M4-100 Mitigates the Pathogenicity of Escherichia coli Strain HMLN-1 in an Intestinal Epithelial Model and Modulates Host Cell Gene Expression. Microorganisms. 2025; 13(6):1428. https://doi.org/10.3390/microorganisms13061428
Chicago/Turabian StyleAsgari, Behnoush, Georgia Bradford, Eva Hatje, Anna Kuballa, and Mohammad Katouli. 2025. "Limosilactobacillus reuteri M4-100 Mitigates the Pathogenicity of Escherichia coli Strain HMLN-1 in an Intestinal Epithelial Model and Modulates Host Cell Gene Expression" Microorganisms 13, no. 6: 1428. https://doi.org/10.3390/microorganisms13061428
APA StyleAsgari, B., Bradford, G., Hatje, E., Kuballa, A., & Katouli, M. (2025). Limosilactobacillus reuteri M4-100 Mitigates the Pathogenicity of Escherichia coli Strain HMLN-1 in an Intestinal Epithelial Model and Modulates Host Cell Gene Expression. Microorganisms, 13(6), 1428. https://doi.org/10.3390/microorganisms13061428






