Taxonomic Diversity and Antimicrobial Potential of Thermophilic Bacteria from Two Extreme Algerian Hot Springs
Abstract
1. Introduction
2. Materials and Methods
2.1. Environmental Sampling and Site Description
2.2. Isolation of Thermophilic Bacteria
2.3. Phenotypic and Physiological Characterization of Isolates
2.4. Proteomic Identification Using MALDI-TOF Mass Spectrometry
2.5. Molecular Identification
2.5.1. Genomic DNA Extraction and Quality Assessment
2.5.2. Genomic Fingerprinting via rep-PCR ((GTG)5-PCR, BOX-PCR, ERIC-PCR)
2.5.3. 16S rRNA Gene Amplification
2.5.4. 16S rRNA Sequencing Analysis
2.5.5. Phylogenetic Analysis
2.6. Screening for Antimicrobial Activity
2.7. Optimization of Antimicrobial Metabolite Production
2.8. Data Processing and Statistical Analysis
3. Results
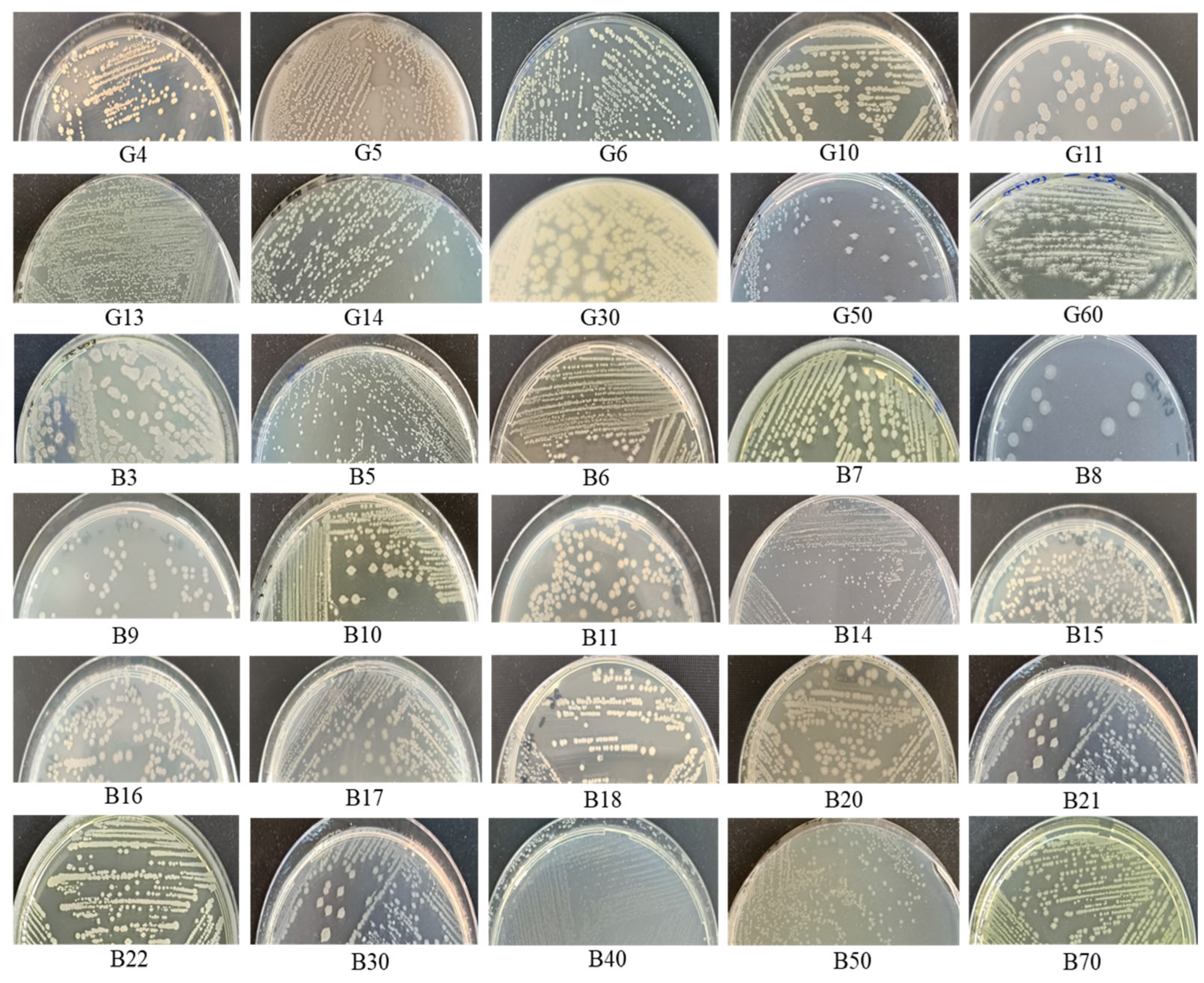
3.1. Environmental Sampling and Bacterial Isolation
3.2. Phenotypic Characterization and Optimum Physiological Parameters
3.3. MALDI-TOF MS Identification
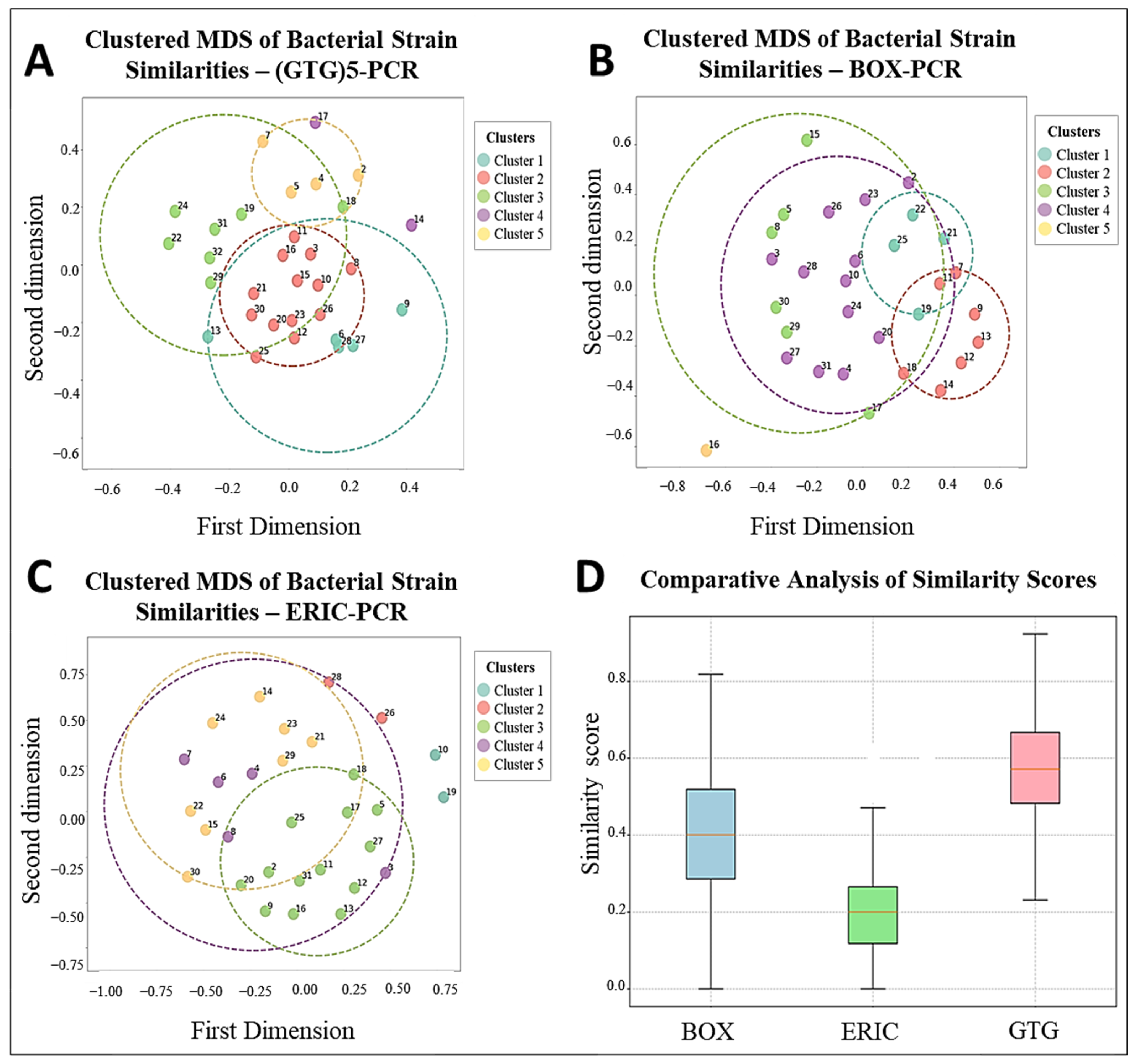
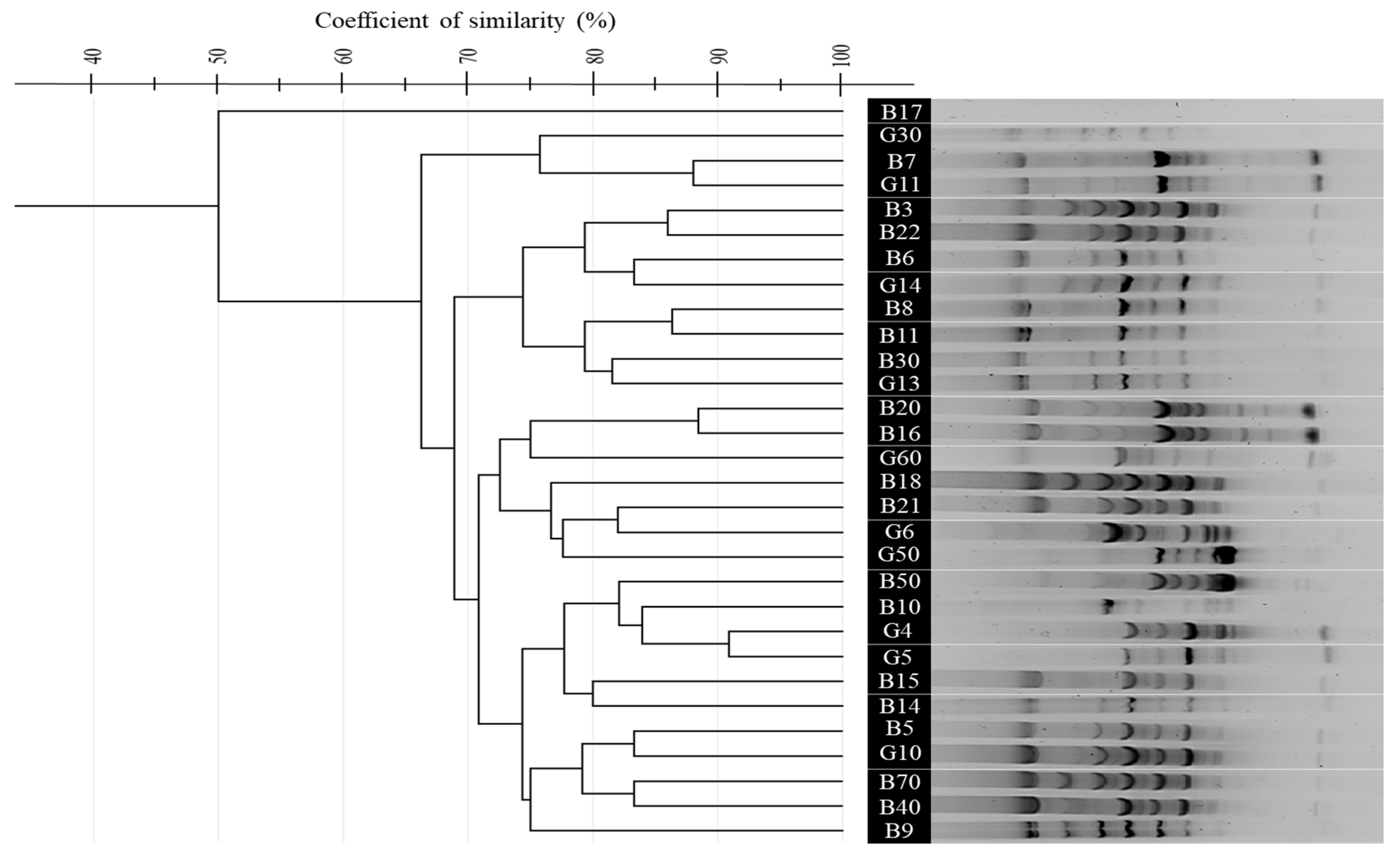
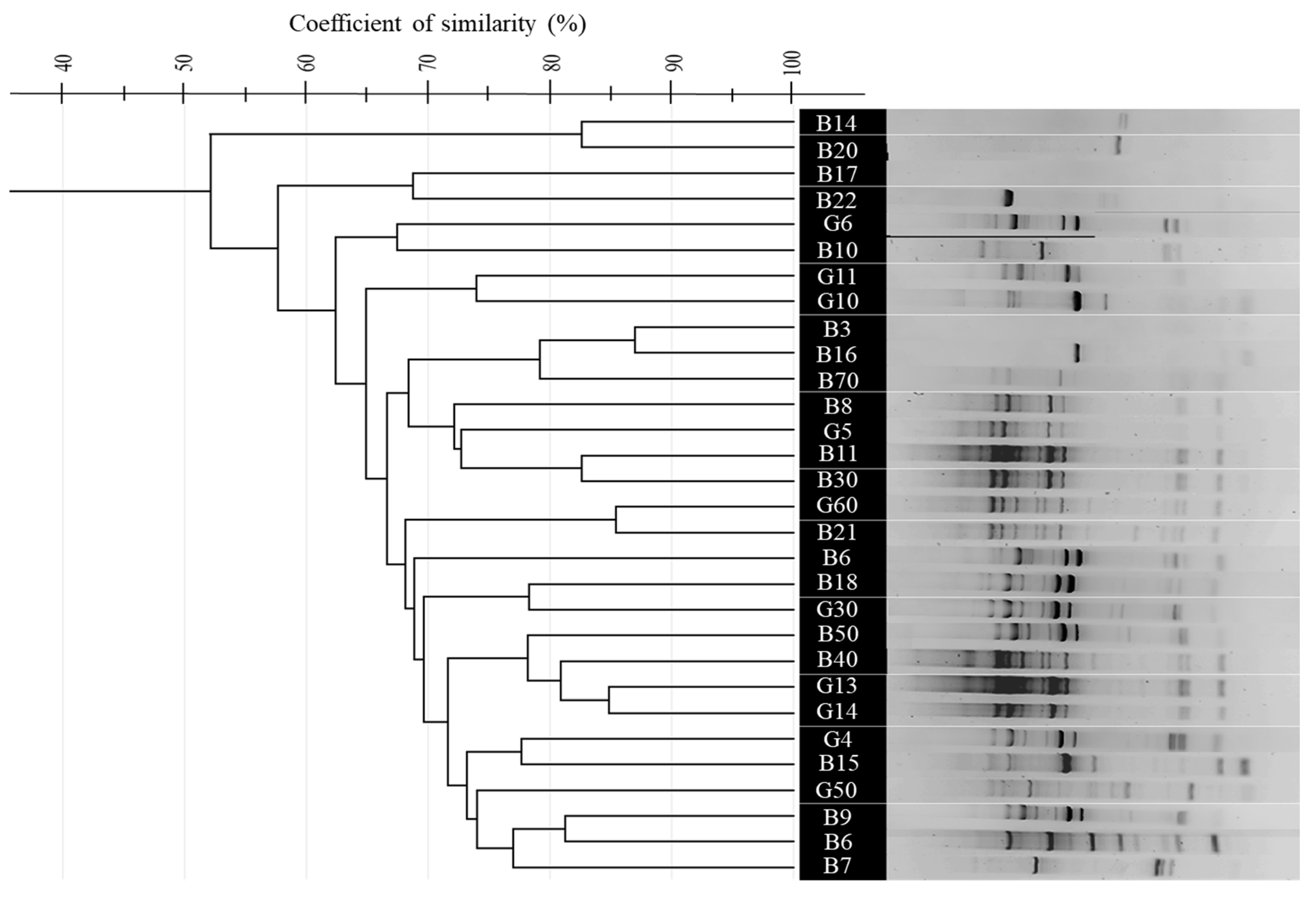
3.4. Comparative Analysis of BOX, ERIC, and (GTG)5-PCR Fingerprinting Profiles
3.5. Sequencing Analysis and Phylogenetic Analysis
3.6. Antimicrobial Activity and Optimization of Culture Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ando, N.; Barquera, B.; Bartlett, D.H.; Boyd, E.; Burnim, A.A.; Byer, A.S.; Colman, D.; Gillilan, R.E.; Gruebele, M.; Makhatadze, G.; et al. The Molecular Basis for Life in Extreme Environments. Annu. Rev. Biophys. 2021, 50, 343–372. [Google Scholar] [CrossRef]
- de la Haba, R.R.; Antunes, A.; Hedlund, B.P. Editorial: Extremophiles: Microbial Genomics and Taxogenomics. Front. Microbiol. 2022, 13, 984632. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Trivedi, P.; Kumar Mishra, A.; Pandey, A.; Palni, L.M.S. Microbial Diversity of Soil from Two Hot Springs in Uttaranchal Himalaya. Microbiol. Res. 2004, 159, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Somayaji, A.; Dhanjal, C.R.; Lingamsetty, R.; Vinayagam, R.; Selvaraj, R.; Varadavenkatesan, T.; Govarthanan, M. An Insight into the Mechanisms of Homeostasis in Extremophiles. Microbiol. Res. 2022, 263, 127115. [Google Scholar] [CrossRef]
- Rawat, M.; Chauhan, M.; Pandey, A. Extremophiles and Their Expanding Biotechnological Applications. Arch. Microbiol. 2024, 206, 247. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Capalash, N.; Sharma, P. Communication Mechanisms in Extremophiles: Exploring Their Existence and Industrial Applications. Microbiol. Res. 2019, 221, 15–27. [Google Scholar] [CrossRef]
- Salwan, R.; Sharma, V. Genomics of Prokaryotic Extremophiles to Unfold the Mystery of Survival in Extreme Environments. Microbiol. Res. 2022, 264, 127156. [Google Scholar] [CrossRef]
- Kaushik, S.; Alatawi, A.; Djiwanti, S.R.; Pande, A.; Skotti, E.; Soni, V. Potential of Extremophiles for Bioremediation. In Microbial Rejuvenation of Polluted Environment: Volume 1; Panpatte, D.G., Jhala, Y.K., Eds.; Springer: Singapore, 2021; pp. 293–328. ISBN 978-981-15-7447-4. [Google Scholar]
- Rafiee, Z.; Jalili Tabaii, M.; Moradi, M.; Harirchi, S. Unveiling Antibacterial Potential and Physiological Characteristics of Thermophilic Bacteria Isolated from a Hot Spring in Iran. Microorganisms 2024, 12, 834. [Google Scholar] [CrossRef]
- Todorov, S.D.; de Almeida, B.M.; Lima, E.M.F.; Fabi, J.P.; Lajolo, F.M.; Hassimotto, N.M.A. Phenolic Compounds and Bacteriocins: Mechanisms, Interactions, and Applications in Food Preservation and Safety. Mol. Nutr. Food Res. 2025, 69, e202400723. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Kuisiene, N.; Jasinskyte, D.; Rutiene, K.; Saleikiene, J.; Chitavichius, D. Novel Bacteriocins Produced by Geobacillus stearothermophilus. Open Life Sci. 2009, 4, 196–203. [Google Scholar] [CrossRef]
- Koniuchovaitė, A.; Petkevičiūtė, A.; Bernotaitė, E.; Gricajeva, A.; Gegeckas, A.; Kalėdienė, L.; Kaunietis, A. Novel Leaderless Bacteriocin Geobacillin 6 from Thermophilic Bacterium Parageobacillus thermoglucosidasius. Front. Microbiol. 2023, 14, 1207367. [Google Scholar] [CrossRef]
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.-S.; Yang, H.; Sun, L. Antimicrobial Peptides for Combating Drug-Resistant Bacterial Infections. Drug Resist. Updates 2023, 68, 100954. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.; Singh, S.P.; Zhang, C. Microorganisms under Extreme Environments and Their Applications. Curr. Res. Microb. Sci. 2022, 3, 100141. [Google Scholar] [CrossRef]
- Burkhardt, C.; Baruth, L.; Neele Meyer-Heydecke; Klippel, B.; Margaryan, A.; Paloyan, A.; Panosyan, H.H.; Antranikian, G. Mining Thermophiles for Biotechnologically Relevant Enzymes: Evaluating the Potential of European and Caucasian Hot Springs. Extremophiles 2023, 28, 5. [Google Scholar] [CrossRef]
- Benammar, L.; İnan Bektaş, K.; Menasria, T.; Beldüz, A.O.; Güler, H.I.; Bedaida, I.K.; Gonzalez, J.M.; Ayachi, A. Diversity and Enzymatic Potential of Thermophilic Bacteria Associated with Terrestrial Hot Springs in Algeria. Braz. J. Microbiol. 2020, 51, 1987–2007. [Google Scholar] [CrossRef] [PubMed]
- Boukhenfouf, W.; Boucenna, A. Comparison of the Radio-Isotopic Composition between Hot Spring Water of Hammam Debagh and Its Associated Deposits. Radiat. Prot. Dosim. 2019, 187, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Naimi, M.N.; Cherif, A. The Hottest African Thermal Spring (Hammam Chellala, Algeria). Int. J. Earth Sci. (Geol. Rundsch.) 2021, 110, 651–652. [Google Scholar] [CrossRef]
- Belhai, M.; Fujimitsu, Y.; Bouchareb-Haouchine, F.Z.; Nishijima, J. Geology, Geothermometry, Isotopes and Gas Chemistry of the Northern Algerian Geothermal System. In Proceedings of the World Geothermal Congress 2015, Melbourne, Australia, 19–25 April 2015. [Google Scholar]
- Benziada, M. Hydrogeochemical Study of Northern Algeria Hot Springs for Thermal Treatments. JCCE 2019, 13, 47–54. [Google Scholar] [CrossRef]
- Valenzuela, B.; Solís-Cornejo, F.; Araya, R.; Zamorano, P. Isolation of Thermophilic Bacteria from Extreme Environments in Northern Chile. Microorganisms 2024, 12, 473. [Google Scholar] [CrossRef]
- Saikia, R.; Gogoi, D.K.; Mazumder, S.; Yadav, A.; Sarma, R.K.; Bora, T.C.; Gogoi, B.K. Brevibacillus laterosporus Strain BPM3, a Potential Biocontrol Agent Isolated from a Natural Hot Water Spring of Assam, India. Microbiol. Res. 2011, 166, 216–225. [Google Scholar] [CrossRef]
- Kochhar, N.; I․K, K.; Shrivastava, S.; Ghosh, A.; Rawat, V.S.; Sodhi, K.K.; Kumar, M. Perspectives on the Microorganism of Extreme Environments and Their Applications. Curr. Res. Microb. Sci. 2022, 3, 100134. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Villar, R.; Escalante, A.; Astudillo-Melgar, F.; Lizárraga-Mendiola, L.; Vázquez-Rodríguez, G.A.; Hidalgo-Lara, M.E.; Coronel-Olivares, C. Isolation and Characterization of Thermophilic Bacteria from a Hot Spring in the State of Hidalgo, Mexico, and Geochemical Analysis of the Thermal Water. Microorganisms 2024, 12, 1066. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.-S.; Huang, L.-N. Microbial Diversity in Extreme Environments. Nat. Rev. Microbiol. 2022, 20, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Saghatelyan, A.; Margaryan, A.; Panosyan, H.; Birkeland, N.-K. Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review. Microorganisms 2021, 9, 1473. [Google Scholar] [CrossRef]
- Rahi, P.; Prakash, O.; Shouche, Y.S. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass-Spectrometry (MALDI-TOF MS) Based Microbial Identifications: Challenges and Scopes for Microbial Ecologists. Front. Microbiol. 2016, 7, 1359. [Google Scholar] [CrossRef]
- Strejcek, M.; Smrhova, T.; Junkova, P.; Uhlik, O. Whole-Cell MALDI-TOF MS Versus 16S rRNA Gene Analysis for Identification and Dereplication of Recurrent Bacterial Isolates. Front. Microbiol. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Cihan, A.C. Taxonomic Classification of Anoxybacillus Isolates from Geothermal Regions in Turkey by 16S rRNA Gene Sequences and ARDRA, ITS-PCR, Rep-PCR Analyses. Pol. J. Microbiol. 2013, 62, 149–163. [Google Scholar] [CrossRef]
- Selim, S.; Sherif, M.E.; El-Alfy, S.; Hagagy, N. Genetic Diversity among Thermophilic Bacteria Isolated from Geothermal Sites by Using Two PCR Typing Methods. Geomicrobiol. J. 2014, 31, 161–170. [Google Scholar] [CrossRef]
- Hussin, N.; Ismail, A.; Balakrishnan, V.; Lachimanan, Y.; Sasidharan, S. Identification of Appropriate Sample and Culture Method for Isolation of New Thermophilic Bacteria from Hot Spring. Afr. J. Microbiol. Res. 2011, 5, 217–221. [Google Scholar]
- Guta, M.; Abebe, G.; Bacha, K.; Cools, P. Screening and Characterization of Thermostable Enzyme-Producing Bacteria from Selected Hot Springs of Ethiopia. Microbiol. Spectr. 2024, 12, e03710-23. [Google Scholar] [CrossRef]
- Abussaud, M.J.; Alanagreh, L.; Abu-Elteen, K. Isolation, Characterization and Antimicrobial Activity of Streptomyces Strains from Hot Spring Areas in the Northern Part of Jordan. Afr. J. Biotechnol. 2013, 12, 7124–7132. [Google Scholar]
- Moyes, R.B.; Reynolds, J.; Breakwell, D.P. Differential Staining of Bacteria: Gram Stain. Curr. Protoc. Microbiol. 2009, 15, A.3C.1–A.3C.8. [Google Scholar] [CrossRef]
- Iwase, T.; Tajima, A.; Sugimoto, S.; Okuda, K.; Hironaka, I.; Kamata, Y.; Takada, K.; Mizunoe, Y. A Simple Assay for Measuring Catalase Activity: A Visual Approach. Sci. Rep. 2013, 3, 3081. [Google Scholar] [CrossRef] [PubMed]
- McElvania TeKippe, E.; Shuey, S.; Winkler, D.W.; Butler, M.A.; Burnham, C.-A.D. Optimizing Identification of Clinically Relevant Gram-Positive Organisms by Use of the Bruker Biotyper Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry System. J. Clin. Microbiol. 2013, 51, 1421–1427. [Google Scholar] [CrossRef]
- Gekenidis, M.-T.; Studer, P.; Wüthrich, S.; Brunisholz, R.; Drissner, D. Beyond the Matrix-Assisted Laser Desorption Ionization (MALDI) Biotyping Workflow: In Search of Microorganism-Specific Tryptic Peptides Enabling Discrimination of Subspecies. Appl. Env. Microbiol. 2014, 80, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Merzoug, M.; Bendida, K.; Aireche, M.; Zater, Z.Y.; Brakna, C.N.; Hammadi, A.I.; Saidi, Y.; Todorov, S.D.; Saidi, D. Isolation and Characterization of Enterocin-Producing Enterococcus faecium Strains from Algerian Traditional Food “Dried Figs Marinated in Olive Oil”: Functional and Safety Evaluations. Foods 2025, 14, 766. [Google Scholar] [CrossRef]
- Gautam, A. DNA Isolation by Lysozyme and Proteinase K. In DNA and RNA Isolation Techniques for Non-Experts; Gautam, A., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 85–88. ISBN 978-3-030-94230-4. [Google Scholar]
- Adiguzel, A.; Ozkan, H.; Baris, O.; Inan, K.; Gulluce, M.; Sahin, F. Identification and Characterization of Thermophilic Bacteria Isolated from Hot Springs in Turkey. J. Microbiol. Methods 2009, 79, 321–328. [Google Scholar] [CrossRef]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ—A Tool for Analyzing DNA Fingerprint Gel Images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef]
- Hassan, W.M.; Wang, S.Y.; Ellender, R.D. Methods To Increase Fidelity of Repetitive Extragenic Palindromic PCR Fingerprint-Based Bacterial Source Tracking Efforts. Appl. Environ. Microbiol. 2005, 71, 512–518. [Google Scholar] [CrossRef]
- Heras, J.; Dominguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. A Survey of Tools for Analysing DNA Fingerprints. Brief. Bioinform. 2015, 17, 903–911. [Google Scholar] [CrossRef]
- Kuisiene, N.; Jomantiene, R.; Valiunas, D.; Chitavichius, D. Characterization of Thermophilic Proteolytic Spore-Forming Bacteria from a Geothermal Site in Lithuania Based on 16S rDNA RFLP and ITS-PCR Analyses1. Microbiology 2002, 71, 712–716. [Google Scholar] [CrossRef]
- Zater, Z.Y.; Merzoug, M.; Baltaci, M.O.; Todorov, S.D.; Adiguzel, A.; Roudj, S. Impact of a Novel Caseinolytic Protease Single Mutation on Lactiplantibacillus pentosus Growth Performance. Process. Biochem. 2024, 145, 145–152. [Google Scholar] [CrossRef]
- Versalovic, J.; Koeuth, T.; Lupski, J.R. Distribution of Repetitive DNA Sequences in Eubacteria and Application to Fingerprinting of Bacterial Genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J. Genomic Fingerprinting of Bacteria Using Repetitive Sequence-Based Polymerase Chain Reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Bell, J.R. A Simple Way to Treat PCR Products Prior to Sequencing Using ExoSAP-IT®. BioTechniques 2008, 44, 834. [Google Scholar] [CrossRef] [PubMed]
- Alzohairy, A.M. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Madden, T. The BLAST Sequence Analysis Tool. 2002. Available online: https://www.google.com/url?sa=t&source=web&rct=j&opi=89978449&url=http://www.binf.gmu.edu/vaisman/binf630/madden_blast_ch16.pdf&ved=2ahUKEwi0tce0qPWNAxXXSGwGHVixINYQFnoECBcQAQ&usg=AOvVaw1BpGZ_djgQshEN3DBz7V6A (accessed on 12 April 2025).
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Saitou, N. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Beerli, P. Pairwise Distance Methods. 2005. Available online: https://www.google.com/url?sa=t&source=web&rct=j&opi=89978449&url=https://people.sc.fsu.edu/~pbeerli/BSC-5936/10-26-05/lecture_15.pdf&ved=2ahUKEwijhfXwp_WNAxVMS3ADHVarB0UQFnoECBAQAQ&usg=AOvVaw0d7yR6wCTCjgu42mMVyFwJ (accessed on 12 April 2025).
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K.; Battistuzzi, F.U. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Casida, L.E. GELRITE as a Gelling Agent in Media for the Growth of Thermophilic Microorganisms. Appl. Environ. Microbiol. 1984, 47, 427–429. [Google Scholar] [CrossRef]
- Saad, M.; Abdelsamei, H.; Ibrahim, E.; Abdou, A.; El Sohaimy, S. Effect of pH, Heat Treatments and Proteinase K Enzyme on the Activity of Lactobacillus acidophilus Bacteriocin. Benha Vet. Med. J. 2015, 28, 210–215. [Google Scholar] [CrossRef]
- Haque, M.U.; Rahman, M.A.; Haque, M.A.; Sarker, A.K.; Islam, M.A.U. Modulation of Antibacterial Activity of Actinomycetes by Co-Culture with Pathogenic Bacteria. Bangladesh Pharm. J. 2015, 18, 61–65. [Google Scholar] [CrossRef]
- Harris, C.R. Array Programming with NumPy. Nature 2020, 585, 357–362. Available online: https://www.nature.com/articles/s41586-020-2649-2 (accessed on 10 June 2025). [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. JOSS 2021, 6, 3021. [Google Scholar] [CrossRef]
- Virtanen, P. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python|Nature Methods. Nat. Methods 2020, 17, 261–272. [Google Scholar]
- Pedregosa, F.; Pedregosa, F.; Varoquaux, G.; Varoquaux, G.; Org, N.; Gramfort, A.; Gramfort, A.; Michel, V.; Michel, V.; Fr, L.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ishii, M.; Igarashi, Y.; Kodama, T. Colony Formation of Hydrogeno-bacter thermophilus on a Plate Solidified with GELRITE. Agric. Biol. Chem. 1987, 51, 3139–3141. [Google Scholar] [CrossRef]
- Kazemi, M.J.; Kargar, M.; Nowroozi, J.; Sepahi, A.A.; Doosti, A.; Manafi, Z. Optimization and Comparison of Different Procedures of Preparation GELRITE and Starch as Solidifying Agents for Culturing Indigenous Thermoacidophilc Microorganisms and How to Keep Them at the Temperature of 70 °C. Biosci. Biotechnol. Res. Asia 2017, 14, 63–68. [Google Scholar] [CrossRef]
- Raddadi, N.; Cherif, A.; Daffonchio, D.; Neifar, M.; Fava, F. Biotechnological Applications of Extremophiles, Extremozymes and Extremolytes. Appl. Microbiol. Biotechnol. 2015, 99, 7907–7913. [Google Scholar] [CrossRef]
- Sun, J.; He, X.; Le, Y.; Al-Tohamy, R.; Ali, S.S. Potential Applications of Extremophilic Bacteria in the Bioremediation of Extreme Environments Contaminated with Heavy Metals. J. Environ. Manag. 2024, 352, 120081. [Google Scholar] [CrossRef]
- Timperio, A.M.; Gorrasi, S.; Zolla, L.; Fenice, M. Evaluation of MALDI-TOF Mass Spectrometry and MALDI BioTyper in Comparison to 16S rDNA Sequencing for the Identification of Bacteria Isolated from Arctic Sea Water. PLoS ONE 2017, 12, e0181860. [Google Scholar] [CrossRef] [PubMed]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of Adaptation of Microorganisms of the Three Domains of Life to High Salt Concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Verma, J.; Sourirajan, A.; Dev, K. Bacterial Diversity in 110 Thermal Hot Springs of Indian Himalayan Region (IHR). 3 Biotech 2022, 12, 238. [Google Scholar] [CrossRef]
- Panosyan, H.; Margaryan, A.; Birkeland, N.-K. Geothermal Springs in Armenia and Nagorno-Karabakh: Potential Sources of Hydrolase-Producing Thermophilic Bacilli. Extremophiles 2020, 24, 519–536. [Google Scholar] [CrossRef]
- Mathan Kumar, R.; Jani, K.; Parvathi, J.R.; Thomas, B.M.; Raja, S.S.S.; Pandey, A.; Sharma, A. Bacterial Diversity of Geochemically Distinct Hot Springs Located in Maharashtra, India. Arch. Microbiol. 2022, 204, 110. [Google Scholar] [CrossRef]
- Kopcakova, A.; Stramova, Z.; Kvasnova, S.; Godany, A.; Perhacova, Z.; Pristas, P. Need for Database Extension for Reliable Identification of Bacteria from Extreme Environments Using MALDI TOF Mass Spectrometry. Chem. Pap. 2014, 68, 1435–1442. [Google Scholar] [CrossRef]
- Kedaid, F.-Z.; Mesbah, M. Geochemical Approach to the Bou Hadjar Hydrothermal System (NE Algeria). Geothermics 1996, 25, 249–257. [Google Scholar] [CrossRef]
- Amarouche-Yala, S.; Benouadah, A.; El Ouahab Bentabet, A.; Moulla, A.S.; Ouarezki, S.A.; Azbouche, A. Physicochemical, Bacteriological, and Radiochemical Characterization of Some Algerian Thermal Spring Waters. Water Qual. Expo. Health 2015, 7, 233–249. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Ganbat, D.; Oh, D.; Kim, H.; Jeong, G.E.; Cha, I.-T.; Kim, S.-B.; Nam, G.; Jung, Y.-J.; Lee, S.-J. Isolation and Characterization of Thermophilic Bacteria from Hot Springs in Republic of Korea. Microorganisms 2022, 10, 2375. [Google Scholar] [CrossRef] [PubMed]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic Study of Aerobic Thermophilic Bacilli: Descriptions of Geobacillus Subterraneus Gen. Nov., Sp. Nov. and Geobacillus Uzenensis Sp. Nov. from Petroleum Reservoirs and Transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the New Combinations G. Stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, T.L.; Gugliandolo, C.; Caccamo, D.; Stackebrandt, E. Three Novel Halotolerant and Thermophilic Geobacillus Strains from Shallow Marine Vents. Syst. Appl. Microbiol. 2002, 25, 450–455. [Google Scholar] [CrossRef]
- Chan, C.S.; Chan, K.-G.; Tay, Y.-L.; Chua, Y.-H.; Goh, K.M. Diversity of Thermophiles in a Malaysian Hot Spring Determined Using 16S rRNA and Shotgun Metagenome Sequencing. Front. Microbiol. 2015, 6, 177. [Google Scholar] [CrossRef]
- Rowe, L.; Dowd, S.E.; Davidson, K.; Kovarik, C.; VanAken, M.; Jarabek, A.; Taylor, C. Comparing Microbial Populations from Diverse Hydrothermal Features in Yellowstone National Park: Hot Springs and Mud Volcanoes. Front. Microbiol. 2024, 15, 1409664. [Google Scholar] [CrossRef]
- Efe, D.; Ahmet, A. Molecular Typing of Thermophilic Bacilli Isolated from Different Hot Springs of Turkey. Res. J. Biotechnol. 2014, 9, 83–88. [Google Scholar]
- Wani, A.K.; Akhtar, N.; Sher, F.; Navarrete, A.A.; Américo-Pinheiro, J.H.P. Microbial Adaptation to Different Environmental Conditions: Molecular Perspective of Evolved Genetic and Cellular Systems. Arch. Microbiol. 2022, 204, 144. [Google Scholar] [CrossRef]
- Oztas Gulmus, E.; Gormez, A. Identification and Characterization of Novel Thermophilic Bacteria from Hot Springs, Erzurum, Turkey. Curr. Microbiol. 2020, 77, 979–987. [Google Scholar] [CrossRef]
- Savas, S.; Adiguzel, A.; Inan, K.; Ozkan, H.; Gulluce, M.; Sahin, F. Molecular Characterization of Thermophilic Bacteria Isolated from Van City Ercis Town Hasanabdal Hot Spring. Rom. Biotech. Lett. 2009, 14, 4445–4454. [Google Scholar]
- Park, C.; Kim, S.B.; Choi, S.H.; Kim, S. Comparison of 16S rRNA Gene Based Microbial Profiling Using Five Next-Generation Sequencers and Various Primers. Front. Microbiol. 2021, 12, 715500. [Google Scholar] [CrossRef]
- Soy, S.; Lakra, U.; Prakash, P.; Suravajhala, P.; Nigam, V.K.; Sharma, S.R.; Bayal, N. Exploring Microbial Diversity in Hot Springs of Surajkund, India through 16S rRNA Analysis and Thermozyme Characterization from Endogenous Isolates. Sci. Rep. 2023, 13, 14221. [Google Scholar] [CrossRef]
- Najar, I.N.; Sherpa, M.T.; Das, S.; Das, S.; Thakur, N. Microbial Ecology of Two Hot Springs of Sikkim: Predominate Population and Geochemistry. Sci. Total Environ. 2018, 637–638, 730–745. [Google Scholar] [CrossRef]
- Wang, J.; Goh, K.M.; Salem, D.R.; Sani, R.K. Genome Analysis of a Thermophilic Exopolysaccharide-Producing Bacterium—Geobacillus Sp. WSUCF1. Sci. Rep. 2019, 9, 1608. [Google Scholar] [CrossRef]
- Ulucay, O.; Gormez, A.; Ozic, C. Identification, Characterization and Hydrolase Producing Performance of Thermophilic Bacteria: Geothermal Hot Springs in the Eastern and Southeastern Anatolia Regions of Turkey. Antonie Van Leeuwenhoek 2022, 115, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Peña, A.; Teeling, H.; Huerta-Cepas, J.; Santos, F.; Yarza, P.; Brito-Echeverría, J.; Lucio, M.; Schmitt-Kopplin, P.; Meseguer, I.; Schenowitz, C.; et al. Fine-Scale Evolution: Genomic, Phenotypic and Ecological Differentiation in Two Coexisting Salinibacter Ruber Strains. ISME J. 2010, 4, 882–895. [Google Scholar] [CrossRef]
- Mwamburi, S.M.; Islam, S.I.; Dinh-Hung, N.; Dangsawat, O.; Sowanpreecha, R.; Khang, L.T.P.; Montha, N.; Therdtatha, P.; Dwinanti, S.H.; Permpoonpattana, P.; et al. Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential. Microorganisms 2024, 12, 2476. [Google Scholar] [CrossRef] [PubMed]
- Aktypis, A.; Kalantzopoulos, G. Purification and Characterization of Thermophilin ST-1, a Novel Bacteriocin Produced by Streptococcus Thermophilus ACA-DC 0001. Lait 2003, 83, 365–378. [Google Scholar] [CrossRef]
- Kabuki, T.; Uenishi, H.; Watanabe, M.; Seto, Y.; Nakajima, H. Characterization of a Bacteriocin, Thermophilin 1277, Produced by Streptococcus Thermophilus SBT1277. J. Appl. Microbiol. 2007, 102, 971–980. [Google Scholar] [CrossRef]
- Rossi, F.; Marzotto, M.; Cremonese, S.; Rizzotti, L.; Torriani, S. Diversity of Streptococcus Thermophilus in Bacteriocin Production; Inhibitory Spectrum and Occurrence of Thermophilin Genes. Food Microbiol. 2013, 35, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Egan, K.; Field, D.; Ross, R.P.; Cotter, P.D.; Hill, C. In Silico Prediction and Exploration of Potential Bacteriocin Gene Clusters Within the Bacterial Genus Geobacillus. Front. Microbiol. 2018, 9, 2116. [Google Scholar] [CrossRef] [PubMed]
- Kita, K.; Yoshida, S.; Masuo, S.; Nakamura, A.; Ishikawa, S.; Yoshida, K. Genes Encoding a Novel Thermostable Bacteriocin in the Thermophilic Bacterium Aeribacillus Pallidus PI8. J. Appl. Microbiol. 2023, 134, lxad293. [Google Scholar] [CrossRef] [PubMed]
- Salini, F.; Iacumin, L.; Comi, G.; Dicks, L.M.T. Thermophilin 13: In Silico Analysis Provides New Insight in Genes Involved in Bacteriocin Production. Microorganisms 2023, 11, 611. [Google Scholar] [CrossRef]
- Etayash, H.; Azmi, S.; Dangeti, R.; Kaur, K. Peptide Bacteriocins—Structure Activity Relationships. Curr. Top. Med. Chem. 2016, 16, 220–241. [Google Scholar] [CrossRef]
- Alkhalili, R.N.; Bernfur, K.; Dishisha, T.; Mamo, G.; Schelin, J.; Canbäck, B.; Emanuelsson, C.; Hatti-Kaul, R. Antimicrobial Protein Candidates from the Thermophilic Geobacillus Sp. Strain ZGt-1: Production, Proteomics, and Bioinformatics Analysis. Int. J. Mol. Sci. 2016, 17, 1363. [Google Scholar] [CrossRef]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite Induction via Microorganism Co-Culture: A Potential Way to Enhance Chemical Diversity for Drug Discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Hockett, K.L.; Baltrus, D.A. Use of the Soft-Agar Overlay Technique to Screen for Bacterially Produced Inhibitory Compounds. JoVE 2017, 119, 55064. [Google Scholar] [CrossRef]
- Bassler, B.L.; Greenberg, E.P.; Stevens, A.M. Cross-Species Induction of Luminescence in the Quorum-Sensing Bacterium Vibrio harveyi. J. Bacteriol. 1997, 179, 4043–4045. [Google Scholar] [CrossRef]
- Messaoudi, S.; Manai, M.; Kergourlay, G.; Prévost, H.; Connil, N.; Chobert, J.-M.; Dousset, X. Lactobacillus salivarius: Bacteriocin and Probiotic Activity. Food Microbiol. 2013, 36, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-Z.; Zhang, W.-Q.; Hu, P.-F.; Yang, Q.-Q.; Molnár, I.; Xu, P.; Zhang, B.-B. Harnessing Microbial Co-Culture to Increase the Production of Known Secondary Metabolites. Nat. Prod. Rep. 2025, 42, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Nie, R.; Zhu, Z.; Qi, Y.; Wang, Z.; Sun, H.; Liu, G. Bacteriocin Production Enhancing Mechanism of Lactiplantibacillus paraplantarum RX-8 Response to Wickerhamomyces Anomalus Y-5 by Transcriptomic and Proteomic Analyses. Front. Microbiol. 2023, 14, 1111516. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Dicks, L.M.T. Bacteriocin Production by Pediococcus pentosaceus Isolated from Marula (Scerocarya Birrea). Int. J. Food Microbiol. 2009, 132, 117–126. [Google Scholar] [CrossRef]
- Chanos, P.; Mygind, T. Co-Culture-Inducible Bacteriocin Production in Lactic Acid Bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 4297–4308. [Google Scholar] [CrossRef]


| Category | Target Gene | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) | Amplicon Size (bp) | Tm (°C) | Reference |
|---|---|---|---|---|---|---|
| Molecular identification | 16S rRNA | AGAGTTTGATCCTGGCTCAG | ACGGCTACCTTGTTACGACTT | 1485 | 56 | [44,45] |
| Genotyping | ERIC | ATGTAAGCTCCTGGGGATTCAC | AAGTAAGTGACTGGGGTGAGCG | / | 52 | [46] |
| (GTG)5 | GTGGTGGTGGTGGTG | / | / | 45 | [47] | |
| BOX | CTACGGCAAGGCGACGCTGACG | / | / | 53 | [47] |
| Isolate Code |
Proteomic Identification
(MALDI-TOF MS) | MALDI-TOF Score |
Molecular Identification
(16S rRNA Gene Sequencing) | Similarity Rate (%) * |
Closest Phylogenetic Relative
(GenBank Accession Number) ** |
|---|---|---|---|---|---|
| G4 | Brevibacillus aydinogluensis | 2.23 | Brevibacillus aydinogluensis | 99.32% | Brevibacillus aydinogluensis (NR_117986.1) |
| G5 | Brevibacillus aydinogluensis | 2.28 | Brevibacillus aydinogluensis | 99.32% | Brevibacillus aydinogluensis (NR_117986.1) |
| G6 | Brevibacillus aydinogluensis | 2.24 | Brevibacillus aydinogluensis | 98.64% | Brevibacillus aydinogluensis (NR_117986.1) |
| G10 | Geobacillus kaustophilus | 2.32 | Geobacillus kaustophilus | 98.49% | Geobacillus kaustophilus (NR_115285.2) |
| G11 | Geobacillus kaustophilus | 1.98 | Geobacillus proteiniphilus | 100% | Geobacillus proteiniphilus (NR_178413.1) |
| G13 | Geobacillus stearothermophilus | 2.07 | Geobacillus stearothermophilus | 99.40% | Geobacillus stearothermophilus (NR_115284.2) |
| G14 | Geobacillus stearothermophilus | 2.1 | Geobacillus stearothermophilus | 99.64% | Geobacillus stearothermophilus (NR_115284.2) |
| G30 | Bacillus licheniformis | 2.01 | Bacillus licheniformis | 99.45% | Bacillus licheniformis (NR_118996.1) |
| G50 | Brevibacillus aydinogluensis | 1.85 | Brevibacillus thermoruber | 99.74% | Brevibacillus thermoruber (NR_112213.1) |
| G60 | Aneurinibacillus thermoaerophilus | 2.31 | Aneurinibacillus thermoaerophilus | 99.59% | Aneurinibacillus thermoaerophilus (NR_112216.1) |
| B3 | Bacillus licheniformis | 1.93 | Bacillus paralicheniformis | 100% | Bacillus paralicheniformis (NR_137421.1) |
| B5 | Geobacillus kaustophilus | 2.14 | Geobacillus kaustophilus | 99.75% | Geobacillus kaustophilus (NR_114089.1) |
| B6 | Geobacillus kaustophilus | 2.23 | Geobacillus kaustophilus | 99.87% | Geobacillus kaustophilus (NR_114089.1) |
| B7 | Geobacillus kaustophilus | 2.13 | Geobacillus kaustophilus | 99.80% | Geobacillus kaustophilus (NR_114089.1) |
| B8 | Geobacillus thermoleovorans | 2.24 | Geobacillus thermoleovorans | 99.62% | Geobacillus thermoleovorans (NR_115286.2) |
| B9 | Geobacillus kaustophilus | 2.01 | Geobacillus kaustophilus | 99.49% | Geobacillus kaustophilus (NR_114089.1) |
| B10 | Geobacillus kaustophilus | 2.37 | Geobacillus kaustophilus | 99.86% | Geobacillus kaustophilus (NR_115285.2) |
| B11 | Geobacillus kaustophilus | 1.97 | Geobacillus proteiniphilus | 100% | Geobacillus proteiniphilus (NR_178413.1) |
| B14 | Geobacillus kaustophilus | 2.04 | Geobacillus kaustophilus | 99.32% | Geobacillus kaustophilus (NR_115285.2) |
| B15 | Geobacillus kaustophilus | 2.23 | Geobacillus kaustophilus | 97.96% | Geobacillus kaustophilus (NR_114089.1) |
| B16 | Geobacillus kaustophilus | 2.14 | Geobacillus kaustophilus | 97.32% | Geobacillus kaustophilus (NR_114089.1) |
| B17 | Geobacillus kaustophilus | 1.83 | Geobacillus proteiniphilus | 100% | Geobacillus proteiniphilus (NR_178413.1) |
| B18 | Geobacillus jurassicus | 1.98 | Geobacillus vulcani | 99.55% | Geobacillus vulcani (NR_025426.1) |
| B20 | Geobacillus kaustophilus | 2.18 | Geobacillus proteiniphilus | 99.88% | Geobacillus proteiniphilus (NR_178413.1) |
| B21 | Geobacillus kaustophilus | 2.17 | Geobacillus kaustophilus | 99.40% | Geobacillus kaustophilus (NR_114089.1) |
| B22 | Geobacillus kaustophilus | 2.27 | Geobacillus kaustophilus | 98.92% | Geobacillus kaustophilus (NR_114089.1) |
| B30 | Geobacillus jurassicus | 1.77 | Geobacillus thermoleovorans | 100% | Geobacillus thermoleovorans (NR_115286.2) |
| B40 | Aeribacillus pallidus | 2.15 | Aeribacillus pallidus | 99.68% | Aeribacillus pallidus (NR_026515.1) |
| B50 | Aeribacillus pallidus | 2.39 | Aeribacillus pallidus | 99.84% | Aeribacillus pallidus (NR_026515.1) |
| B70 | Geobacillus kaustophilus | 2.17 | Geobacillus kaustophilus | 99.92% | Geobacillus kaustophilus (NR_115285.2) |
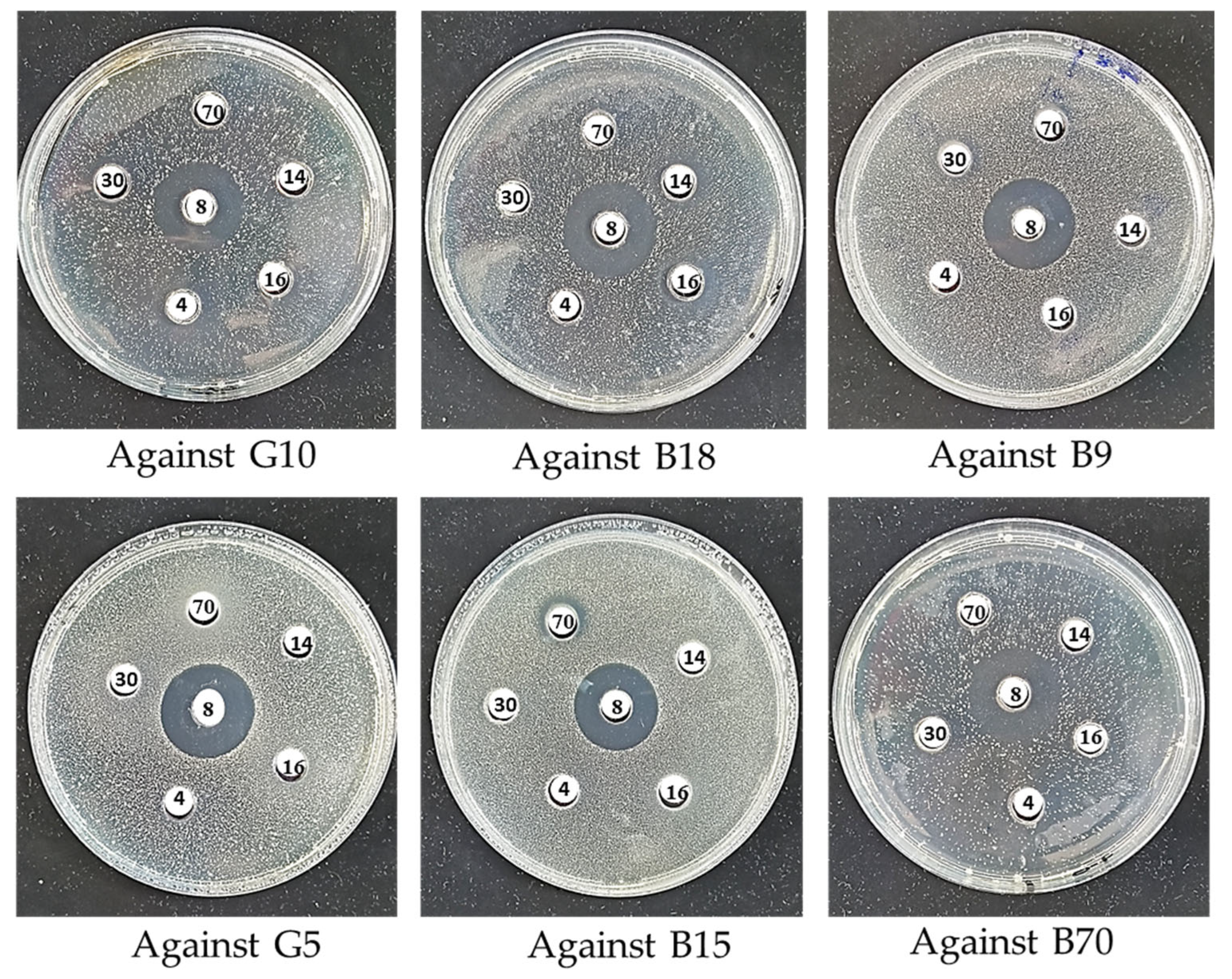
| Target Isolates | Inhibitory Isolates | |||||
|---|---|---|---|---|---|---|
| G4 | B8 | G14 | B16 | B30 | B70 | |
| G10 | 14 | 23 | - | 11 | 11 | - |
| B18 | - | 24 | 14 | 16 | - | - |
| B9 | - | 23 | - | - | 11 | - |
| G5 | - | 23.5 | - | - | - | - |
| B15 | - | 22.5 | - | - | - | 13 |
| B70 | - | 22.5 | - | - | 12 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aireche, M.; Merzoug, M.; Hammadi, A.I.; Zater, Z.Y.; Bendida, K.; Brakna, C.N.; Berrazeg, M.; Aireche, A.Y.; Saidi, Y.; Todorov, S.D.; et al. Taxonomic Diversity and Antimicrobial Potential of Thermophilic Bacteria from Two Extreme Algerian Hot Springs. Microorganisms 2025, 13, 1425. https://doi.org/10.3390/microorganisms13061425
Aireche M, Merzoug M, Hammadi AI, Zater ZY, Bendida K, Brakna CN, Berrazeg M, Aireche AY, Saidi Y, Todorov SD, et al. Taxonomic Diversity and Antimicrobial Potential of Thermophilic Bacteria from Two Extreme Algerian Hot Springs. Microorganisms. 2025; 13(6):1425. https://doi.org/10.3390/microorganisms13061425
Chicago/Turabian StyleAireche, Marwa, Mohamed Merzoug, Amaria Ilhem Hammadi, Zohra Yasmine Zater, Keltoum Bendida, Chaimaa Naila Brakna, Meryem Berrazeg, Ahmed Yassine Aireche, Yasmine Saidi, Svetoslav Dimitrov Todorov, and et al. 2025. "Taxonomic Diversity and Antimicrobial Potential of Thermophilic Bacteria from Two Extreme Algerian Hot Springs" Microorganisms 13, no. 6: 1425. https://doi.org/10.3390/microorganisms13061425
APA StyleAireche, M., Merzoug, M., Hammadi, A. I., Zater, Z. Y., Bendida, K., Brakna, C. N., Berrazeg, M., Aireche, A. Y., Saidi, Y., Todorov, S. D., Arabet, D., & Saidi, D. (2025). Taxonomic Diversity and Antimicrobial Potential of Thermophilic Bacteria from Two Extreme Algerian Hot Springs. Microorganisms, 13(6), 1425. https://doi.org/10.3390/microorganisms13061425







