Abstract
Here, we report the characterization of two Cupriavidus strains, NOV2-1 and OV2-1, isolated from an iron-oxide deposit in an underground tunnel of the Naica mine in Mexico. This unique biotope, characterized by its high temperature (≈50 °C) and the presence of heavy metals, is no longer available for sampling at this time. The genomes of NOV2-1 and OV2-1 comprised two replicons: a chromosome of 3.58 and 3.53 Mb, respectively, and a chromid of 2.1 Mb in both strains. No plasmids were found. The average nucleotide identity and the core genome phylogeny showed that NOV2-1 and OV2-1 belonged to the Cupriavidus gilardii species. NOV2-1 and OV2-1 grew up to 48 °C, with an optimal temperature of 42 °C. Discrete differences were observed between C. gilardii CCUG38401T, NOV2-1, and OV2-1 in the biochemical tests. NOV2-1 and OV2-1 presented resistance to zinc, lead, copper, cadmium, nickel, and cobalt. Several complete and incomplete gene clusters related to the resistance to these heavy metals (ars, czc, cop 1, sil-cop 2, cup, mmf, and mer) were detected in the genome of these strains. Although further studies are needed to determine the origin and role of the detected gene clusters, it is suggested that the czc system may have been mobilized by horizontal gene transfer. This study expands the extreme biotopes where Cupriavidus strains can be retrieved.
1. Introduction
The Naica mine, located in the state of Chihuahua in the northwestern part of Mexico, is globally renowned for its extraordinary geological formations, particularly the Cave of the Crystals, which harbors the world’s largest gypsum crystals [1]. In addition to being a natural geological wonder, Naica was one of the most important lead and zinc mines in Mexico before it closed its operations for an indefinite time in 2015 due to a flood [2]. At present, the mine and caves are inaccessible. Naica’s water originates from magmatic bodies as well as meteoric infiltrations, and the water that springs in the mine galleries has a temperature between 50 and 60 °C and an almost neutral pH [3]. This water has been classified as calcium–sulfate type and has a low NaCl content and significant amounts of metals, mainly zinc (199.3 μg/L) and lead (78.9 μg/L) [4]. Additionally, high concentrations of arsenic (up to 156.54 ppb) have been detected in waters of the Meoqui–Delicias aquifer, which, given its geographical positioning, is a recharge source of Naica’s water [5].
Since the discovery of the Cave of the Crystals in 2000, research conducted on Naica and its caves has predominantly been centered on the genesis of the crystals and other mineralogical, microclimatological, and paleoenvironmental aspects [1,3,6]. Although it has been suggested that microorganisms present in the Naica thermal aquifer might have participated in crystal genesis [7], only three microbiological studies on Naica have been reported in the literature [8,9,10]. This singular subsurface biotope, combining hot conditions and the presence of heavy metal, is of particular interest both for basic studies on microbial adaptation to extreme environments and for potential environmental applications of the microorganisms residing there.
To predict the type of microorganisms that might have participated in the formation of the giant crystals, [10] studied the microbial diversity in subsurface hydrothermal water springing at 60 °C in the deepest mine galleries (−700/760 m) by cloning and sequencing 16S rRNA amplicons obtained by nested PCR. Microorganisms belonging to the Thaumarchaeota, Euryarchaeota, Betaproteobacteria, Candidate Division OP3 bacteria, Firmicutes, and Alphaproteobacteria were detected in this study. According to these authors, all the retrieved OTUs were autochthonous except a Betaproteobacteria related to a Delftia strain from a wastewater plant and Alcaligenes denitrificans, an opportunistic pathogen. On their part, [9] have isolated Actinobacteria from the crystals and walls of the cave of the Crystals. The phenotypic tests and 16S rRNA gene sequencing data separated these isolates into two subgroups closely related to the Prauserella genus (Pseudonocardiaceae family). Finally, we studied the bacteria associated with minerals and hot water springs from caves and tunnels of the Naica mine using culture-dependent and culture-independent (DGGE) approaches. This study revealed the presence of Firmicutes, Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria in gypsum crystals, iron oxide crusts, and hot springs [8]. These bacteria were likely autochthonous with some allochtonous components due to human intervention. Isolated bacteria included Bacillus, Brevibacillus, Paenibacillus, Schlegelella, Cupriavidus, Pseudoxanthomonas, and Lysobacter.
The study of microorganisms previously isolated in Naica is an opportunity to gather more information on how bacteria adapt to multi-extreme environments. Here, we report the physiological and genomic characterization of two Cupriavidus gilardii strains, NOV2-1 and OV2-1, previously isolated from an iron oxide crust within the Naica mine by our group [8,11]. We show that these strains exhibit both temperature and metal tolerance. The genus Cupriavidus, “lover of copper” belongs to the Betaproteobacteria class, family Burkholderiaceae and comprises a group of Gram-negative, peritrichously flagellated aerobic rods with chemoheterotrophic or chemolithotrophic metabolism, that do not assimilate glucose and can use several amino acids as sole carbon and nitrogen sources [12,13,14]. This genus presently encompasses 20 described species [15], which have been isolated in many places around the world from a variety of natural and anthropogenic environments as soil, water, wastewater, human clinical samples, Agave rhizosphere, Mimosa root nodules, volcanic mudflow, natural asphalt deposit and International Space station [13,16,17,18]. The Cupriavidus metallidurans strain CH34, isolated from a metallurgical plant in Belgium in the 70 s, is the model system for mesophilic bacterial HMR [19]. Related strains have been found in different industrial sites and even in clinical samples [16]. Recently, the C. gilardii species has also gained attention for its HMR and as a potential indicator of heavy metals contamination in tropical environments [18,20]. The present work expands the habitats where Cupriavidus species, such as C. gilardii can be retrieved and again highlights the ability of this genus to adapt and survive under harsh conditions as those found in the singular Naica biotope.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
The Cupriavidus reference strains used in this work, Cupriavidus metallidurans LMG1195/CH34, Cupriavidus necator LMG8453/N-1, Cupriavidus taiwanensis LMG19424, and Cupriavidus gilardii CCUG38401T, were obtained from the ATCC [21,22,23]. The Cupriavidus isolates NOV2-1 and OV2-1 had been previously isolated from a semi-soft iron oxide deposit in a tunnel outside the Cueva de las Velas at a depth of −290 m in the Naica mine and a temperature around 44 °C, as described in [8,11]. The strains were routinely cultured in nutrient broth (BD Bioxon, Becton Dickinson, Mexico) under orbital agitation (200 rpm), at 30 °C for C. metallidurans CH34, C. necator LMG1199, and C. taiwanensis LMG19424 or 42 °C for NOV2-1, OV2-1, and C. gilardii CCUG38401T. The strains were conserved on nutrient broth agar plates at 4 °C for short-term storage, or in glycerol at −80 °C for long-term storage.
2.2. DNA Extraction, Genome Sequencing, Assembly and Annotation
The NOV2-1 and OV2-1 genomic DNA was extracted and purified using the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA) following the manufacturer’s instructions. The quality and concentration of the DNA were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and further confirmed by agarose gel electrophoresis. Whole-genome sequencing using the PacBio Sequel and Illumina HiSeq 4000 platform was performed at Macrogen (Seoul, Republic of Korea). The Illumina sequencing reads were subjected to quality control with FastQC v0.11.8 [24]. Read trimming was performed with Trim Galore v0.6.1 [25]. Hybrid genome assemblies were performed with SPAdes v3.12.0 [26,27] and Unicycler v0.4.8. The obtained assemblies were then merged and optimized with Metassembler v1.5 [28,29]. Genome completeness was determined using CheckM (v1.2.2) [29]. The final consensus assembly was subjected to gene prediction and functional annotation using Prokka v1.12 [30] and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [31] available at “https://www.ncbi.nlm.nih.gov/refseq/annotation_prok/” (accessed on 10 January 2023).
2.3. Physiological Characterization
The effect of temperature on the growth of NOV2-1 and OV2-1 was first estimated by spotting overnight liquid cultures on nutrient agar plates and incubating them at 30, 37, 42, 48, and 50 °C for 24 h. The effect of temperature on the maximum growth rate (μmax) was then evaluated in nutrient broth at 35, 37, 40, 42, 45, and 48 °C. For this, overnight cultures grown at 42 °C were used to inoculate 50 mL of fresh medium at an initial optical density at 600 nm (OD600) of 0.1 in 125 mL Erlenmeyer flasks. The cultures were grown at 200 rpm for 9 h, and samples were taken every hour to measure the OD600 using a BioPhotometer Plus (Eppendorf, Hamburg, Germany) instrument. The μmax at each temperature tested was calculated by linear regression of the data during exponential growth. Biochemical analyses were performed by the VITEK 2 automated system according to the manufacturer’s instructions using Gram-negative bacterium identification cards.
2.4. Evaluation of Heavy Metals Tolerance
The NOV2-1 and OV2-1 strains and the Cupriavidus reference strains were grown overnight in nutrient broth at 42 °C and 30 °C, respectively, with orbital shaking at 200 rpm. The cultures were then washed twice with M9 minimal salts (Sigma-Aldrich, Burlington, MA, USA) 1× and adjusted to an OD600 of 0.2 into M9 minimal salts 1×, serially diluted (10−1–10−5), and spotted (15 μL) onto Tris-buffered LB Lennox agar, with or without metals in square Petri dishes. (The medium was buffered with Tris to maintain the pH neutrality for the metal tolerance experiments). The Tris-buffered LB Lennox agar composition was as follows (for 1 L): tryptone (10 g), yeast extract (5 g), tris-base (6.06 g), and bacteriological agar (15 g). The pH was adjusted to 7.1. Heavy metals stock solutions (CuCl2, CoCl2 6H2O, CdCl2, Pb(N2O6), ZnCl2, and NiCl2) were prepared in Milli-Q water and filter-sterilized. These stock solutions were added to the solid medium to achieve final concentrations of 0.5 to 9 mM.
2.5. Comparative Genomics Analyses
A total of 39 Cupriavidus, 4 Burkholderia, and 2 Ralstonia complete genomes were downloaded from the NCBI GenBank repository (Table S1). The genome sequences of C. gilardii NOV2-1 and OV2-1 can be accessed under the GenBank accession numbers CP083437.1, CP083438.1, CP083735.1, and CP083736.1. The average nucleotide identity (ANI) was evaluated with PYANI v0.2 (average_nucleotide_adeintity.py) [32]. The core genome was obtained with GET_HOMOLOGUES (v11042019) [33]. The orthologous gene clusters produced by GET_HOMOLOGUES were then processed with GET_PHYLOMARKERS (v2.2.5_9May18) [34] to select optimal phylogenetic markers. The phylogenomic reconstruction was performed with RaxML v8.2.10 using the GTR-GAMMA substitution model with 100 bootstrap replicates [35]. The Burkholderia and Ralstonia genera were used as an outgroup for phylogenetic analysis. The presence of HMR gene clusters in the NOV2-1, OV2-1, and other Cupriavidus gilardii strains genomes used for comparison was investigated by using a bidirectional best hit (BBH) approach. Several HMR gene clusters (ars, cup, cop 1, czc, mer, mmf, cop 2, pbr, cnr, and ncc) responsive to the metals studied here have been identified in the genome of the multi-metal-resistant model bacterium C. metallidurans CH34 [36,37]. The proteins encoded by these gene clusters were submitted to reciprocal pairwise sequence comparisons between the predicted protein-coding genes of NOV2-1 and OV2-1, C. gilardii genomes, and C. metallidurans CH34 using BLASTp. Proteins were considered reciprocal best hits and likely orthologs when they presented amino acid identities > 80%, query cover > 80%, e-value near 0, and were part of an operon structure. The pbr, cnr, and ncc clusters, found to be absent in NOV2-1 and OV2-1, were not further investigated in other C. gilardii genomes.
3. Results
3.1. General Properties of the NOV2-1 and OV2-1 Genomes
Table 1 presents the general properties of the NOV2-1 and OV2-1 genomes. The total genome sizes of NOV2-1 and OV2-1 were 5.7 and 5.6 Mb, respectively. The estimated genome completeness was 99.89 and 99.66%, with an estimated contamination of 0.26 and 0.23% for NOV2-1 and OV2-1, respectively, indicating high-quality genomes. The GC contents were 67.5% in both strains. The genomes consisted of two chromosomes of 3.6 Mb and 2.1 Mb for NOV2-1 and, 3.5 Mb and 2.1 Mb for OV2-1. A total of 5024 and 5011 putative coding sequences (CDS) were predicted in NOV2-1 and OV2-1, of which 4095 and 4064 were assigned a function, respectively. Twelve ribosomal RNAs (rRNAs) and 59 transfer RNAs (tRNAs) were predicted in both genomes. Of the rRNAs and tRNAs found in both strains, 9 and 52 were in the largest replicon, respectively.

Table 1.
General genomic features of NOV2-1 and OV2-1.
3.2. Phylogenetic Characterization
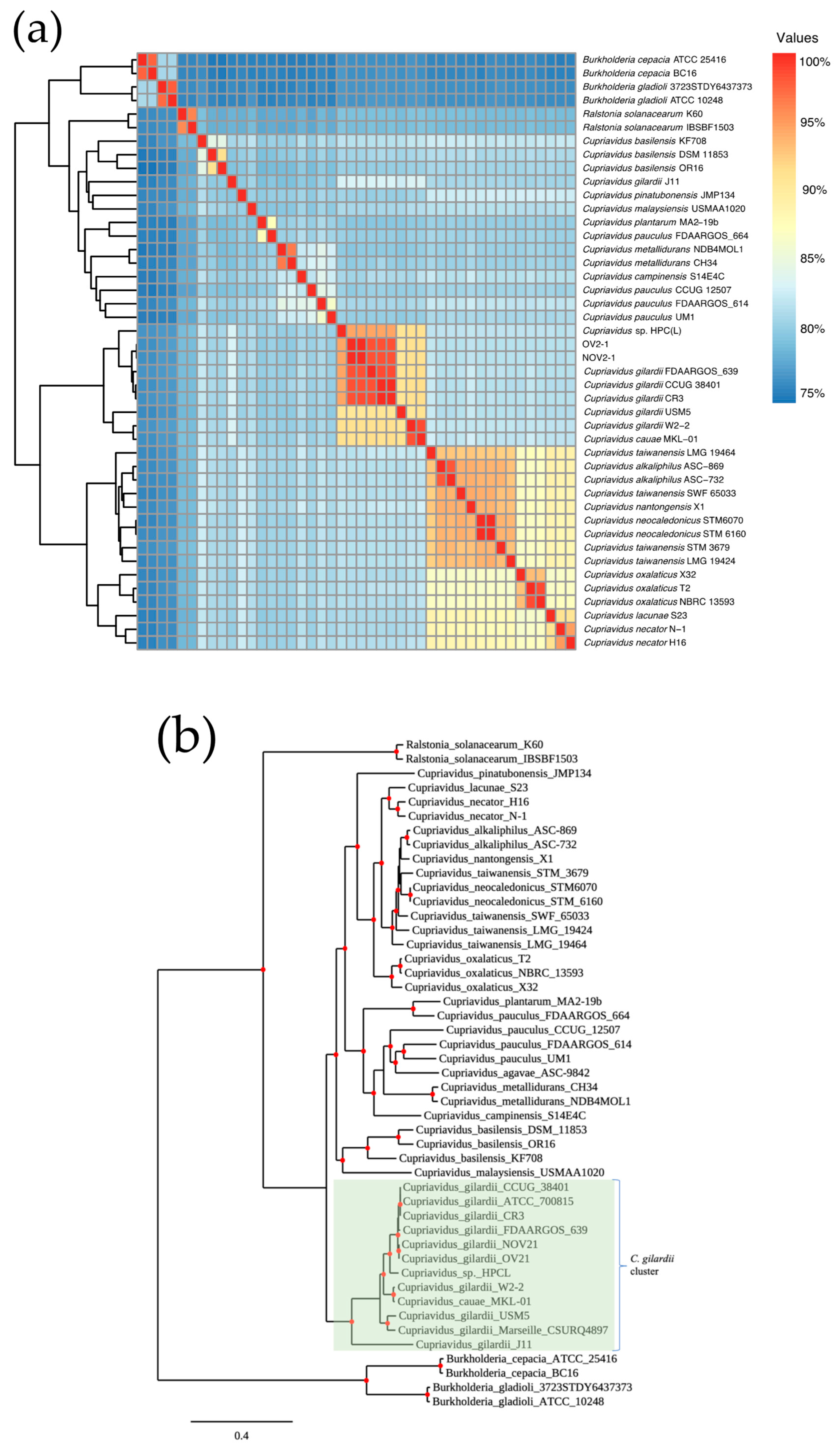
The genomes of NOV2-1 and OV2-1, 36 other Cupriavidus strains from 12 different species, and two unclassified Cupriavidus isolates with available whole genome sequences were analyzed by ANI (Figure 1a and Table S2). The ANI values between NOV2-1 and C. gilardii CCUG 38401T, C. gilardii FDAARGOS_639 (reference genome), and C. gilardii CR3 were 98.31, 98.45, and 98.48%, respectively. For OV2-1, the ANI values were 98.29, 98.42, and 98.45%, respectively. The pangenome analysis based on 38 Cupriavidus genomes and the NOV 2-1 and OV 2-1 genomes allowed for the retrieval of a final set of 510 orthologous genes as phylogenetic markers. The concatenated alignment of these genes was then used to build a phylogenetic tree (Figure 1b) where it can be observed that NOV 2-1 and OV 2-1 grouped with the C. gilardii branch. So, according to ANI percentages and the phylogeny, these strains will be referred to as C. gilardii NOV2-1 and C. gilardii OV2-1. Additionally, the phylogenetic tree also showed that C. gilardii J11 branched separately from C. gilardii and that C. taiwanensis STM 3679 and C. pauculus FDAARGOS 664 should be reclassified.
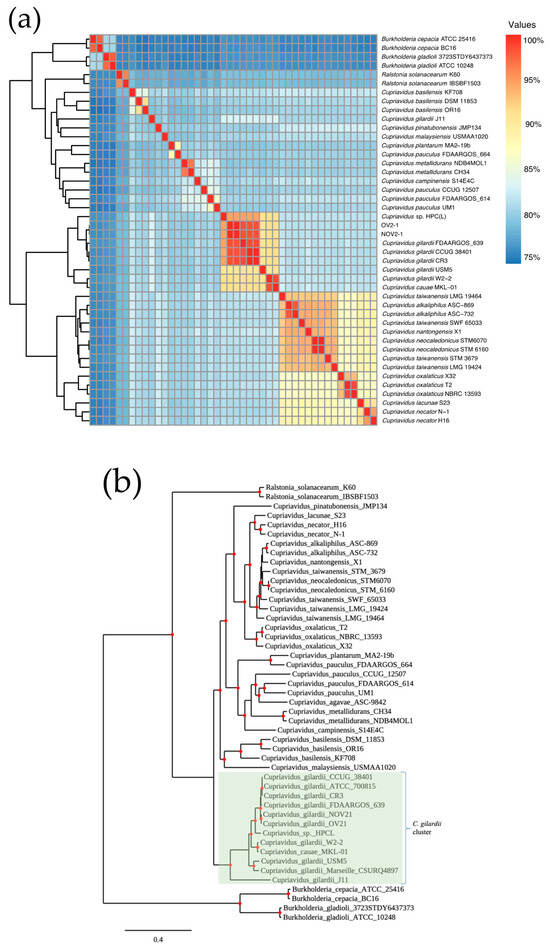
Figure 1.
Pairwise genomic ANI values and maximum-likelihood phylogeny of NOV2-1, OV2-1, and other Cupriavidus strains. (a) Heatmap of ANI. (b) Phylogeny generated from 510 concatenated core genes. The Burkholderia and Ralstonia strains were used as outgroups to root the tree. The scale bar represents the number of substitutions per site.
3.3. Physiological Characteristics of NOV2-1 and OV2-1
NOV2-1 and OV2-1 are fast-growing, Gram-negative, motile, rod-shaped bacteria that form creamy, slightly domed, and slightly mucoid colonies within 1–2 days when grown on nutrient agar at 42 °C. Table 2 presents a summary of the physiological characteristics of strains NOV2-1 and the reference strains used here. As C. gilardii CCUG38401T, NOV2-1 and OV2-1 could grow between 30 and 48 °C and no growth was observed at 50 °C while C. necator LMG8453 and C. taiwanensis LMG19424 only grew up to 37 °C and C. metallidurans CH34 up to 30 °C (Figure S1). The two strains had an optimal growth rate between 42 and 45 °C (Figure S2). Growth was better without NaCl in the medium, and growth was reduced by half when 1% (p/v) of NaCl was added. Concerning pH, both strains exhibited optimal growth at pH 7–8; however, OV2-1 displayed a broader pH tolerance (5.5–9.9) compared to NOV2-1 (7–9.9).

Table 2.
Summary of physiological characteristics of strains NOV2-1 and OV2-1 compared to the Cupriavidus reference strains used in this work. The complete results of the 47 biochemical tests included in the Vitek 2 Gram-negative identification card are shown in Table S4.
The biochemical tests performed on NOV2-1, OV2-1, and the reference strains (C. metallidurans CH34, C. taiwanensis LMG19424, and C. gilardii CCUG38401T) (Table S3) showed that all were positive to L-proline acrylamidase, L-lactate alkalinization, succinate alkalinization, and tyrosine arylamidase. NOV2-1 and OV2-1 were negative for D-glucose and all the other monosaccharides, disaccharides, and sugar alcohols included in the test, as the reference strains. Differences between NOV2-1, OV2-1, and C. gilardii CCUG38401T were observed. For example, C. gilardii CCUG38401T and all the other reference strains were positive for ELLMAN, while both NOV2-1 and OV2-1 were negative for this test. For L-malate and L-lactate assimilation NOV2-1 and OV2-1 were positive as C. necator N-1 and C. taiwanensis LMG19424, whereas C. gilardii CCUG38401T was negative. OV 2-1, C. gilardii CCUG38401T and C. metallidurans CH34 gave a positive result for glutamyl arylamidase pNA, whereas NOV2-1 produced a negative result. OV2-1 was positive for citrate (sodium) utilization as C. necator N-1, C. metallidurans CH34, and C. taiwanensis LMG19424, whereas NOV2-1 and C. gilardii CCUG38401T were negative.
3.4. Tolerance to Heavy Metals
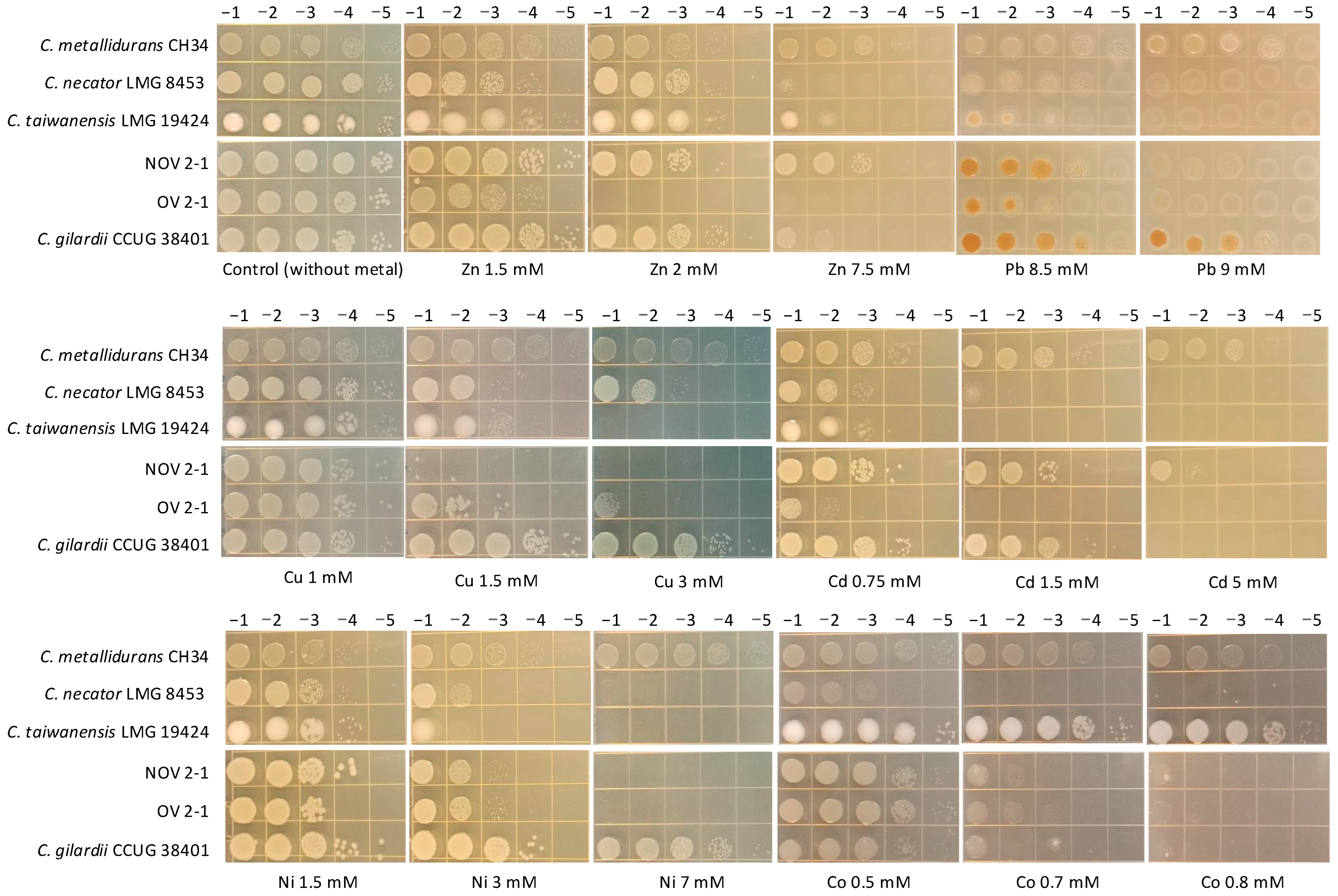
As NOV2-1 and OV2-1 were isolated from a mine, their tolerance to heavy metals was tested. The multi-metal-resistant bacterial model C. metallidurans CH34 and the other Cupriavidus reference strains were included for comparison. NOV2-1, OV2-1, and C. gilardii CCUG38401T were tested at 42 °C, and the other reference strains at 30 °C. The results are presented in Figure 2. For all the metals tested, C. metallidurans CH34 was the highest tolerant strain, as expected. Differences in metal tolerance were observed between the C. gilardii CCUG38401T, NOV2-1, and OV2-1.
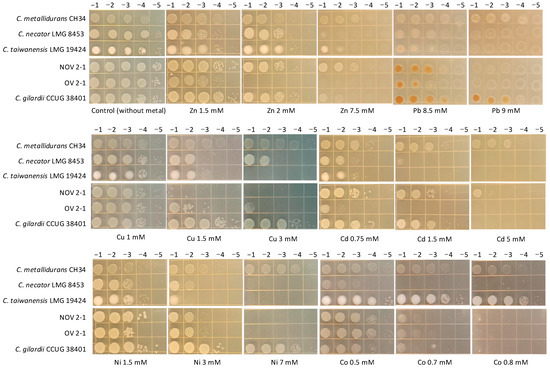
Figure 2.
Heavy metals tolerance profiles of NOV 2-1, OV 2-1, and Cupriavidus reference strains through agar drop plate method. Serial dilutions (top) of overnight cultures were plated on the Tris-buffered LB Lennox agar amended with increasing concentrations of heavy metals (base of the figures).
Concerning Zn, C. metallidurans CH34, NOV2-1, C. taiwanensis LMG19424, and C. gilardii CCUG38401T had the highest tolerance (7.5 mM), followed by C. necator LMG8453 (2 mM), and OV2-1 (1.5 mM). Regarding Pb, NOV2-1, OV2-1, C. taiwanensis LMG19424, and C. necator LMG8453 had similar tolerance levels (8.5 mM). C. gilardii CCUG38401T could grow up to 9 mM of this metal as C. metallidurans CH34. In the C. gilardii strains (NOV2-1, OV2-1, and CCUG38401T) and, to some extent, in C. taiwanensis LMG19424, bacterial tolerance to Pb was characterized by brown colonies, indicating lead precipitation [38]. For Cu, NOV2-1 did not grow at the lowest concentration tested (1.5 mM), while OV2-1 tolerated up to 3 mM, as C. metallidurans CH34 and C. gilardii CCUG38401T, which both showed full growth at this concentration. C. taiwanensis LMG19424 had an intermediate tolerance (1.5 mM of Cu). In the case of Cd, NOV2-1 presented a high tolerance (5 mM), similar to that of C. metallidurans CH34, while C. gilardii CCUG38401T had a lower tolerance (1.5 mM), followed by C. necator LMG 8453, C. taiwanensis LMG19424, and OV2-1 (0.75 mM). OV2-1 presented poor growth at this Cd concentration. C. metallidurans CH34, and C. gilardii CCUG38401T showed full growth up to 7 mM of Ni while the other strains, including NOV2-1 and OV2-1, only tolerated 3 mM of this metal. C. necator LMG8453 and C. taiwanensis LMG19424 had poor growth at this Ni concentration. Finally, C. metallidurans CH34 and C. taiwanensis LMG19424 were the most Co tolerant strains (0.8 mM) followed by C. gilardii CCUG38401T, NOV2-1, OV2-1, and C. necator LMG8453 which poorly grew at 0.5 mM of Co.
3.5. Genomic Analysis of HMR Gene Clusters
3.5.1. ars Cluster
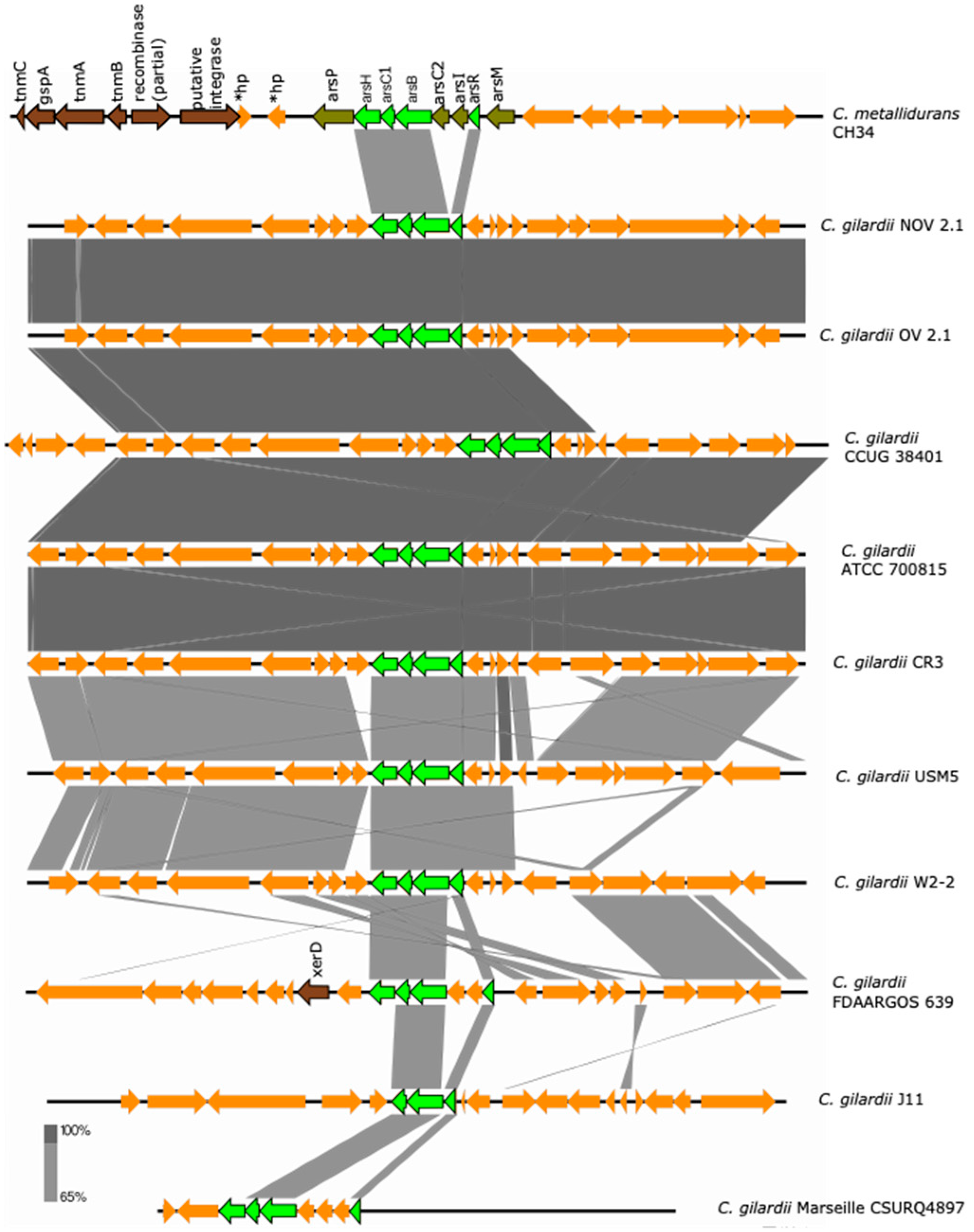
The ars cluster is encoded on chromosome 1 of C. metallidurans CH34 and consists of eight genes (asrP: MFS permease; asrH: NADPH-dependent FMN reductase; arsC1: arsenate reductase; arsB: arsenite efflux pump; asrC2: arsenate reductase; arsI: lactoylglutathione lyase; arsR: transcriptional regulator; asrM: S-adenosyl-L-methionine-dependent methyltransferase) which are maximally activated by the presence of As ions and partially upregulated in the presence of Pb, Zn, Co, Se, Cd and Cu ions [36,39]. An incomplete ars cluster was found in chromosome 1 of NOV2-1 and OV2-1 and in all the other C. gilardii strains analyzed here (Figure 3).
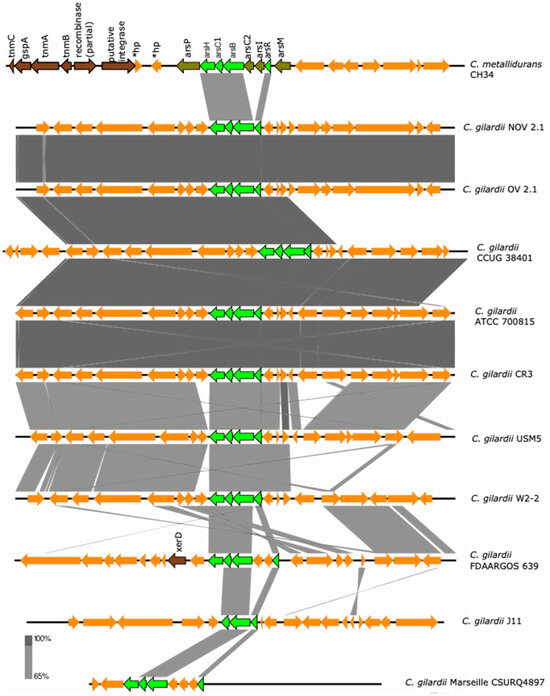
Figure 3.
Map showing the genomic organization of the ars cluster in C. metallidurans CH34, NOV2-1, OV2-1, and other C. gilardii strains. The conserved and non-conserved genes of the cluster are colored in neon and olive green, respectively. Orange arrows indicate neighboring genes. The genes colored in brown indicate parts of horizontal DNA transfer-related sequences. *hp means hypothetical protein. The gray scale indicates the levels of synteny.
The asrH, asrC1, asrB, and asrR were the conserved genes (except in strain J11 where the arsH gene was missing). The arsRBC1H proteins of NOV2-1 and OV2-1 displayed 83.2–87.5% amino acids identity to those of C. metallidurans CH34 (Table S4). The horizontal DNA transfer-related sequences present in C. metallidurans CH34 near the ars cluster were not found in C. gilardii (Figure S3).
3.5.2. czc Cluster
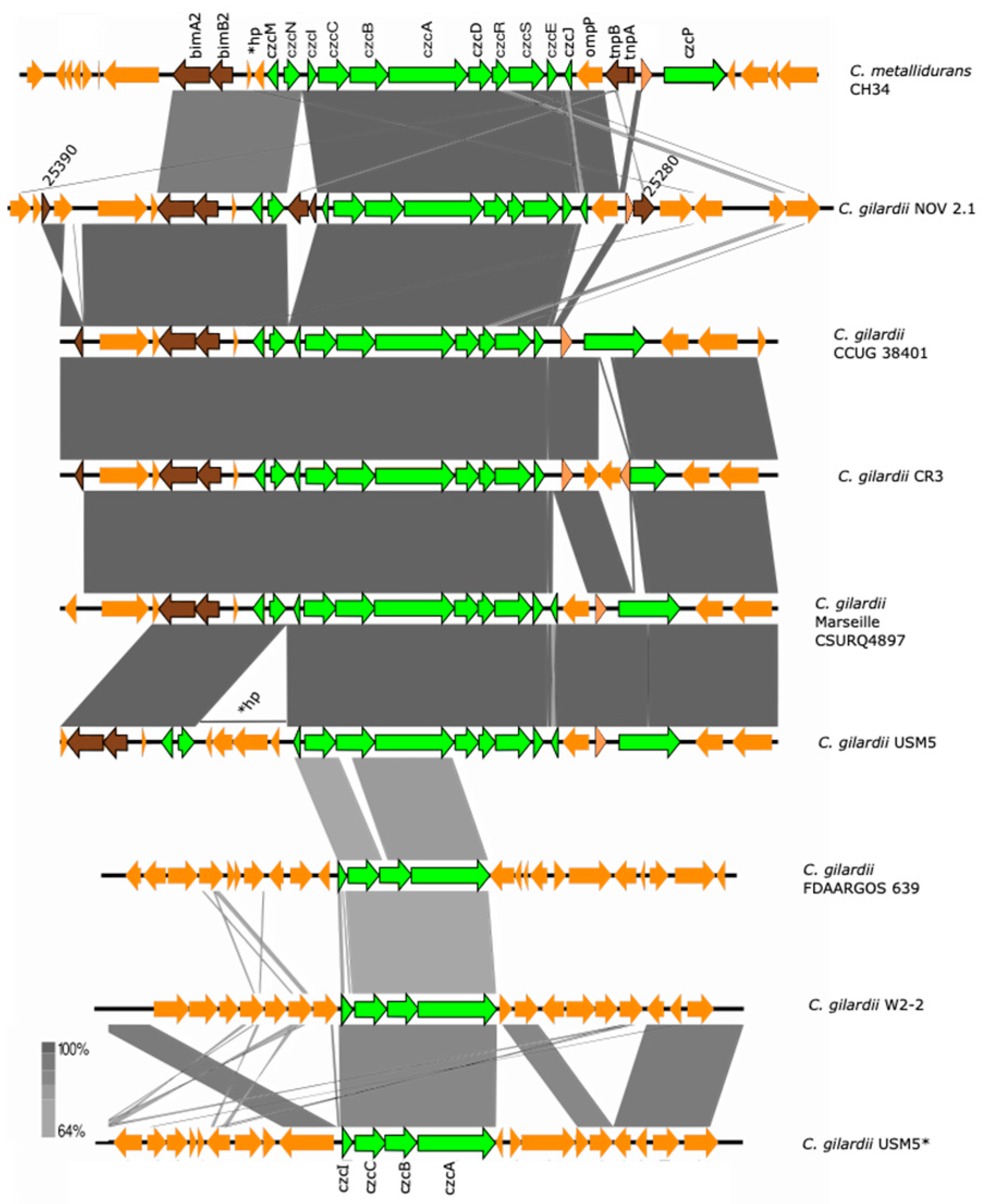
C. metallidurans CH34 has two copies of the czc cluster. The complete and functional czc cluster is encoded on the pMOL30 plasmid and is composed of eleven genes (czcM, MgtC-like Mg(II) transport ATPase; czcN, isoprenylcysteine carboxyl methyltransferase (regulation); czcI, cobalt–zinc–cadmium resistance protein/cation efflux system protein; czcC, outer membrane protein/three components cation proton antiporter efflux system; czcB, membrane fusion protein/three components cation proton antiporter efflux system; czcA, efflux chemiosmotic pump/three components cation proton antiporter efflux system; czcD, cation diffusion facilitator; czcR, two-component regulator; czcS, sensor histidine kinase of the two-component regulatory system; czcE, ion binding protein; czcJ, hypothetical protein; czcP, cation efflux P1-ATPase). This cluster is mainly responsive to Cd, Zn, and Co, although some responses to Pb and Cu have also been detected [36,39].
The complete czc cluster was found in C. gilardii CCUG38401T, CR3, and Marseille CSURQ4897 (Figure 4). An incomplete czc cluster missing the czcP gene and containing an insertion sequence between czcN and czcI was found in chromosome 1 of NOV2-1, while this cluster was completely absent in OV2-1. Only the czcICBA region was found in C. gilardii FDAARGOS 639, W2-2 and USM5. The czc proteins of NOV2-1 had 98.6–100% amino acid identity with those of C. metallidurans CH34 (Table S5). In NOV2-1, the horizontal DNA transfer-related sequences on the right side of the cluster were not conserved to C. metallidurans CH34 (Figure S4).
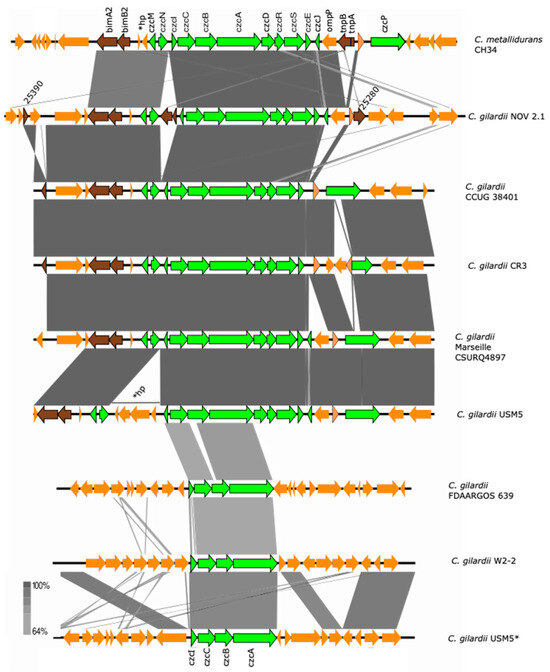
Figure 4.
Map showing the genomic organization of the czc cluster in C. metallidurans CH34, NOV2-1, OV2-1, and other C. gilardii strains. The conserved and non-conserved genes of the cluster are colored in neon and olive green, respectively. Orange arrows indicate neighboring genes. The genes colored in brown indicate parts of horizontal DNA transfer-related sequences. *hp means hypothetical protein. The gray scale indicates the levels of synteny.
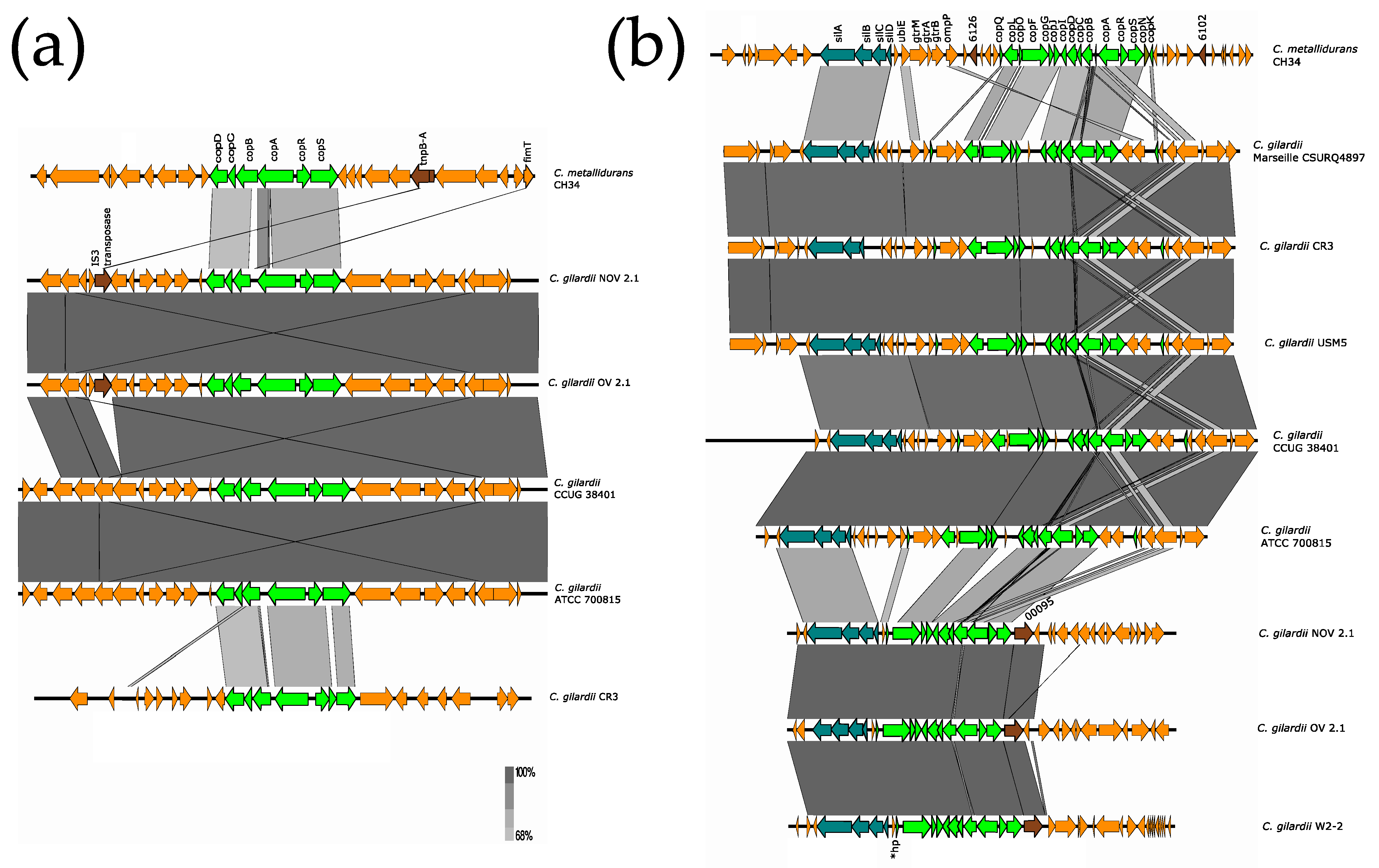
3.5.3. cop Cluster I
The cop cluster 1, encoded on chromosome 2 (chromid) of C. metallidurans CH34, is constituted by six genes (copA, periplasmic multicopper oxidase; copB, periplasmic copper resistance protein B; copC, periplasmic copper resistance C protein precursor; copD, copper resistance protein D transmembrane component; copR, DNA-binding response regulator in two-component regulatory system with CopS; copS, sensory histidine kinase in two-component regulatory system with CopR, senses copper ions). This periplasmic detoxification system appears to be responsive only to Cu ions [36].
NOV2-1 and OV2-1 as well as C. gilardii CCUG 38401, ATTC 700815 and CR3 were found to harbor the complete cop cluster 1 in their chromosome 1 (Figure 5a). This cluster was not detected in the other C. gilardii strains included in the study. The cop proteins of NOV2-1 and OV2-1 had a low amino acid identity to those of C. metallidurans CH34 (from 49.2 to 88.9%) (Table S5). The horizontal DNA transfer sequences at the vicinity of the cop 1 cluster were different in C. gilardii and in C. metallidurans CH34 (Figure S5).
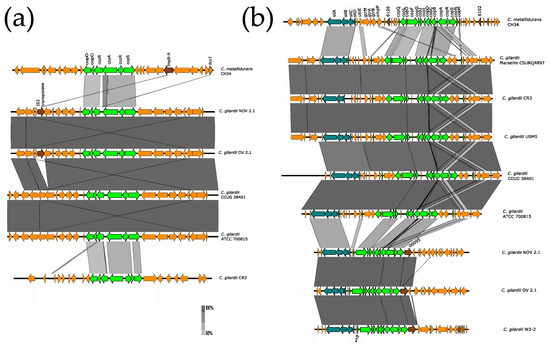
Figure 5.
Map showing the genomic organization of the cop 1 (a) and sil-cop 2 (b) clusters in C. metallidurans CH34, NOV2-1, OV2-1, and other C. gilardii strains. The conserved and non-conserved genes of the cluster are colored in neon and olive green, respectively. Orange arrows indicate neighboring genes. The genes colored in brown indicate parts of horizontal DNA transfer-related sequences. *hp means hypothetical protein. The gray scale indicates the levels of synteny.
3.5.4. sil and cop Cluster 2
The sil and cop cluster 2 are coded contiguously on the pMOL30 plasmid in C. metallidurans CH34.
The sil cluster consists of four genes (silA, proton antiporter metal efflux system; silB, proton antiporter metal efflux system; silC, outer membrane silver efflux system; silD, transmembrane protein). The sil cluster is mainly responsive to Ag, and only the silA gene is upregulated in the presence of Cu [36].
The cop cluster 2 is constituted by the following twenty-one genes: copW (hypothetical protein), copE (copper resistance protein), copH (copper-binding protein), copQ (copper protein), copL (type II enzyme restriction), copO (unknown function), copF (P-type ATPase efflux), copG (thioredoxin-like protein), copJ (putative cytochrome c), copI (putative oxido-reductase protein), copD1 (copper resistance protein D), copC1 (Cu2+-binding protein), copB1 (outer membrane protein), copA1 (multi-copper oxidase protein), copR1 (two-component transcriptional regulator), copS1 (sensor histidine kinase/two-component regulatory system), copN (copper protein), copK (Cu+ binding protein), copM (copper resistance protein), copT (putative cytochrome), and copV (copper resistance protein). The cop cluster 2 responds to Cu, Ag, Cd, Ni, Zn, and Co ions [36].
The sil-cop region was present in chromosome 2 (chromid) of NOV2-1 and OV2-1 and in all the C. gilardii genomes analyzed except in FDAARGOS 639 and J11 (Figure 5b). Only ten cop genes (copABCDFGIJRS) were found in C. gilardii CR3, and the silD gene was absent in this strain. The sil and cop gene products of NOV2-1 and OV2-1 had 68–89.5% amino acid identities to those of C. metallidurans CH34 (Table S5). The horizontal DNA transfer sequences at the vicinity of the sil-cop 2 cluster were different in C. gilardii and in C. metallidurans CH34 (Figure S6).
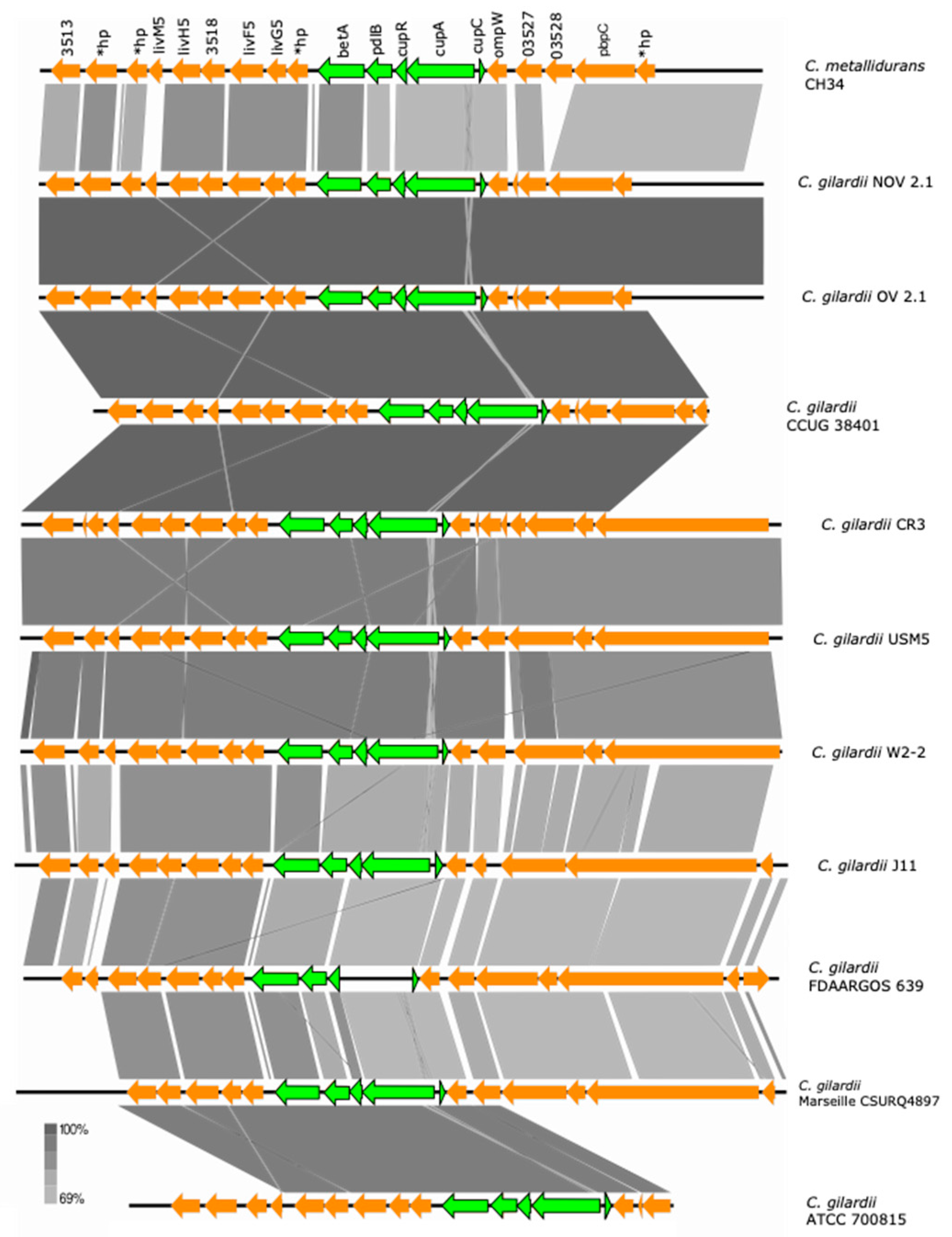
3.5.5. cup Cluster
The cup cluster is located on chromosome 1 of C. metallidurans CH34 and is composed of 3 genes (cupC, copper chaperone, heavy metal ion binding; cupA, P-type ATPase and cupR, DNA-binding transcriptional activator of copper-responsive regulon genes from the MerR family). This cluster exhibits significant upregulation in response to Ag, As, Pb, and Cu ions in C. metallidurans CH34 [36,39]. Two extra genes upstream of cupR, betA2 (encoding a choline dehydrogenase) and pldB (encoding a lysophospholipase) are also thought to belong to the cluster [36].
This cluster and the betA2/pldB adjacent genes were found to be highly conserved in NOV2-1, OV2-1, and other C. gilardii strains except in FDAARGOS639 where the cupA gene was missing (Figure 6). The cup proteins of NOV2-1 and OV2-1 had amino acid identities of 66.4–83.5% to those of C. metallidurans CH34 (Table S5). No horizontal DNA transfer sequences were detected at the proximity of the cup cluster in C. metallidurans CH34 and the C. gilardii strains analyzed here.
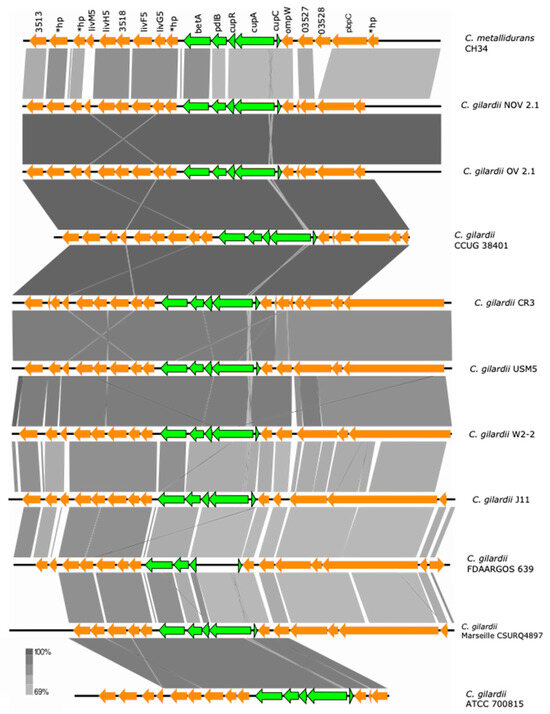
Figure 6.
Map showing the genomic organization of the cup cluster in C. metallidurans CH34, NOV2-1, OV2-1, and other C. gilardii strains. The conserved and non-conserved genes of the cluster are colored in neon and olive green, respectively. Orange arrows indicate neighboring genes. The genes colored in brown indicate parts of horizontal DNA transfer-related sequences. *hp means hypothetical protein. The gray scale indicates the levels of synteny.
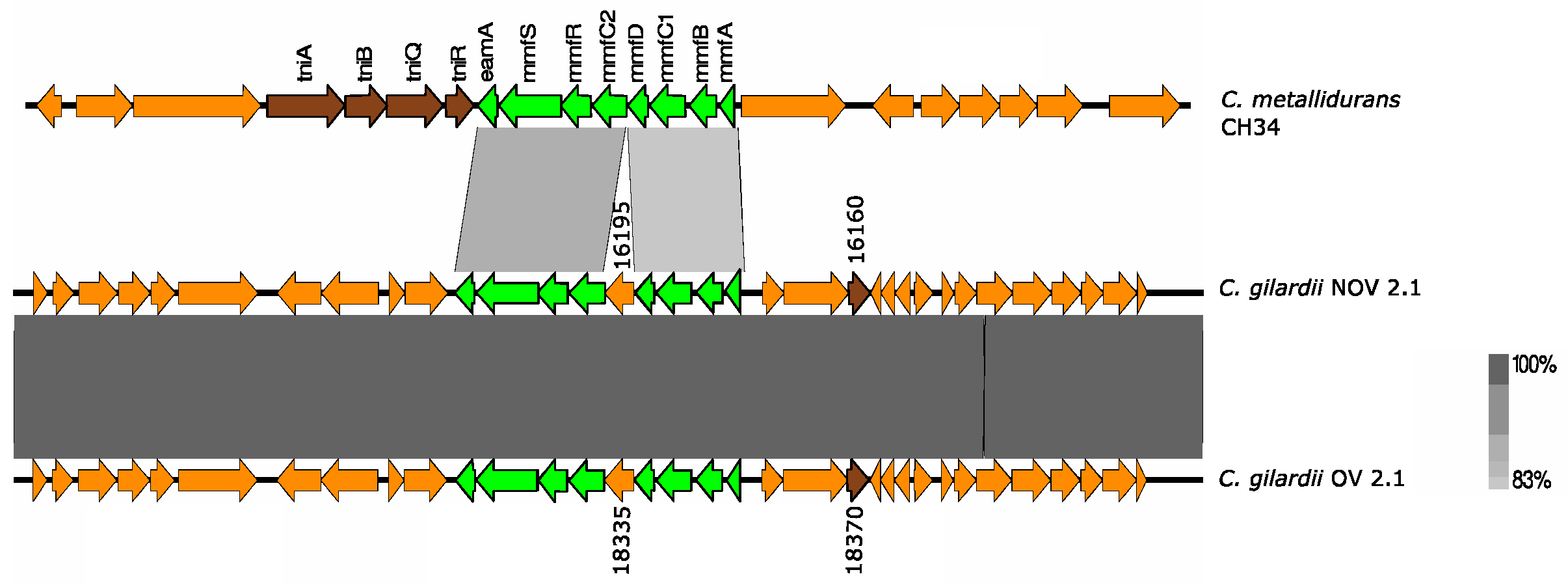
3.5.6. mmf Cluster
C. metallidurans CH34 has three identical copies of the mmf cluster (“multi metal phenotype”), one in chromosome 1 and two in chromosome 2 (chromid). The mmf cluster consists of eight genes contained in the Tn6048 transposon (mmfA: propeptide, PepSY, and peptidase M4 precursor; mmfB: undecaprenyl pyrophosphate phosphatase; mmfC1: putative MFS permease; mmfD: secreted protein; mmfC2: putative MFS permease; mmfR: two-component transcriptional regulator; mmfS: periplasmic sensor signal transduction histidine kinase; eamA: EamA family transporter). This cluster is upregulated in the presence of Pb and Zn [36].
This cluster was only found in NOV2-1 and OV2-1 but not in the other C. gilardii genomes used in this study (Figure 7). The mmf gene products had a higher amino acid sequence identity (74.5–95.6%) than those found on the C. metallidurans CH34 genome (Table S5). The horizontal DNA transfer sequences in the vicinity of the mmf cluster were different in C. gilardii and in C. metallidurans CH34 (Figure S7).
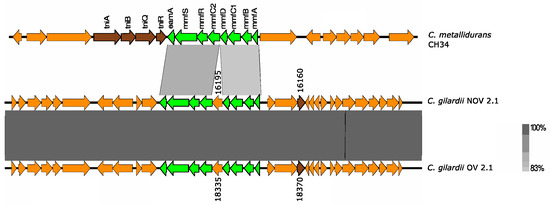
Figure 7.
Map showing the genomic organization of the mmf cluster in C. metallidurans CH34, NOV2-1, OV2-1, and other C. gilardii strains. The conserved and non-conserved genes of the cluster are colored in neon and olive green, respectively. Orange arrows indicate neighboring genes. The genes colored in brown indicate parts of horizontal DNA transfer-related sequences. The gray scale indicates the levels of synteny.
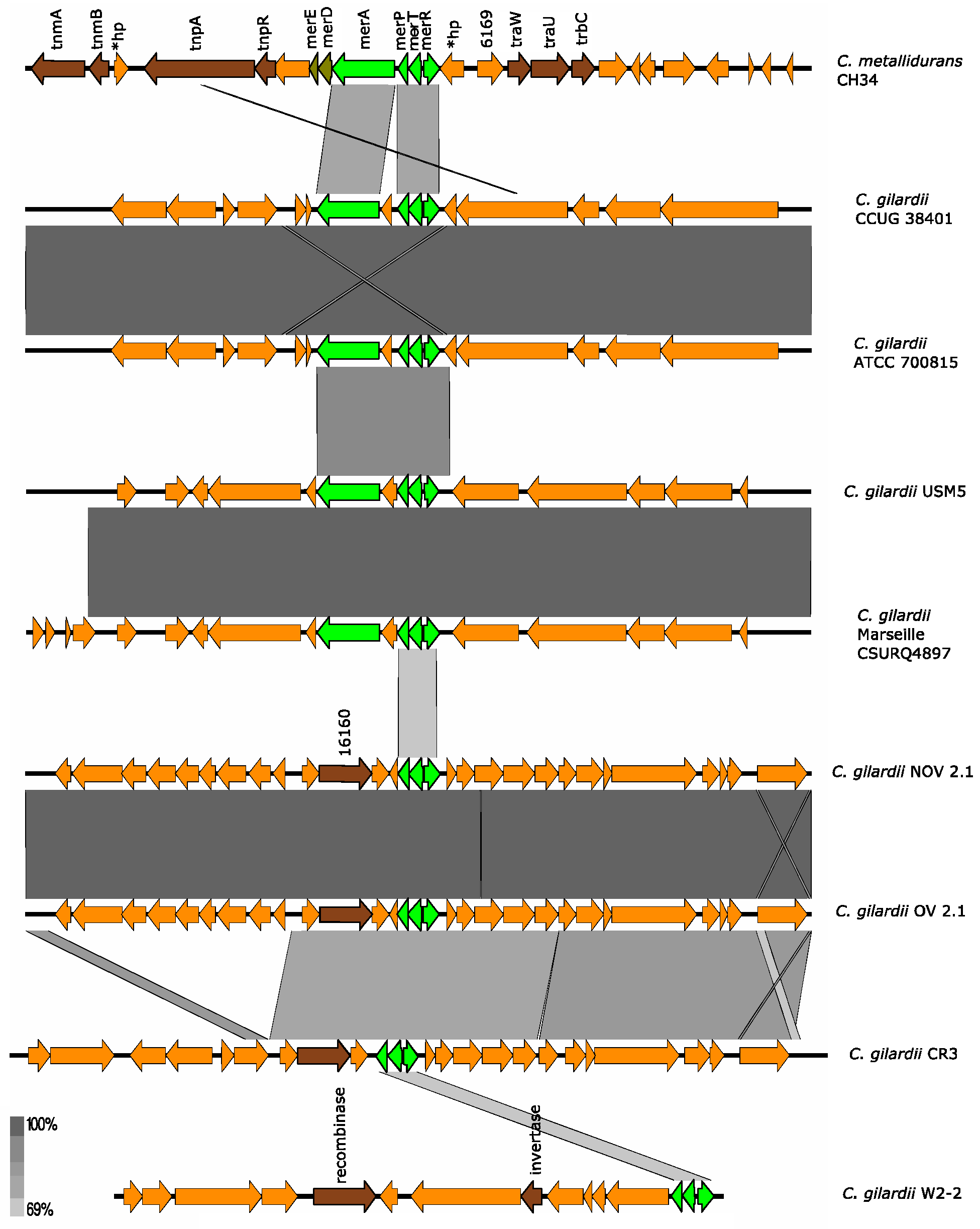
3.5.7. mer Cluster
C. metallidurans CH34 has three copies of the mer cluster, one copy in each plasmid (in the Tn4378 transposon of pMOL28 and the Tn4380 transposon of pMOL30) and one copy in chromosome 1. The mer cluster on chromosome 1 consists of 3 genes (merT: mercury ion transport protein; merP: periplasmic mercury ion-binding protein; merA: mercury reductase). The merA gene encodes a reductase that participates in the demethylation of organic mercury compounds [40].
On pMOL28 and pMOL30, the cluster contains two additional genes, merD and merE, which encode, respectively, a regulatory protein involved in Hg resistance and a broad spectrum organic and inorganic Hg transporter [36,40]. In all cases, a merR gene encoding a regulatory protein is found upstream merT; however, this gene may not be part of the mer cluster [36]. The mer genes (except merR) are fully induced by Hg and partially activated by Cd, Zn, and Pb [36].
None of the C. gilardii genomes used here harbored the complete mer cluster. In C. gilardii CCUG38401, ATCC 700815, USM5, and Marseille CSURQ4897 only the merP, merT, merR, and merA genes were found while only merP, merT, and merR were found in NOV2-1, OV2-1, C. gilardii CR3, and W2-2 (Figure 8). The mer cluster was completely absent in C. gilardii FDAARGOS 639 and J11. The amino acid identity of the mer gene products of NOV2-1 and OV2-1 were 62-71% (Table S5).
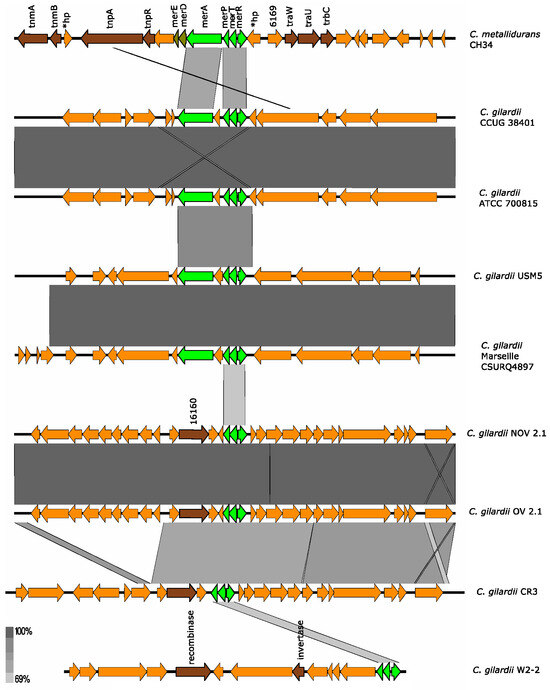
Figure 8.
Map showing the genomic organization of the mer cluster in C. metallidurans CH34, NOV2-1, OV2-1, and other C. gilardii strains. The conserved and non-conserved genes of the cluster are colored in neon and olive green, respectively. Orange arrows indicate neighboring genes. The genes colored in brown indicate parts of horizontal DNA transfer-related sequences. *hp means hypothetical protein. The gray scale indicates the levels of synteny.
4. Discussion
C. gilardii was first retrieved from environmental and clinical samples [41]. Initially named Wautersia gilardii and then Ralstonia gilardii, W. gilardii, together with all the other members of the genus Wautersia were later reclassified into Cupriavidus [12]. Since then, C. gilardii has been isolated from water, agricultural soil contaminated with the 2,4-dichlorophenoxyacetic acid herbicide, soils contaminated with heavy metals and asphalts, as well as human clinical samples [18,42,43]. C. gilardii has potential biotechnological applications in the detoxification of heavy metals and biodegradation of recalcitrant compounds, such as naphthenic acids and 2,4-dichlorophenoxyacetic acid [18,44,45,46]. It is also considered an emerging multidrug-resistant pathogen found in many environments [47].
Here, we sequenced the genomes of two C. gilardii strains, NOV2-1 and OV2-1, isolated from an iron oxide crust sampled in a tunnel of the Naica mine [8,11]. Unique characteristics of this biotope are the thermophilic environment (45–55 °C) and the presence of heavy metals [1]. NOV2-1 and OV2-1 had initially been identified as C. taiwanensis by partial 16S rRNA gene sequencing [8,11]. The standard ANI cut-off value for considering genomes to be from the same species is 95–96% [48]. For Cupriavidus, ANI cut-off values of 90% have been proposed [49]. As NOV2-1 and OV2-1 showed ANI values > 98% with C. gilardii CCUG 38401T (Figure 1a and Table S2), it can be concluded that these strains belong to the C. gilardii species. The phylogenetic tree based on 510 concatenated core genes also clearly showed that NOV2-1 and OV2-1 grouped with C. gilardii species rather than with C. taiwanensis (Figure 1b). The phylogenetic tree and the ANI values for Cupriavidus cauae USM5 (90.62% of ANI with CCUG 38401) and Cupriavidus sp. HPC(L) (94.4% of ANI with CCUG 38401) indicated that these strains should be reclassified as C. gilardii. The reclassification of Cupriavidus sp. HPC(L) into the C. gilardii species had already been proposed from a previous comparative genomic analysis [47]. C. gilardii J11, which was located on a separate branch in the phylogenetic tree and showed an ANI value of 82.84% with CCUG 3840T, may be a new Cupriavidus species. These results again highlight the complexity of the Cupriavidus taxonomy [12,13].
The genomes of NOV2-1 and OV2-1 consisted of two chromosomes (chr1: 3.58 Mb and chr2: 2.1 Mb for NOV2-1 and chr1: 3.53 Mb and chr2: 2.1 Mb for OV2-1) summing, respectively, 5.67 and 5.64 Mb (Table 1). These values as well as the GC and CDS content fall in the range reported for other C. gilardii complete genomes (Table S1). No plasmids were found in NOV2-1 and OV2-1, although small replicons of 33,330 and 87,261 bp have been reported in other C. gilardii strains, such as FDAARGOS_639 and QJ1. The chromosome 1 of both strains displayed dnaA-dnaN-gyrB gene structures (locus tags: K6V71_17740-50 in NOV2-1 and K7A44_16775-85 in OV2-1) while chromosome 2 presented a conserved repA-parA-parB region indicating a plasmid-like replication and partitioning mechanisms (locus tags: K6V71_04650-65 in NOV2-1 and K7A44_04730-45 in OV2-1). Additionally, 24 of the 510 core genes were located on chromosome 2 in both strains (Table S6) of which 13 were related to motility (chemotaxis proteins and flagellum synthesis) and the other to amino acids catabolism, gluconate utilization (permease and Entner-Doudoroff enzyme), cell wall biosynthesis and breakdown of aromatic compounds. The combination of a few core genes and plasmid-like replication origin indicates that chromosome 2 is a chromid [50,51]. This is not surprising since Cupriavidus and Ralstonia are known to harbor a secondary chromosome designated a chromid, i.e., a replicon that is neither a chromosome nor a plasmid [49,52,53,54]. These results are consistent with those obtained by [55], which analyzed the chromids present in nine Cupriavidus strains from several species and proposed that these secondary replicons allow the cells to expand their genome content through horizontal gene transfer and adapt to novel environments.
As the Naica mine is a thermophilic biotope, the influence of temperature on the growth of NOV2-1 and OV2-1 was evaluated. These strains had their maximum specific growth rate (μmax) at 42 °C and showed growth capacity up to 48 °C as C. gilardii CCUG 38401T (Figures S1 and S2). C. metallidurans CH34, C. necator LMG 8453, and C. taiwanensis LMG19424 could not grow at temperatures above 37 °C. These results indicate that the capacity to grow at higher temperatures (>40 °C) could be a phenotypic characteristic of the C. gilardii species, although more studies with other C. gilardii strains are needed to confirm this assumption. Concerning temperature, C. metallidurans CH34 displays an intriguing temperature-induced mutagenesis and mortality phenotype when temperature is shifted from 30 °C to 37 °C [56]. This phenotype has been recently attributed to a “genetic defect” at the lysA locus, which encodes the diaminopimelate decarboxylase gene, part of the peptidoglycan biosynthetic pathway [57]. Using the C. metallidurans CH34 Rmet_6588- lysA sequence as the query in a Blastn search against C. gilardii CR3, OV2-1, and NOV2-1, we found that the CH34 sequence presented important differences with its C. gilardii counterparts, specifically gaps and inversions in the 120 bp upstream the ATG codon of the Rmet_6588 gene. This gene putatively encodes a lipoprotein. These differences might influence the expression of the lipoprotein and the synthesis of peptidoglycan and explain the growth of C. gilardii strains above 37 °C. Detailed genomic studies and experiments are needed to study this phenotype.
The Cupriavidus strains used in this study were unable to assimilate five or six carbon carbohydrates and sugar alcohols (Table S3). These results are consistent with the description of other members of the Cupriavidus genus [12,16,23,58]. Such a carbohydrate utilization pattern has been found advantageous for the microbial detoxification of lignocellulosic biomass hydrolysates where the detoxifying microorganism selectively metabolizes lignocellulose-derived microbial inhibitors without compromising the fermentable sugars fraction [59]. This strategy has been reported with Cupriavidus basilensis for the removal of furans and phenolic compounds in acid-pretreated Miscanthus giganteus hydrolysates [60].
Regarding heavy metals tolerance, C. metallidurans CH34 was the most tolerant bacteria, which is consistent with this strain, the model organism to study heavy metal resistance and tolerance [61]. Several gene clusters involved in heavy metal homeostasis (ars, czc, cop 1, sil-cop 2, cup, mmf, and mer) were detected, fully or partially conserved, in NOV2-1, OV2-1, and other C. gilardii genomes (Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8). The presence of HMR clusters in the NOV2-1 and OV2-1 genomes were related to the heavy metal tolerance assay, where NOV2-1 and OV2-1 presented a high tolerance to heavy metals (Ni, Co, Cu, Cd, Zn, and Pb ions) at the millimolar levels in a LB-Lennox buffered solid medium (Figure 2), indicating that these strains can orchestrate a protective process against high levels of these metal ions.
OV2-1 was more sensitive to Cd and Zn than NOV2-1 and C. gilardii CCUG 38401T. This could be because the czc cluster was not found on the genome of OV2-1 contrary to NOV 2-1 and C. gilardii CCUG 38401T, which possessed an almost complete czc cluster (czcABCDEIJMNRS) (Figure 4) with a 98-100% of amino acid identity with the C. metallidurans CH34 czc gene products (Table S5). This cluster consists of eleven genes, including czcMNICBADRSEJP, which have been reported to be important for Co, Zn, and Cd tolerance in C. metallidurans CH34 [37]. It encodes three different efflux systems: (1) a cation diffusion facilitator CzcD, (2) a heavy metal Resistance–Nodulation–Division (RND)-driven system CzcCBA, and (3) a cation efflux P1B4-type ATPase CzcP that transports ions from the cytoplasm to the periplasm [37,62,63]. This system shields the cytoplasm against high extracellular concentrations of Zn, Co, and Cd ions [64]. The deletion of the czcA and czcB genes results in the complete loss of ions efflux and sensitivity to Cd, Co, and Zn in C. metallidurans [37]. The czcP gene was absent in NOV2-1 and no differences in tolerance in Zn, Cd, and Co tolerance were observed between this strain and C. gilardii CCUG 38401T (Figure 2), which supports the role of CzcP as a metal resistance enhancer [37].
Other ATPases from the P1B2 type, such as CadA and ZntA, are also involved in Zn and Co resistance in C. metallidurans CH34 [65]. Using the C. metallidurans CH34 ZntA sequence as the query in a Blastp search, possible homologs of this protein were found in the NOV2-1 genome (locus tag K6V71_07115, coverage: 91%, E-value: 0, identity: 74.8%) and in the OV2-1 genome (locus tag K7A44_07215, coverage: 89%, E-value: 0, identity: 75%). No homologues of CadA were found in NOV2-1 and OV2-1. The absence of homologs of CadA, CzcP, and the Pbr system in NOV2-1 and OV2-1 could be the limiting factor for these strains to tolerate higher concentrations of Cd, Zn, and Pb. However, detailed transcriptomic and proteomic analyses are needed to confirm this hypothesis.
In C. metallidurans, the czc cluster is flanked by tyrosine-type recombinase/integrase (upstream) and IS3 transposase family (downstream) sequences related to the mobility of this cluster [66]. These sequences were partially conserved in NOV2-1, however two additional and identical IS3-related insertion sequences were also detected, one inside the cluster between the czcI and czcN genes (locus tag K6V71_25340) and one on the right side of the cluster (locus tag K6V71_25285) (Figure S4). A nucleotide blast analysis showed that this insertion sequence was 100% identical to a sequence in the genome of a Cupriavidus campinensis strain (MJ1). The high degree of sequence identity of the czc proteins and the presence of conserved sequences related to horizontal DNA transfer suggests a horizontal transmission of this cluster among the Cupriavidus genus.
The cop 1, cop 2, and cup clusters were found in NOV2-1, OV2-1, and other C. gilardii genomes (Figure 5 and Figure 6). These clusters are the three major Cu detoxification systems in C. metallidurans CH34. Among them, the cop cluster 1 only responds to Cu ions, while the cop cluster 2 responds to Cd, Cu, Ni, Zn, Pb, Ag, and Co. Even though NOV2-1 had homologs of these clusters in its genome, this strain was slightly more sensitive to Cu than OV2-1 and CCUG38401T indicating that some genes or proteins in these clusters may not be functional or that other genes are needed for optimal Cu resistance. Surprisingly, a homologous mmf cluster was found both in C. gilardii NOV2-1 and OV2-1 but not in the other C. gilardii strains used in this study (Figure 7). Nevertheless, no growth differences were observed in the presence of Cd, Co, and Zn between NOV2-1 and CCUG38401T; these results could indicate that the function of this cluster can be substituted by other HMR proteins and systems. Although this cluster is strongly induced in the presence of Zn and Pb, its function is still unknown [67]. Furthermore, an incomplete mer cluster was found in NOV2-1, OV2-1, and other C. gilardii genomes, being the merP (periplasmic binding protein), merT (mercuric ion transport) and merR (transcriptional regulator) conserved genes (Figure 8). However, the functionality of these incomplete clusters during heavy metal exposure in C. gilardii NOV2-1, and OV2-1, and other C. gilardii strains has to be verified. Different or no horizontal DNA transfer sequences were present around these HMR gene clusters in C. gilardii compared to C. metallidurans, their role in the mobilization and expression of HMR genes in the Cupriavidus genus requires additional studies (Figures S5–S8).
Finally, an incomplete ars cluster homologue, lacking the arsC2 (arsenate reductase), arsI (lactoylglutathione lyase), arsM (methyltransferase), and arsP (permease) genes, was found in NOV2 -1, OV2-1, and other C. gilardii genomes (Figure 3). From a physiological point of view, the main genes for arsenic detoxification are present: arsR (transcriptional regulator), arsB (efflux transporter), arsC1 (reductase), and arsH (resistance protein). Although As was not studied here, the presence of this metal has been reported in Naica’s water and in the Meoqui-Delicias aquifer [4,5]. The presence of this cluster could reflect the presence of As; however, it has been reported that ars genes are also present in microorganisms isolated in As-free habitats [68]. A recent survey of arsenic-related genes in soil microbiomes has revealed a potential vertical and horizontal transfer history of the ars genes [69].
5. Major Implications and Future Perspectives
The findings from this study have important implications for understanding the genetic and physiological characteristics of the C. gilardii species, especially in the context of environmental adaptability and bioremediation potential. The genomic analysis of the strains NOV2-1 and OV2-1, isolated from the thermophilic and heavy metal containing environment of the Naica mine, revealed the capacity of this species to withstand both high temperature and the presence of metals. It has been shown that metals availability and, therefore, toxicity increases as temperature rises [70]. The high tolerance of these strains to multiple heavy metals, such as nickel, cobalt, copper, cadmium, zinc, and lead, suggests that C. gilardii could be an important player in the bioremediation of contaminated sites at elevated temperatures, especially in the context of climate change. Research, both at the genomic and experimental level, on the origin, evolution, mobility, and expression of HMR gene clusters in the Cupriavidus genus should continue, taking advantage of the increasing number of strains and completely sequenced genomes from different environments. This work also raises new research opportunities on temperature adaptation mechanisms with the Cupriavidus taxon and its relationship with HMR.
6. Conclusions
This study clarifies the taxonomic classification of two novel Cupriavidus strains, NOV2-1 and OV2-1, isolated from a unique, thermophilic, heavy metal-rich environment within the Naica mine located in Chihuahua, Mexico. Through comparative genomic analyses, we conclusively reclassified these strains as C. gilardii. Moreover, the reclassification of other Cupriavidus strains is also proposed here, underscoring the complexity of the Cupriavidus taxonomy. Furthermore, our genomic analysis revealed the presence of a secondary chromosome, or chromid, in both NOV2-1 and OV2-1, a characteristic feature of the Cupriavidus genus. Physiologically, these strains exhibit thermotolerance at temperatures up to 48 °C, a trait potentially characteristic of C. gilardii. Nutritionally, they share a similar carbohydrate utilization pattern with other Cupriavidus species, potentially advantageous in lignocellulosic biomass detoxification. Finally, our study provides insights into the genetic basis of heavy metal tolerance in these strains. The presence and differential conservation of HMR gene clusters, including czc, cop, cup, mmf, mer, and ars, correlate with the observed tolerance levels to various heavy metals. Specifically, the absence of a complete czc cluster in OV2-1 likely contributes to its increased sensitivity to Cd and Zn. The presence of mobile genetic elements surrounding these HMR clusters suggests horizontal gene transfer as a key mechanism for the dissemination and expression of HMR capacity within Cupriavidus populations. This study also highlights the potential of these strains for biotechnological applications in bioremediation, heavy metal detoxification, and biomass hydrolysate detoxification.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13040809/s1, Figure S1: Growth of Cupriavidus Reference Strains and the NOV2-1 and OV2-1 Strains at Different Temperatures; Figure S2: Maximum Specific Growth Rate (μmax) of strains NOV2-1 and OV2-1 at different temperatures; Figure S3: Sequences Related to Horizontal DNA Transfer at the Viscinity of the ars Cluster in C. metallidurans CH34, NOV2-1 and OV2-1; Figure S4: Sequences Related to Horizontal DNA Transfer at the Viscinity of of the czc Cluster in CH34, NOV2-1 and OV2-1; Figure S5: Sequences Related to Horizontal DNA Transfer at the Viscinity of the cop 1 Cluster; Figure S6: Sequences Related to Horizontal DNA Transfer at the Viscinity of the cop 2 Cluster; Figure S7: Sequences Related to Horizontal DNA Transfer at the Viscinity of the mmf Cluster; Figure S8: Sequences Related to Horizontal DNA Transfer at the Viscinity of the mer Cluster; Table S1: Genome Structure Comparison Between NOV2-1, OV2-1, and other C. gilardii strains; Table S2: Strains and Isolates Used in Comparative Genomic Analyses; Table S3: Average Percentage Nucleotide Identity (ANI) Between Pairs of Cupriavidus genomes; Table S4: Results of Carbon Source Utilization, Inhibition and Resistance, and Enzymatic Activities Tests of C. gilardii NOV2-1, C. gilardii OV2-1 and Cupriavidus Reference Strains; Table S5: HMR Gene Clusters Detected in C. gilardii NOV2.1 and OV2.1; Table S6: Core Genes Found in the NOV2.1 and OV2.1 Chromids.
Author Contributions
Conceptualization, methodology, review, and editing, all authors; Data obtention and curation, A.G.-S., L.L.-A., M.L.-S. and G.J.-F.; bacterial collection, S.L.B.; resources and funding, S.L.B.; original draft preparation, S.L.B. and A.G.-S.; supervision, S.L.B., A.G.-d.L.S. and M.A.C. All authors have read and agreed to the published version of the manuscript.
Funding
Antonio González-Sánchez received a postdoctoral scholarship from the “Universidad Autónoma Metropolitana” in the “Doctorado en Ciencias Biológicas y de la Salud” recipient program (2018–2020). This work was funded through the DCNI project “Microbiología de bioprocesos”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The chromosomes and chromids sequences of C. gilardii NOV2-1 and OV2-1 can be accessed under the GenBank accession number CP083437.1, CP083438.1, CP083735.1 and CP083736.1 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_025643215.1/ and https://www.ncbi.nlm.nih.gov/datasets/taxonomy/82541/, accessed on 22 January 2025). The sequencing data of PacBio and Illumina reads are available in the NCBI SRA database via the BioProject accession number PRJNA757420 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA757420/, accessed on 22 January 2025).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gázquez, F.; Calaforra, J.M.; Forti, P.; Badino, G. The Caves of Naica: A Decade of Research. Bol. Geol. Y Min. 2016, 127, 147–163. [Google Scholar]
- Peñoles Naica. Available online: https://www.penoles.com.mx/nuestras-operaciones/unidades-mineras/naica.html (accessed on 12 February 2025).
- Carreño-Márquez, I.J.A.; Castillo-Sandoval, I.; Pérez-Cázares, B.E.; Fuentes-Cobas, L.E.; Esparza-Ponce, H.E.; Menéndez-Méndez, E.; Fuentes-Montero, M.E.; Montero-Cabrera, M.E. Evolution of the Astonishing Naica Giant Crystals in Chihuahua, Mexico. Minerals 2021, 11, 292. [Google Scholar] [CrossRef]
- Briceño Prieto, S.B. Caracterización Geoquímica de Los Megacristales de Yeso de Naica, Chihuahua y Su Relación Con Los Procesos de Interacción Agua-Roca. Master’s Thesis, Universidad Nacional Autónoma de México, Ciudad de Mexico, Mexico, 2011. [Google Scholar]
- Bencomo-Calderón, M.; Herrera-Peraza, E.F.; Villalobos-Aragón, A. As and Pb Presence within the Meoqui-Delicias Aquifer, Chihuahua, Mexico. Water 2024, 16, 2538. [Google Scholar] [CrossRef]
- Otálora, F.; García-Ruiz, J. Nucleation and Growth of the Naica Giant Gypsum Crystals. Chem. Soc. Rev. 2014, 43, 2013–2026. [Google Scholar]
- Forti, P.; Sanna, L. The Naica Project—A Multidisciplinary Study of the Largest Gypsum Crystals of the World. Episodes 2010, 33, 23–32. [Google Scholar] [CrossRef]
- Espino del Castillo, A.; Beraldi-Campesi, H.; Amador-Lemus, P.; Beltrán, H.I.; Le Borgne, S. Bacterial Diversity Associated with Mineral Substrates and Hot Springs from Caves and Tunnels of the Naica Underground System (Chihuahua, Mexico). Int. J. Speleol. 2018, 47, 213–227. [Google Scholar] [CrossRef]
- Quintana, E.T.; Badillo, R.F.; Maldonado, L.A. Characterisation of the First Actinobacterial Group Isolated from a Mexican Extremophile Environment. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013, 104, 63–70. [Google Scholar] [CrossRef]
- Ragon, M.; Van Driessche, A.E.S.; García-Ruíz, J.M.; Moreira, D.; López-García, P. Microbial Diversity in the Deep-Subsurface Hydrothermal Aquifer Feeding the Giant Gypsum Crystal-Bearing Naica Mine, Mexico. Front. Microbiol. 2013, 4, 37. [Google Scholar] [CrossRef]
- Espino del Castillo Rodríguez, A. Caracterización de La Comunidad Bacteriana Presente En Diferentes Superficies Minerales Del Yacimiento de La Mina de Naica (Chihuahua, México) y Su Participación Como Agente Biogeoquímico. Ph.D. Thesis, Universidad Autónoma Metropolitana, Ciudad de Mexico, Mexico, 2019. [Google Scholar]
- Vandamme, P.; Coenye, T. Taxonomy of the Genus Cupriavidus: A Tale of Lost and Found. Int. J. Syst. Evol. Microbiol. 2004, 54, 2285–2289. [Google Scholar] [CrossRef]
- Coenye, T. The Family Burkholderiaceae. In The Prokaryotes, 4th ed.; Section II Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 759–776. [Google Scholar] [CrossRef]
- Vaneechoutte, M.; Kämpfer, P.; De Baere, T.; Falsen, E.; Verschraegen, G. Wautersia Gen. Nov., a Novel Genus Accomodating the Phylogenetic Lineage Including Ralstonia eutropha and Related Species, and Proposal of Ralstonia [Pseudomonas] Syzygii (Roberts et al. 1990) Comb. Nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 317–327. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of Prokaryotic Names with Standing in Nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M. The History of Cupriavidus metallidurans Strains Isolated from Anthropogenic Environments. In Metal Response in Cupriavidus metallidurans: Volume I: From Habitats to Genes and Proteins; Mergeay, M., Van Houdt, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–19. [Google Scholar] [CrossRef]
- Arroyo-Herrera, I.; Maldonado-Hernández, J.; Rojas-Rojas, F.U.; Meza-Radilla, G.; Larios-Serrato, V.; Vásquez-Murrieta, M.S.; Whitman, W.B.; De Los Santos, P.E. Cupriavidus agavae Sp. Nov., a Species Isolated from Agave L. Rhizosphere in Northeast Mexico. Int. J. Syst. Evol. Microbiol. 2020, 70, 4165–4170. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, M.; Xiao, J.; Hao, L.; Crowley, D.E.; Zhang, Z.; Yu, J.; Huang, N.; Huo, M.; Wu, J. Genome Sequence Analysis of the Naphthenic Acid Degrading and Metal Resistant Bacterium Cupriavidus gilardii CR3. PLoS ONE 2015, 10, e0132881. [Google Scholar] [CrossRef] [PubMed]
- von Rozycki, T.; Nies, D.H. Cupriavidus metallidurans: Evolution of a Metal-Resistant Bacterium. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2009, 96, 115–139. [Google Scholar] [CrossRef]
- De La Rosa- Acosta, M.; Jiménez-Collazo, J.; Maldonado-Román, M.; Malavé-Llamas, K.; Carlos Musa-Wasil, J. Bacteria as Potential Indicators of Heavy Metal Contamination in a Tropical Mangrove and the Implications on Environmental and Human Health. J. Trop. Life Sci. 2015, 5, 110–116. [Google Scholar] [CrossRef]
- Makkar, N.S.; Casipa, L.E. Cupriavidus necator gen. nov., sp. nov.; A Nonobligate Bacterial Predator of Bacteria in Soil. Int. J. Syst. Evol. Microbiol. 1987, 37, 323–326. [Google Scholar] [CrossRef]
- Chen, W.M.; Laevens, S.; Lee, T.M.; Coenye, T.; De Vos, P.; Mergeay, M.; Vandamme, P. Ralstonia taiwanensis Sp. Nov., Isolated from Root Nodules of Mimosa Species and Sputum of a Cystic Fibrosis Patient. Int. J. Syst. Evol. Microbiol. 2001, 51, 1729–1735. [Google Scholar] [CrossRef]
- Goris, J.; De Vos, P.; Coenye, T.; Hoste, B.; Jansses, D.; Brim, H.; Diels, L.; Mergeay, M.; Kersters, K.; Vandamme, P. Classification of Metal-Resistant Bacteria from Industrial Biotopes as Ralstonia campinensis Sp. Nov., Ralstonia metallidurans Sp. Nov. and Ralstonia Basilensis Steinle et al. 1998 Emend. Int. J. Syst. Evol. Microbiol. 2001, 51, 1773–1782. [Google Scholar] [CrossRef]
- Andrews, S. FastQC—A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 August 2022).
- Krueger, F. A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 15 February 2025).
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Wences, A.H.; Schatz, M.C. Metassembler: Merging and Optimizing de Novo Genome Assemblies. Genome Biol. 2015, 16, 207. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a Versatile Software Package for Scalable and Robust Microbial Pangenome Analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef]
- Vinuesa, P.; Ochoa-Sánchez, L.E.; Contreras-Moreira, B. GET_PHYLOMARKERS, a Software Package to Select Optimal Orthologous Clusters for Phylogenomics and Inferring Pan-Genome Phylogenies, Used for a Critical Geno-Taxonomic Revision of the Genus Stenotrophomonas. Front. Microbiol. 2018, 9, 771. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Monsieurs, P.; Moors, H.; Van Houdt, R.; Janssen, P.J.; Janssen, A.; Coninx, I.; Mergeay, M.; Leys, N. Heavy Metal Resistance in Cupriavidus metallidurans CH34 Is Governed by an Intricate Transcriptional Network. BioMetals 2011, 24, 1133–1151. [Google Scholar] [CrossRef]
- Monsieurs, P.; Hobman, J.; Vandenbussche, G.; Mergeay, M.; Van Houdt, R. Response of Cupriavidus metallidurans CH34 to Metals. In Metal Response in Cupriavidus metallidurans: Volume I: From Habitats to Genes and Proteins; Mergeay, M., Van Houdt, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 21–44. [Google Scholar] [CrossRef]
- Utami, U.; Harianie, L.; Dunyana, N.R. Romaidi Lead-Resistant Bacteria Isolated from Oil Wastewater Sample for Bioremediation of Lead. Water Sci. Technol. 2020, 81, 2244–2249. [Google Scholar] [CrossRef]
- Maertens, L.; Leys, N.; Matroule, J.Y.; Van Houdt, R. The Transcriptomic Landscape of Cupriavidus metallidurans CH34 Acutely Exposed to Copper. Genes. 2020, 11, 1049. [Google Scholar] [CrossRef]
- Abbaszade, G.; Szabó, A.; Vajna, B.; Farkas, R.; Szabó, C.; Tóth, E. Whole Genome Sequence Analysis of Cupriavidus Campinensis S14E4C, a Heavy Metal Resistant Bacterium. Mol. Biol. Rep. 2020, 47, 3973–3985. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Falsent, E.; Hostef, B.; W Govant, J.R.; Kersters, K.; Vandamme, P. Classification of Alcaligenes faecalis-like Isolates from the Environment and Human Clinical Samples as Ralstonia gilardii Sp. Nov. Int. J. Syst. Evol. Microbiol. 1999, 49, 405–413. [Google Scholar]
- Sichtig, H.; Minogue, T.; Yan, Y.; Stefan, C.; Hall, A.; Tallon, L.; Sadzewicz, L.; Nadendla, S.; Klimke, W.; Hatcher, E.; et al. FDA-ARGOS Is a Database with Public Quality-Controlled Reference Genomes for Diagnostic Use and Regulatory Science. Nat. Commun. 2019, 10, 3313. [Google Scholar] [CrossRef]
- Zhang, Z.; Deng, W.; Wang, S.; Xu, L.; Yan, L.; Liao, P. First Case Report of Infection Caused by Cupriavidus gilardii in a Non-Immunocompromised Chinese Patient. IDCases 2017, 10, 127–129. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, M.; Zhou, D.; Fan, W.; Wang, X.; Huo, M. Bioremoval of Cu2+ from CMP Wastewater by a Novel Copper-Resistant Bacterium Cupriavidus gilardii CR3: Characteristics and Mechanisms. RSC Adv. 2017, 7, 18793–18802. [Google Scholar] [CrossRef]
- Wu, X.; Wang, W.; Liu, J.; Pan, D.; Tu, X.; Lv, P.; Wang, Y.; Cao, H.; Wang, Y.; Hua, R. Rapid Biodegradation of the Herbicide 2,4-Dichlorophenoxyacetic Acid by Cupriavidus gilardii T-1. J. Agric. Food Chem. 2017, 65, 3711–3720. [Google Scholar] [CrossRef]
- Pérez-Pantoj, D.; Leiva-Novoa, P.; Donoso, R.A.; Little, C.; Godoy, M.; Pieper, D.H.; Gonzáleza, B. Hierarchy of Carbon Source Utilization in Soil Bacteria: Hegemonic Preference for Benzoate in Complex Aromatic Compound Mixtures Degraded by Cupriavidus pinatubonensis Strain JMP134. Appl. Environ. Microbiol. 2015, 81, 3914–3924. [Google Scholar] [CrossRef]
- Ruiz, C.; McCarley, A.; Espejo, M.L.; Cooper, K.K.; Harmon, D.E. Comparative Genomics Reveals a Well-Conserved Intrinsic Resistome in the Emerging Multidrug-Resistant Pathogen Cupriavidus gilardii. mSphere 2019, 4, e00631-19. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Moriuchi, R.; Dohra, H.; Kanesaki, Y.; Ogawa, N. Complete Genome Sequence of 3-Chlorobenzoate-Degrading Bacterium Cupriavidus necator NH9 and Reclassification of the Strains of the Genera Cupriavidus and Ralstonia Based on Phylogenetic and Whole-Genome Sequence Analyses. Front. Microbiol. 2019, 10, 133. [Google Scholar] [CrossRef] [PubMed]
- Fournes, F.; Val, M.E.; Skovgaard, O.; Mazel, D. Replicate Once per Cell Cycle: Replication Control of Secondary Chromosomes. Front. Microbiol. 2018, 9, 1833. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Lower, R.P.J.; Kim, N.K.D.; Young, J.P.W. Introducing the Bacterial “Chromid”: Not a Chromosome, Not a Plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Janssen, P.J.; van Houdt, R.; Moors, H.; Monsieurs, P.; Morin, N.; Michaux, A.; Benotmane, M.A.; Leys, N.; Vallaeys, T.; Lapidus, A.; et al. The Complete Genome Sequence of Cupriavidus metallidurans Strain CH34, a Master Survivalist in Harsh and Anthropogenic Environments. PLoS ONE 2010, 5, e10433. [Google Scholar] [CrossRef]
- Lykidis, A.; Pérez-Pantoja, D.; Ledger, T.; Mavromatis, K.; Anderson, I.J.; Ivanova, N.N.; Hooper, S.D.; Lapidus, A.; Lucas, S.; González, B.; et al. The Complete Multipartite Genome Sequence of Cupriavidus necator JMP134, a Versatile Pollutant Degrader. PLoS ONE 2010, 5, e9729. [Google Scholar] [CrossRef]
- Mergeay, M.; Van Houdt, R. Plasmids as Secondary Chromosomes. In Molecular Life Sciences an Encyclopedic Reference; Wells, R.D., Bond, J.S., Klinman, J., Masters, B.S.S., Eds.; Springer: New York, NY, USA, 2018; pp. 961–964. [Google Scholar] [CrossRef]
- Dicenzo, G.C.; Mengoni, A.; Perrin, E. Chromids Aid Genome Expansion and Functional Diversification in the Family Burkholderiaceae. Mol. Biol. Evol. 2019, 36, 562–574. [Google Scholar] [CrossRef]
- Van Houdt, R.; Vandecraen, J.; Heylen, W.; Leys, N.; Monsieurs, P.; Provoost, A.; Aertsen, A. Phenotypic and Genetic Characterization of Temperature-Induced Mutagenesis and Mortality in Cupriavidus metallidurans. Front. Microbiol. 2021, 12, 698330. [Google Scholar] [CrossRef]
- Arroua, B.; Bellanger, X.; Guilloteau, H.; Mathieu, L.; Merlin, C. Atypical Stress Response to Temperature and NaOCl Exposure Leading to Septation Defect during Cell Division in Cupriavidus metallidurans CH34. FEMS Microbiol. Lett. 2014, 353, 32–39. [Google Scholar] [CrossRef]
- Florentino, L.A.; Jaramillo, P.M.D.; Silva, K.B.; da Silva, J.S.; de Oliveira, S.M.; de Souza Moreira, F.M. Physiological and Symbiotic Diversity of Cupriavidus necator Strains Isolated from Nodules of Leguminosae Species. Sci. Agric. 2012, 69, 247–258. [Google Scholar] [CrossRef]
- Ujor, V.C.; Okonkwo, C.C. Microbial Detoxification of Lignocellulosic Biomass Hydrolysates: Biochemical and Molecular Aspects, Challenges, Exploits and Future Perspectives. Front. Bioeng. Biotechnol. 2022, 10, 1061667. [Google Scholar] [CrossRef]
- Agu, C.V.; Ujor, V.; Gopalan, V.; Ezeji, T.C. Use of Cupriavidus Basilensis-Aided Bioabatement to Enhance Fermentation of Acid-Pretreated Biomass Hydrolysates by Clostridium beijerinckii. J. Ind. Microbiol. Biotechnol. 2016, 43, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M.; Van Houdt, R. Cupriavidus metallidurans CH34, a Historical Perspective on Its Discovery, Characterization and Metal Resistance. FEMS Microbiol. Ecol. 2021, 97, fiaa247. [Google Scholar] [CrossRef] [PubMed]
- Scherer, J.; Nies, D.H. CzcP Is a Novel Efflux System Contributing to Transition Metal Resistance in Cupriavidus metallidurans CH34. Mol. Microbiol. 2009, 73, 601–621. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.T.; Ross, M.O.; Hoffman, B.M.; Rosenzweig, A.C. Metal Selectivity of a Cd-, Co-, and Zn-Transporting P1B-Type ATPase. Biochemistry 2017, 56, 85–95. [Google Scholar] [CrossRef]
- Schulz, V.; Schmidt-Vogler, C.; Strohmeyer, P.; Weber, S.; Kleemann, D.; Nies, D.H.; Herzberg, M. Behind the Shield of Czc: ZntR Controls Expression of the Gene for the Zinc-Exporting P-Type ATPase ZntA in Cupriavidus metallidurans. J. Bacteriol. 2021, 203, e00052-21. [Google Scholar] [CrossRef]
- Legatzki, A.; Grass, G.; Anton, A.; Rensing, C.; Nies, D.H. Interplay of the Czc System and Two P-Type ATPases in Conferring Metal Resistance to Ralstonia metallidurans. J. Bacteriol. 2003, 185, 4354–4361. [Google Scholar] [CrossRef]
- Ali, M.M.; Provoost, A.; Maertens, L.; Leys, N.; Monsieurs, P.; Charlier, D.; Van Houdt, R. Genomic and Transcriptomic Changes That Mediate Increased Platinum Resistance in Cupriavidus metallidurans. Genes 2019, 10, 63. [Google Scholar] [CrossRef]
- Van Houdt, R.; Mergeay, M. Genomic Context of Metal Response Genes in Cupriavidus metallidurans with a Focus on Strain CH34. In Metal Response in Cupriavidus metallidurans: Volume I: From Habitats to Genes and Proteins; Mergeay, M., Van Houdt, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 45–89. [Google Scholar] [CrossRef]
- Fekih, I.B.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of Arsenic Resistance Genes in Prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar]
- Dunivin, T.K.; Yeh, S.Y.; Shade, A. A Global Survey of Arsenic-Related Genes in Soil Microbiomes. BMC Biol. 2019, 17, 45. [Google Scholar] [CrossRef]
- Banaee, M.; Zeidi, A.; Mikušková, N.; Faggio, C. Assessing Metal Toxicity on Crustaceans in Aquatic Ecosystems: A Comprehensive Review. Biol. Trace Elem. Res. 2024, 202, 5743–5761. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).