Genome Analysis of the Multidrug-Resistant Campylobacter coli BCT3 of the Sequence Type (ST) 872 Isolated from a Pediatric Diarrhea Case
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation and Antimicrobial Susceptibility Testing
2.2. Genome Sequencing and Bioinformatics Analysis
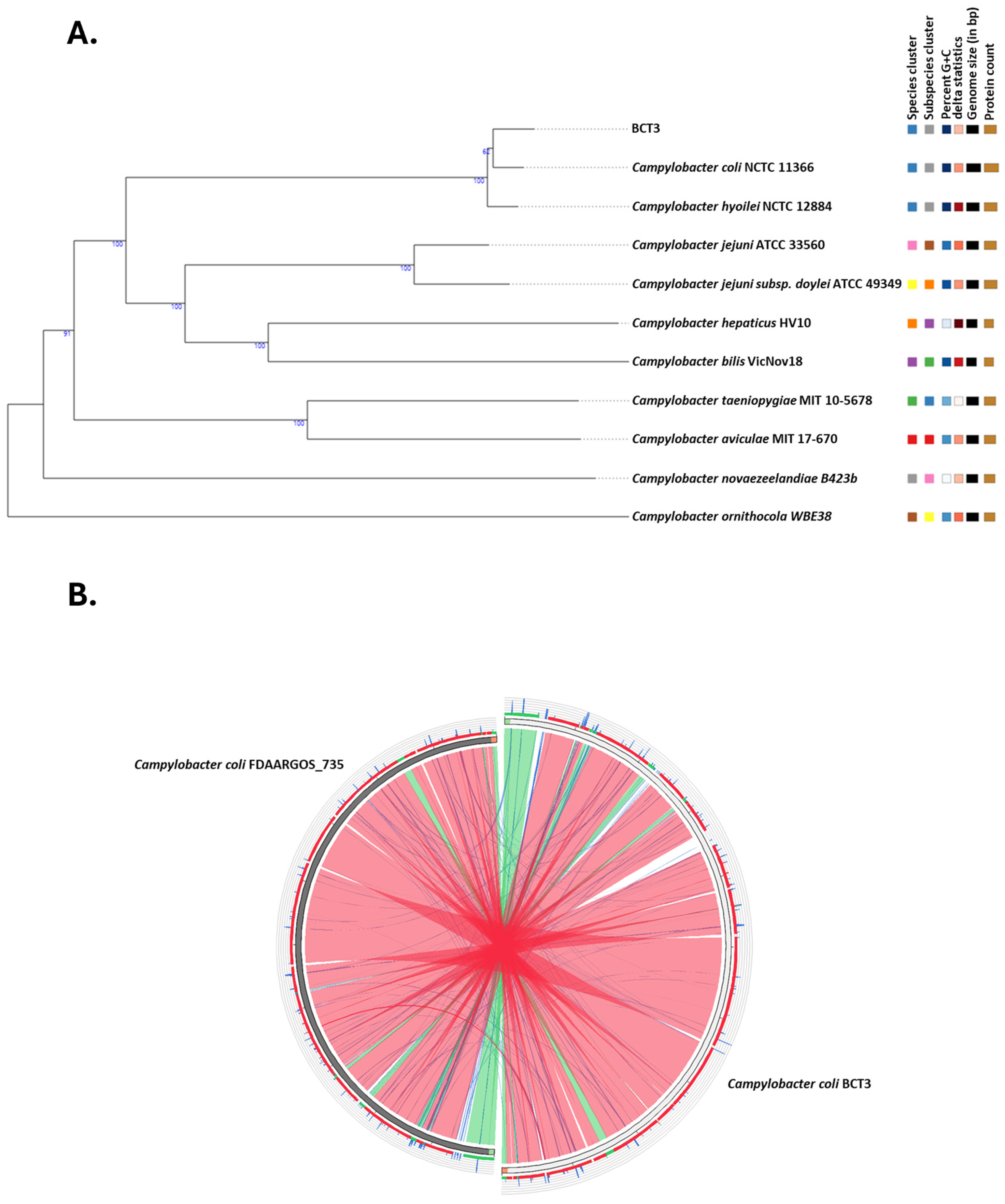
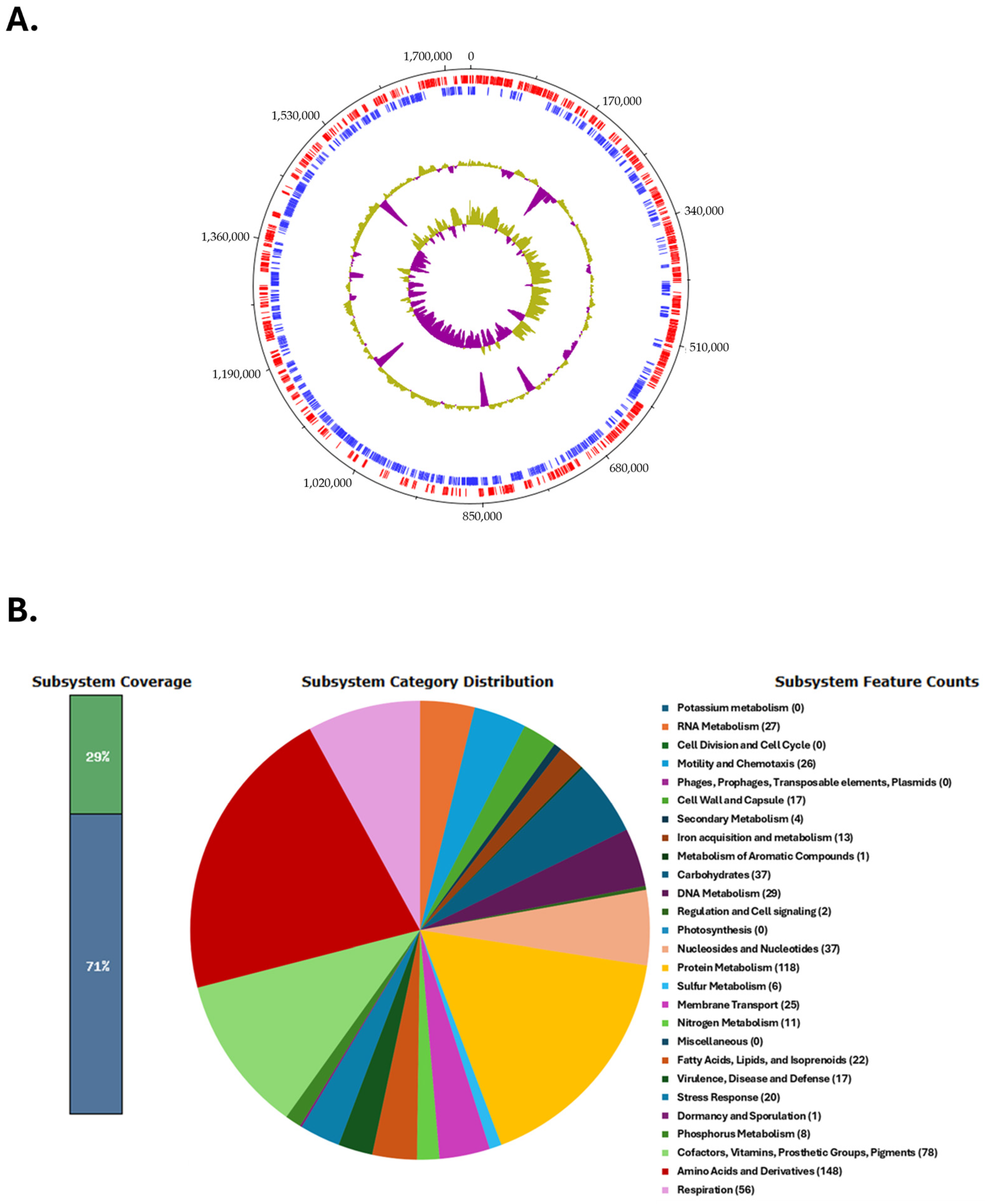
3. Results and Discussion
3.1. General Characteristics of the Genome of Campylobacter coli Strain BCT3
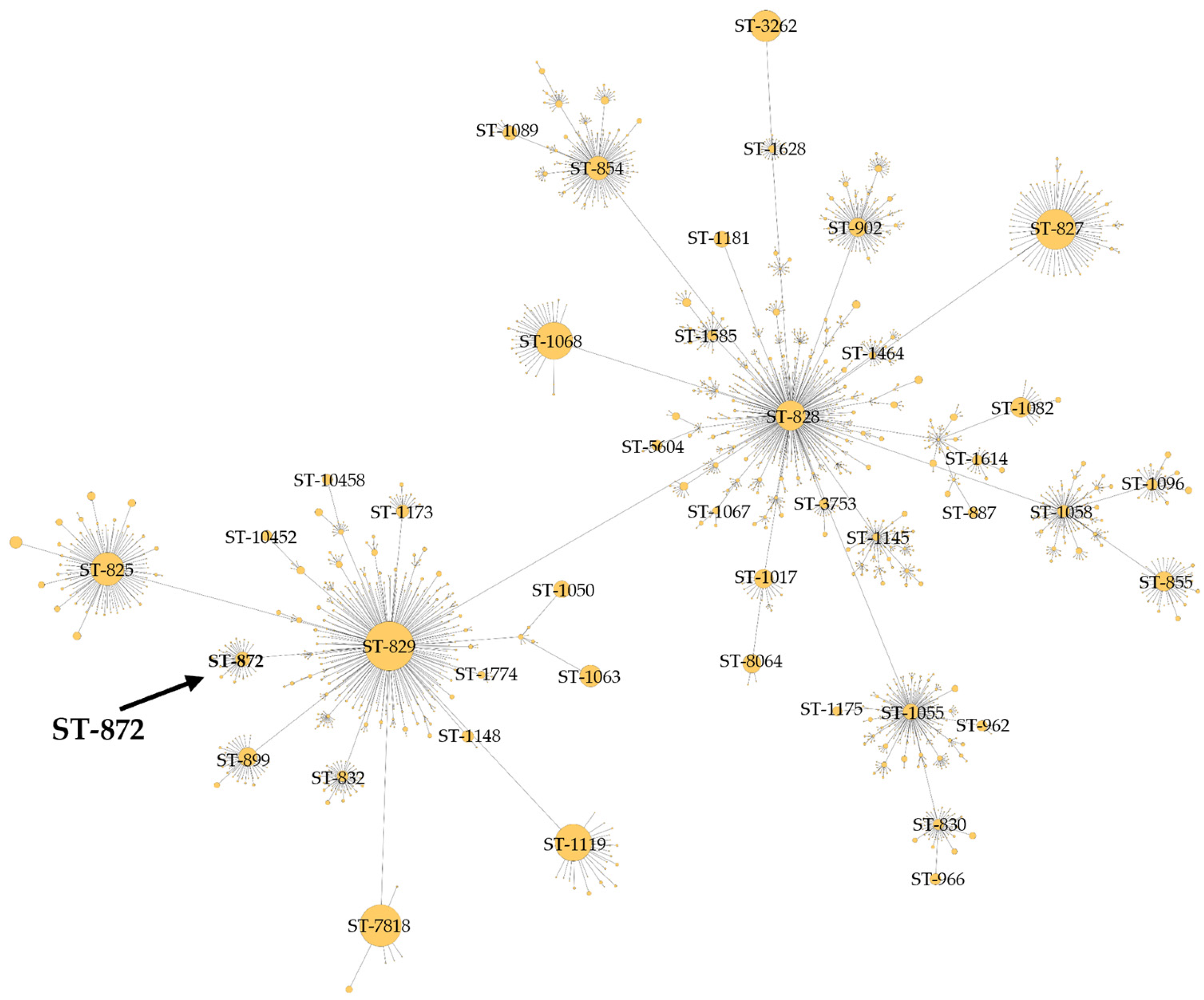
3.2. Multilocus Sequence Typing
3.3. Virulence Factors
| Gene | Function | Ref. |
|---|---|---|
| cadF | Outer membrane fibronectin-binding protein | [57,74] |
| pebA | Binding protein of an ABC transporter system | [77,94] |
| ciaB | Invasion antigen | [72,74] |
| ciaC | Invasion antigen | [72,74] |
| flgB | Flagellar basal body rod protein | [79,94] |
| flgC | Flagellar basal body rod protein | [79] |
| flgD | Flagellar basal body rod modification protein | [79] |
| flgE | Flagellar hook protein | [72,79,95] |
| flgF | Flagellar basal body rod protein | [80] |
| flgG | Flagellar basal body rod protein | [74] |
| flgH | Flagellar L-ring protein precursor | [80] |
| flgI | Flagellar P-ring protein precursor | [96] |
| flgJ | Flagellar protein involved in peptidoglycan hydrolysis | [96] |
| flgK | Flagellar hook-associated protein | [79] |
| flgM | Negative regulator of flagellin synthesis | [97] |
| flgP | Flagellar motility protein | [72] |
| flgQ | Flagellar motility protein | [75] |
| flgR | σ54—associated transcriptional activator | [74] |
| flgS | Signal transduction histidine kinase | [74] |
| flhA | Flagellar biosynthesis protein | [72,74] |
| flhB | Flagellar biosynthesis protein | [74] |
| flhF | Flagellar biosynthesis regulator | [79] |
| flhG | ATP-binding protein | [79] |
| flaC | Secreted flagellin | [72] |
| flaD | Flagellar protein | [81] |
| flaG | Flagellar filament length control | [72,97] |
| fliA | Flagellar biosynthesis sigma factor | [72] |
| fliD | Flagellar hook-associated protein | [79,98] |
| fliE | Flagellar hook–basal body complex protein | [79] |
| fliF | Flagellar M-ring protein | [74,79] |
| fliG | Flagellar motor switch protein | [74,79] |
| fliI | Flagellum-specific ATPse | [79] |
| fliL | Flagellar basal body protein | [79] |
| fliM | Flagellar motor switch protein | [79] |
| fliN | Flagellar motor switch protein | [79] |
| fliP | Flagellar biosynthesis protein | [74] |
| fliQ | Flagellar biosynthesis protein | [74] |
| fliR | Flagellar biosynthetic protein | [74] |
| fliS | Flagellar secretion chaperone | [72,78,79] |
| fliW | Flagellar assembly protein | [72,78] |
| fliY | Flagellar motor switch protein | [74] |
| rpoN | RNA polymerase factor σ54 | [84] |
| pflA | Paralysed flagellum protein | [74] |
| motA | Flagellar motor protein | [72,74] |
| cheA | Histidine kinase sensor | [72,74,82,83] |
| cheV | Phosphotransferase | [72,74] |
| cheW | Phosphotransferase | [72,74] |
| cheY | Cytoplasmic response regulator | [72,74] |
| pseA | Pseudaminic acid biosynthesis protein | [87] |
| pseB | UDP-N-acetylglucosamine 4,6-dehydratase | [87,99] |
| pseC | UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine transaminase | [87] |
| pseD | Motility accessory factor | [87] |
| pseE | Motility accessory factor | [79,87] |
| pseF | Acylneuraminate cytidylyltransferase | [87] |
| pseG | UDP-2,4-diacetamido-2,4,6-trideoxy-beta-L-altropyranose hydrolase | [87] |
| pseH | UDP-4-amino-4, 6-dideoxy-N-acetyl-beta-L-altrosamine N-acetyltransferase | [87] |
| pseI | N-acetylneuraminic acid synthetase | [100] |
| eptC | Phosphoethanolamine transferase | [59] |
| ptmA | Flagellin modification protein | [79,86] |
| ptmB | Acylneuraminate cytidylyltransferase | [79,86] |
| gmhA | Phosphoheptose isomerase | [91,92] |
| gmhB | DD-heptose 17-bisphosphate phosphatase | [91] |
| waaC | Heptosyltransferase I | [90,92] |
| waaF | Heptosyltransferase II | [92] |
| waaV | Glucosyltransferase | [92,93] |
| hldD | ADP-glyceromanno-heptose 6-epimerase | [59] |
| hldE | Bifunctional D-beta-D-heptose 7-phosphate kinase/D-beta-D-heptose 1-phosphate adenylyltransferase | [59] |
| kpsD | Capsule polysaccharide export system periplasmic protein | [72,92] |
| kpsF | D-arabinose 5-phosphate isomerase | [92] |
| kpsM | Capsule polysaccharide export system inner membrane protein | [72,92] |
| kpsS | Capsule polysaccharide modification protein | [92] |
| kpsT | Capsule polysaccharide export ATP-binding protein | [92] |
3.4. Antimicrobial Resistance
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaakoush, N.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed]
- Esan, O.B.; Pearce, M.; van Hecke, O.; Roberts, N.; Collins, D.R.J.; Violato, M.; McCarthy, N.; Perera, R.; Fanshawe, T.R. Factors associated with sequelae of Campylobacter and non-typhoidal Salmonella infections: A systematic review. EBioMedicine 2017, 15, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of colorectal cancer in terms of epidemiology, risk factors, development, symptoms and diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef] [PubMed]
- Leonhard, S.E.; van der Eijk, A.A.; Andersen, H.; Antonini, G.; Arends, S.; Attarian, S.; Barroso, F.A.; Bateman, K.J.; Batstra, M.R.; Benedetti, L.; et al. An international perspective on preceding infections in Guillain-Barré syndrome: The IGOS-1000 cohort. Neurology 2022, 99, e1299–e1313. [Google Scholar] [CrossRef]
- WHO. Campylobacter. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/campylobacter (accessed on 21 November 2024).
- Veronese, P.; Dodi, I. Campylobacter jejuni/coli infection: Is it still a concern? Microorganisms 2024, 12, 2669. [Google Scholar] [CrossRef]
- Quino, W.; Caro-Castro, J.; Hurtado, V.; Flores-León, D.; Gonzalez-Escalona, N.; Gavilan, R.G. Genomic analysis and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli in Peru. Front. Microbiol. 2022, 12, 802404. [Google Scholar] [CrossRef]
- Rukambile, E.; Sintchenko, V.; Muscatello, G.; Kock, R.; Alders, R. Infection, colonization and shedding of Campylobacter and Salmonella in animals and their contribution to human disease: A review. Zoonoses Public Health 2019, 66, 562–578. [Google Scholar] [CrossRef]
- Popa, S.A.; Morar, A.; Ban-Cucerzan, A.; Imre, K. Last decade mini-review of the scientific progresses in the monitoring of the occurrence and antimicrobial susceptibility profile of poultry origin Campylobacter spp. within the European Union countries. Rev. Rom. Med. Vet. 2022, 32, 75–82. [Google Scholar]
- Popa, S.A.; Morar, A.; Ban-Cucerzan, A.; Tîrziu, E.; Herman, V.; Imre, M.; Florea, T.; Morar, D.; Pătrînjan, R.-T.; Imre, K. First study in the frequency of isolation and phenotypic antimicrobial resistance profiles of pig and cattle origin Campylobacter strains in Romania. Vet. Res. Commun. 2024, 48, 2621–2627. [Google Scholar] [CrossRef]
- Popa, S.A.; Herman, V.; Tîrziu, E.; Morar, A.; Ban-Cucerzan, A.; Imre, M.; Pătrînjan, R.-T.; Imre, K. Public health risk of Campylobacter spp. isolated from slaughterhouse and retail poultry meat: Prevalence and antimicrobial resistance profiles. Pathogens 2025, 14, 316. [Google Scholar] [CrossRef]
- Natsos, G.; Mouttotou, N.K.; Magiorkinis, E.; Ioannidis, A.; Rodi-Burriel, A.; Chatzipanagiotou, S.; Koutoulis, K.C. Prevalence of and risk factors for Campylobacter spp. colonization of broiler chicken flocks in Greece. Foodborne Pathog. Dis. 2020, 17, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Bunduruș, I.A.; Balta, I.; Ștef, L.; Ahmadi, M.; Peț, I.; McCleery, D.; Corcionivoschi, N. Overview of virulence and antibiotic resistance in Campylobacter spp. livestock isolates. Antibiotics 2023, 12, 402. [Google Scholar] [CrossRef]
- Elmi, A.; Nasher, F.; Dorrell, N.; Wren, B.; Gundogdu, O. Revisiting Campylobacter jejuni virulence and fitness factors: Role in sensing, adapting, and competing. Front. Cell. Infect. Microbiol. 2021, 10, 607704. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, K.; Wołkowicz, T.; Osek, J. Antimicrobial resistance and virulence-associated traits of Campylobacter jejuni isolated from poultry food chain and humans with diarrhea. Front. Microbiol. 2018, 9, 1508. [Google Scholar] [CrossRef] [PubMed]
- Ioannidou, V.; Ioannidis, A.; Magiorkinis, E.; Bagos, P.; Nicolaou, C.; Legakis, N.; Chatzipanagiotou, S. Multilocus sequence typing (and phylogenetic analysis) of Campylobacter jejuni and Campylobacter coli strains isolated from clinical cases in Greece. BMC Res. Notes 2013, 6, 359. [Google Scholar] [CrossRef]
- Magana, M.; Chatzipanagiotou, S.; Burriel, A.R.; Ioannidis, A. Inquiring into the gaps of Campylobacter surveillance methods. Vet. Sci. 2017, 4, 36. [Google Scholar] [CrossRef]
- Aguilar, C.; Jiménez-Marín, Á.; Martins, R.P.; Garrido, J.J. Interaction between Campylobacter and intestinal epithelial cells leads to a different proinflammatory response in human and porcine host. Vet. Immunol. Immunopathol. 2014, 162, 14–23. [Google Scholar] [CrossRef]
- Ikejiofor, O.K.; Tion, M.T.; Shima, F.K. Campylobacter: Virulence factors and pathogenesis. In Recent Advances in Bacterial Biofilm Studies—Formation, Regulation, and Eradication in Human Infections; IntechOpen: London, UK, 2024. [Google Scholar] [CrossRef]
- Kovač, J.; Čadež, N.; Stessl, B.; Stingl, K.; Gruntar, I.; Ocepek, M.; Trkov, M.; Wagner, M.; Smole Možina, S. High genetic similarity of ciprofloxacin-resistant Campylobacter jejuni in central Europe. Front. Microbiol. 2015, 6, 1169. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Q.; Tang, M.; Pu, J.; Zhang, J.; Lu, J.; Zhang, Y.; Gao, Y. Aminoglycoside resistance and possible mechanisms in Campylobacter spp. isolated from chicken and swine in Jiangsu, China. Front. Microbiol. 2021, 12, 716185. [Google Scholar] [CrossRef]
- Nunes, A.; Oleastro, M.; Alves, F.; Liassine, N.; Lowe, D.M.; Benejat, L.; Ducounau, A.; Jehanne, Q.; Borges, V.; Gomes, J.P.; et al. Recurrent Campylobacter jejuni infections with in vivo selection of resistance to macrolides and carbapenems: Molecular characterization of resistance determinants. Microbiol. Spectr. 2023, 11, e0107023. [Google Scholar] [CrossRef]
- Ghimpețeanu, O.M.; Pogurschi, E.N.; Popa, D.C.; Dragomir, N.; Drăgotoiu, T.; Mihai, O.D.; Petcu, C.D. Antibiotic use in livestock and residues in food—A public health threat: A review. Foods 2022, 11, 1430. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control (ECDC). Campylobacteriosis Annual Epidemiological Report for 2022. 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/campylobacteriosis-annual-epidemiological-report-2022 (accessed on 24 October 2024).
- EFSA; European Centre for Disease Prevention and Control (ECDC). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2021–2022. EFSA J. 2024, 22, e8583. [Google Scholar] [CrossRef]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing Disk Diffusion Test Methodology. 2024. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology (accessed on 15 March 2024).
- EUCAST. European Committee on Antimicrobial Susceptibility Testing, Clinical Breakpoints—Breakpoints and Guidance. Version 14.0. 2024. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 15 March 2024).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 20 July 2024).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Raney, B.; Paten, B.; Pham, S. Ragout—A reference-assisted assembly tool for bacterial genomes. Bioinformatics 2014, 30, i302–i309. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing genomic data quality and beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef]
- Darzentas, N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads. 2015. Available online: https://github.com/tseemann/snippy (accessed on 20 July 2024).
- Dingle, K.E.; Colles, F.M.; Wareing, D.R.; Ure, R.; Fox, A.J.; Bolton, F.E.; Bootsma, H.J.; Willems, R.J.; Urwin, R.; Maiden, M.C. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 2001, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G.; On, S.L.W.; Wang, G.; Fontanoz, S.; Lastovica, A.J.; Mandrell, R.E. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 2005, 43, 2315–2329. [Google Scholar] [CrossRef]
- Cody, A.J.; Bray, J.E.; Jolley, K.A.; McCarthy, N.D.; Maiden, M.C.J. Core genome multilocus sequence typing scheme for stable, comparative analyses of Campylobacter jejuni and C. coli human disease isolates. J. Clin. Microbiol. 2017, 55, 2086–2097. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Seemann, T. Abricate. Github. 2014. Available online: https://github.com/tseemann/abricate (accessed on 20 July 2024).
- Ortega-Sanz, I.; Barbero-Aparicio, J.A.; Canepa-Oneto, A.; Rovira, J.; Melero, B. CamPype: An open-source workflow for automated bacterial whole-genome sequencing analysis focused on Campylobacter. BMC Bioinform. 2023, 24, 291. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2019, 48, D561–D569. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Zarske, M.; Luu, H.Q.; Deneke, C.; Knüver, M.-T.; Thieck, M.; Hoang, H.T.T.; Bretschneider, N.; Pham, N.T.; Huber, I.; Stingl, K. Identification of knowledge gaps in whole-genome sequence analysis of multi-resistant thermotolerant Campylobacter spp. BMC Genom. 2024, 25, 156. [Google Scholar] [CrossRef]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef]
- Tedersoo, T.; Roasto, M.; Mäesaar, M.; Kisand, V.; Ivanova, M.; Meremäe, K. The prevalence, counts, and MLST genotypes of Campylobacter in poultry meat and genomic comparison with clinical isolates. Poult. Sci. 2022, 101, 101703. [Google Scholar] [CrossRef]
- Tang, Y.; Jiang, Q.; Tang, H.; Wang, Z.; Yin, Y.; Ren, F.; Kong, L.; Jiao, X.; Huang, J. Characterization and prevalence of Campylobacter spp. from broiler chicken rearing period to the slaughtering process in Eastern China. Front. Vet. Sci. 2020, 7, 227. [Google Scholar] [CrossRef]
- Elhadidy, M.; Miller, W.G.; Arguello, H.; Álvarez-Ordóñez, A.; Dierick, K.; Botteldoorn, N. Molecular epidemiology and antimicrobial resistance mechanisms of Campylobacter coli from diarrhoeal patients and broiler carcasses in Belgium. Transbound. Emerg. Dis. 2019, 66, 463–475. [Google Scholar] [CrossRef]
- Phu, D.H.; Wongtawan, T.; Wintachai, P.; Nhung, N.T.; Yen, N.T.P.; Carrique-Mas, J.; Turni, C.; Omaleki, L.; Blackall, P.J.; Thomrongsuwannakij, T. Molecular characterization of Campylobacter spp. isolates obtained from commercial broilers and native chickens in Southern Thailand using whole genome sequencing. Poult. Sci. 2024, 103, 103485. [Google Scholar] [CrossRef] [PubMed]
- Audu, B.J.; Norval, S.; Bruno, L.; Meenakshi, R.; Marion, M.; Forbes, K.J. Genomic diversity and antimicrobial resistance of Campylobacter spp. from humans and livestock in Nigeria. J. Biomed. Sci. 2022, 29, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, X.; Chen, X.; Li, Y.; Guo, J.; Gao, J.; Jiao, X.; Tang, Y.; Huang, J. Prevalence and genetic characterization of Campylobacter from clinical poultry cases in China. Microbiol. Spectr. 2023, 11, e00797-23. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, F.; Liu, T.; Ouyang, C.; Wang, X.; Li, M.; Huang, Z.; Huang, J.; Wang, L.; Wang, X. Identification of a multidrug resistance genomic island harboring a nonfunctional optrA gene in Campylobacter coli of chicken origin. Vet. Microbiol. 2024, 293, 110083. [Google Scholar] [CrossRef]
- Bai, Y.; Ma, J.; Li, F.; Yang, B.; Ren, X.; Wang, Y.; Hu, Y.; Dong, Y.; Wang, W.; Zhang, J.; et al. Antimicrobial resistance and genomic characterization of Campylobacter jejuni and Campylobacter coli isolated from retail chickens in Beijing, China. Microorganisms 2024, 12, 1601. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, X.; Liu, Y.; Jiang, J.; Shen, Z.; Chen, Q.; Ma, X. Multilocus sequence types and antimicrobial resistance of Campylobacter jejuni and C. coli isolates of human patients from Beijing, China, 2017–2018. Front. Microbiol. 2020, 11, 554784. [Google Scholar] [CrossRef]
- Mossong, J.; Mughini-Gras, L.; Penny, C.; Devaux, A.; Olinger, C.; Losch, S.; Cauchie, H.M.; van Pelt, W.; Ragimbeau, C. Human Campylobacteriosis in Luxembourg, 2010–2013: A case-control study combined with multilocus sequence typing for source attribution and risk factor analysis. Sci. Rep. 2016, 6, 20939. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, X.; Liu, Y.; Cui, Q.; Qin, X.; Niu, Y.; Wang, C.; Wang, T.; Chen, Q.; Ding, S.; et al. Genomic insights into the increased occurrence of Campylobacteriosis caused by antimicrobial-resistant Campylobacter coli. mBio 2022, 13, e02835-22. [Google Scholar] [CrossRef]
- Gao, F.; Tu, L.; Chen, M.; Chen, H.; Zhang, X.; Zhuang, Y.; Luo, J.; Chen, M. Erythromycin resistance of clinical Campylobacter jejuni and Campylobacter coli in Shanghai, China. Front. Microbiol. 2023, 14, 1145581. [Google Scholar] [CrossRef]
- Kim, S.Y.; An, D.; Jeong, H.; Kim, J. Antimicrobial susceptibility patterns and genetic diversity of Campylobacter spp. isolates from patients with diarrhea in South Korea. Microorganisms 2024, 12, 94. [Google Scholar] [CrossRef]
- Jehanne, Q.; Bénéjat, L.; Ducournau, A.; Aptel, J.; Pivard, M.; Gillet, L.; Jauvain, M.; Lehours, P. Increasing rates of erm(B) and erm(N) in human Campylobacter coli and Campylobacter jejuni erythromycin-resistant isolates between 2018 and 2023 in France. Antimicrob. Agents Chemother. 2025, 69, e01668-24. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.K.; Dallas, J.F.; Wilson, D.J.; Strachan, N.J.C.; McCarthy, N.D.; Jolley, K.A.; Colles, F.M.; Rotariu, O.; Ogden, I.D.; Forbes, K.J.; et al. Evolution of an agriculture-associated disease causing Campylobacter coli clade: Evidence from national surveillance data in Scotland. PLoS ONE 2010, 5, e15708. [Google Scholar] [CrossRef] [PubMed]
- Asakura, H.; Sakata, J.; Nakamura, H.; Yamamoto, S.; Murakami, S. Phylogenetic diversity and antimicrobial resistance of Campylobacter coli from humans and animals in Japan. Microbes Environ. 2019, 34, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Tegtmeyer, N.; Sharafutdinov, I.; Harrer, A.; Soltan Esmaeili, D.; Linz, B.; Backert, S. Campylobacter virulence factors and molecular host–pathogen interactions. Curr. Top. Microbiol. Immunol. 2021, 431, 169–202. [Google Scholar] [CrossRef]
- Guerry, P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007, 15, 456–461. [Google Scholar] [CrossRef]
- Kreling, V.; Falcone, F.H.; Kehrenberg, C.; Hensel, A. Campylobacter sp.: Pathogenicity factors and prevention methods—New molecular targets for innovative antivirulence drugs? Appl. Microbiol. Biotechnol. 2020, 104, 10409–10436. [Google Scholar] [CrossRef]
- Bolton, D.J. Campylobacter virulence and survival factors. Food Microbiol. 2015, 48, 99–108. [Google Scholar] [CrossRef]
- Konkel, M.E.; Talukdar, P.K.; Negretti, N.M.; Klappenbach, C.M. Taking control: Campylobacter jejuni binding to fibronectin sets the stage for cellular adherence and invasion. Front. Microbiol. 2020, 11, 564. [Google Scholar] [CrossRef]
- Pina-Mimbela, R.; Madrid, J.A.; Kumar, A.; Torrelles, J.B.; Rajashekara, G. Polyphosphate kinases modulate Campylobacter jejuni outer membrane constituents and alter its capacity to invade and survive in intestinal epithelial cells in vitro. Emerg. Microbes Infect. 2015, 4, e77. [Google Scholar] [CrossRef]
- Radomska, K.A.; Wösten, M.M.S.M.; Ordoñez, S.R.; Wagenaar, J.A.; van Putten, J.P.M. Importance of Campylobacter jejuni FliS and FliW in flagella biogenesis and flagellin secretion. Front. Microbiol. 2017, 8, 1060. [Google Scholar] [CrossRef]
- Dias, T.S.; Panzenhagen, P.; Figueira, A.A.; Costa, G.A.; Rossi, D.A.; de Melo, R.T.; Pereira, V.L.A.; de Aquino, M.H.C. Genomic characterisation of Campylobacter jejuni Cj26: A high-level ciprofloxacin/erythromycin-resistant strain isolated from a poultry carcass in southern Brazil. J. Glob. Antimicrob. Resist. 2023, 34, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Karki, A.B.; Khatri, B.; Fakhr, M.K. Transcriptome analysis of Campylobacter jejuni and Campylobacter coli during cold stress. Pathogens 2023, 12, 960. [Google Scholar] [CrossRef] [PubMed]
- Admasie, A.; Wei, X.; Johnson, B.; Burns, L.; Pawar, P.; Aurand-Cravens, A.; Voloshchuk, O.; Dudley, E.G.; Sisay Tessema, T.; Zewdu, A.; et al. Genomic diversity of Campylobacter jejuni and Campylobacter coli isolated from the Ethiopian dairy supply chain. PLoS ONE 2024, 19, e0305581. [Google Scholar] [CrossRef] [PubMed]
- Marchant, J.; Wren, B.; Ketley, J. Exploiting genome sequence: Predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 2002, 10, 155–159. [Google Scholar] [CrossRef]
- Zautner, A.E.; Tareen, A.M.; Groß, U.; Lugert, R. Chemotaxis in Campylobacter jejuni. Eur. J. Microbiol. Immunol. 2012, 2, 24–31. [Google Scholar] [CrossRef]
- Wösten, M.M.S.M.; Wagenaar, J.A.; van Putten, J.P.M. The FlgS/FlgR Two-component signal transduction system regulates the fla Regulon in Campylobacter jejuni. J. Biol. Chem. 2004, 279, 16214–16222. [Google Scholar] [CrossRef]
- Salah Ud-Din, A.I.M.; Roujeinikova, A. Flagellin glycosylation with pseudaminic acid in Campylobacter and Helicobacter: Prospects for development of novel therapeutics. Cell Mol. Life Sci. 2018, 75, 1163–1178. [Google Scholar] [CrossRef]
- Guerry, P.; Doig, P.; Alm, R.A.; Burr, D.H.; Kinsella, N.; Trust, T.J. Identification and characterization of genes required for post-translational modification of Campylobacter coli VC167 flagellin. Mol. Microbiol. 1996, 19, 369–378. [Google Scholar] [CrossRef]
- Guerry, P.; Ewing, C.P.; Schirm, M.; Lorenzo, M.; Kelly, J.; Pattarini, D.; Majam, G.; Thibault, P.; Logan, S. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 2006, 60, 299–311. [Google Scholar] [CrossRef]
- Oldfield, N.J.; Moran, A.P.; Millar, L.A.; Prendergast, M.M.; Ketley, J.M. Characterization of the Campylobacter jejuni heptosyltransferase ii gene, waaf, provides genetic evidence that extracellular polysaccharide is lipid a core independent. J. Bacteriol. 2002, 184, 2100–2107. [Google Scholar] [CrossRef]
- Taylor, P.L.; Blakely, K.M.; de Leon, G.P.; Walker, J.R.; McArthur, F.; Evdokimova, E.; Zhang, K.; Valvano, M.A.; Wright, G.D.; Junop, M.S. Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants. J. Biol. Chem. 2008, 283, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Klena, J.D.; Gray, S.A.; Konkel, M.E. Cloning, sequencing, and characterization of the lipopolysaccharide biosynthetic enzyme heptosyltransferase I gene (waaC) from Campylobacter jejuni and Campylobacter coli. Gene 1998, 222, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Karlyshev, A.V.; Champion, O.L.; Churcher, C.; Brisson, J.-R.; Jarrell, H.C.; Gilbert, M.; Brochu, D.; St Michael, F.; Li, J.; Wakarchuk, W.W.; et al. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 2005, 55, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, N.; Mangan, J.A.; Laing, K.G.; Hinds, J.; Linton, D.; Al-Ghusein, H.; Barrell, B.G.; Parkhill, J.; Stoker, N.G.; Karlyshev, A.V.; et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001, 11, 1706–1715. [Google Scholar] [CrossRef]
- Parker, C.T.; Horn, S.T.; Gilbert, M.; Miller, W.G.; Woodward, D.L.; Mandrell, R.E. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 2005, 43, 2771–2781. [Google Scholar] [CrossRef]
- Poudel, S.; Li, T.; Chen, S.; Zhang, X.; Cheng, W.H.; Sukumaran, A.T.; Kiess, A.S.; Zhang, L. Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter isolated from broilers and broiler meat raised without antibiotics. Microbiol. Spectr. 2022, 10, e0025122. [Google Scholar] [CrossRef]
- Matsunami, H.; Barker, C.S.; Yoon, Y.H.; Wolf, M.; Samatey, F.A. Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat. Commun. 2016, 7, 13425. [Google Scholar] [CrossRef]
- Reid, A.N.; Pandey, R.; Palyada, K.; Naikare, H.; Stintzi, A. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl. Environ. Microbiol. 2008, 74, 1583–1597. [Google Scholar] [CrossRef]
- Inoue, T.; Barker, C.S.; Matsunami, H.; Aizawa, S.I.; Samatey, F.A. The FlaG regulator is involved in length control of the polar flagella of Campylobacter jejuni. Microbiology 2018, 164, 740–750. [Google Scholar] [CrossRef]
- Freitag, C.M.; Strijbis, K.; van Putten, J.P.M. Host cell binding of the flagellar tip protein of Campylobacter jejuni. Cell Microbiol. 2017, 19, e12714. [Google Scholar] [CrossRef]
- Goon, S.; Kelly, J.F.; Logan, S.M.; Ewing, C.P.; Guerry, P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 2003, 50, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Song, W.S.; Park, M.; Ki, D.U.; Yoon, S. Structural analysis of the pseudaminic acid synthase PseI from Campylobacter jejuni. Biochem. Biophys. Res. Commun. 2022, 635, 252–258. [Google Scholar] [CrossRef] [PubMed]
- The National Antimicrobial Resistance Monitoring System. Global Resistome Data. 2025. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/global-resistome-data (accessed on 20 April 2025).
- Li, W.; Atkinson, G.C.; Thakor, N.S.; Allas, U.; Lu, C.C.; Chan, K.Y.; Tenson, T.; Schulten, K.; Wilson, K.S.; Hauryliuk, V.; et al. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O). Nat. Commun. 2013, 4, 1477. [Google Scholar] [CrossRef] [PubMed]
- Benites, C.; Anampa, D.; Torres, D.; Avalos, I.; Rojas, M.; Conte, C.; Lázaro, C. Prevalence, tetracycline resistance and tet(O) gene identification in pathogenic Campylobacter strains isolated from chickens in retail markets of Lima, Peru. Antibiotics 2022, 11, 1580. [Google Scholar] [CrossRef]
- Casagrande Proietti, P.; Guelfi, G.; Bellucci, S.; De Luca, S.; Di Gregorio, S.; Pieramati, C.; Franciosini, M.P. Beta-lactam resistance in Campylobacter coli and Campylobacter jejuni chicken isolates and the association between blaOXA-61 gene expression and the action of β-lactamase inhibitors. Vet. Microbiol. 2020, 241, 108553. [Google Scholar] [CrossRef]
- Gomes, C.N.; Campioni, F.; Barker, D.O.R.; Che, E.V.; Duque, S.S.; Taboada, E.N.; Falcão, J.P. Antimicrobial resistance genotypes and phenotypes of Campylobacter coli isolated from different sources over a 16-year period in Brazil. J. Glob. Antimicrob. Resist. 2023, 33, 109–113. [Google Scholar] [CrossRef]
- Marotta, F.; Janowicz, A.; Romantini, R.; Di Marcantonio, L.; Di Timoteo, F.; Romualdi, T.; Zilli, K.; Barco, L.; D’Incau, M.; Mangone, I.; et al. Genomic and antimicrobial surveillance of Campylobacter population in Italian poultry. Foods 2023, 12, 2919. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Zhang, Q.; Shen, J. Antimicrobial resistance in Campylobacter spp. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Cobo-Díaz, J.F.; González del Río, P.; Álvarez-Ordóñez, A. Whole Resistome Analysis in Campylobacter jejuni and C. coli Genomes Available in Public Repositories. Front. Microbiol. 2021, 12, 662144. [Google Scholar] [CrossRef]
- Ghatak, S.; Milton, A.A.P.; Das, S.; Momin, K.M.; Srinivas, K.; Pyngrope, D.A.; Priya, G.B. Campylobacter coli of porcine origin exhibits an open pan-genome within a single clonal complex: Insights from comparative genomic analysis. Front. Cell. Infect. Microbiol. 2024, 14, 1449856. [Google Scholar] [CrossRef]
- Hormeño, L.; Palomo, G.; Borge, C.; Vadillo, S.; Píriz, S.; Domínguez, L.; Campos, M.J.; Quesada, A. ant(6)-I genes encoding aminoglycoside o-nucleotidyltransferases are widely spread among streptomycin resistant strains of Campylobacter jejuni and Campylobacter coli. Front. Microbiol. 2018, 9, 2515. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Shen, Z.; Wang, Y.; Deng FLiu, D.; Naren, G.; Dai, L.; Su, C.; Wang, B.; Wang, S.; Wu, C.; et al. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. mBio 2016, 7, e01543-16. [Google Scholar] [CrossRef] [PubMed]
- Alfredson, D.A.; Korolik, V. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 2007, 277, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Portes, A.B.; Panzenhagen, P.; dos Santos, A.M.P.; de Jesus, A.C.S.; Ochioni, A.C.; Duque, S.S.; Aburjaile, F.F.; Brenig, B.; Azevedo, V.; Conte Junior, C.A. Draft genomes of multidrug-resistant Campylobacter jejuni and Campylobacter coli strains from Brazil representing novel sequence types. Microbiol. Resour. Announc. 2024, 13, e00524. [Google Scholar] [CrossRef]
- Pinto-Alphandary, H.; Mabilat, C.; Courvalin, P. Emergence of aminoglycoside resistance genes aadA and aadE in the genus Campylobacter. Antimicrob. Agents Chemother. 1990, 34, 1294–1296. [Google Scholar] [CrossRef]
- Lin, J.; Michel, L.O.; Zhang, Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002, 46, 2124–2131. [Google Scholar] [CrossRef]
- Klenotic, P.A.; Moseng, M.A.; Morgan, C.E.; Yu, E.W. Structural and functional diversity of resistance-nodulation-cell division transporters. Chem. Rev. 2021, 121, 5378–5416. [Google Scholar] [CrossRef]
- Bravo, V.; Katz, A.; Porte, L.; Weitzel, T.; Varela, C.; Gonzalez-Escalona, N.; Blondel, C.J. Genomic analysis of the diversity, antimicrobial resistance and virulence potential of clinical Campylobacter jejuni and Campylobacter coli strains from Chile. PLoS Neglected Trop. Dis. 2021, 15, e0009207. [Google Scholar] [CrossRef]
- Grinnage-Pulley, T.; Zhang, Q. Genetic basis and functional consequences of differential expression of the CmeABC efflux pump in Campylobacter jejuni Isolates. PLoS ONE 2015, 10, e0131534. [Google Scholar] [CrossRef]
- Lin, J.; Akiba, M.; Sahin, O.; Zhang, Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 2005, 49, 1067–1075. [Google Scholar] [CrossRef]
- Dai, L.; Sahin, O.; Grover, M.; Zhang, Q. New and alternative strategies for the prevention, control, and treatment of antibiotic-resistant Campylobacter. Transl. Res. 2020, 223, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wang, C.; Ye, Y.; Liu, Y.; Wang, A.; Li, Y.; Zhou, X.; Pan, H.; Zhang, J.; Xu, X. Molecular identification of multidrug-resistant Campylobacter species from diarrheal patients and poultry meat in Shanghai, China. Front. Microbiol. 2018, 9, 1642. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic | Assessment * |
|---|---|
| Ciprofloxacin | R |
| Tetracycline | R |
| Erythromycin | R |
| Azithromycin | R |
| Clarithromycin | R |
| Doxycycline | R |
| Gene | Function | Resistance | Ref. |
|---|---|---|---|
| tet(O) | Tetracycline resistance protein TetO | Tetracycline, doxycycline, minocycline | [61,94,103,106,107,108] |
| blaOXA-61 | Beta-lactamase | Beta-lactam | [104,105,106,108] |
| blaOXA-489 | Beta-lactamase | [108,109] | |
| blaOXA-605 | Beta-lactamase | [61,81,106] | |
| blaOXA-450 | Beta-lactamase | [106] | |
| aad9 | Aminoglycoside nucleotidyltransferase | Aminoglycoside | [61,106] |
| ant(6)-Ia | Aminoglycoside nucleotidyltransferase | [79,107,110] | |
| aadE | Aminoglycoside 6-adenylyltransferase | [61,106,107] | |
| cmeA | RND efflux system membrane fusion protein CmeA | Multidrug-resistance | [74,106,107,111,112] |
| cmeB | Multidrug efflux RND transporter permease subunit CmeB | ||
| cmeC | Multidrug efflux transporter outer membrane subunit CmeC | ||
| cmeD | Multidrug efflux RND transporter outer membrane subunit CmeD | Multidrug-resistance | [74,107,112] |
| cmeE | Multidrug efflux RND transporter periplasmic adaptor subunit CmeE | ||
| cmeF | Multidrug efflux RND transporter permease subunit CmeF | ||
| gyrA T86I | DNA gyrase subunit A (T86I) | Fluoroquinolone | [61,113] |
| 23S A2075G | 23S rRNA mutation (A2075G) | Macrolide | [61,113] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadimitriou, K.; Ioannidis, A.; Slavko, A.; Chronopoulou, G.; Marmaras, N.; Pangalis, A.; Olntasi, E.; Vassilaki, N.; Koufogeorgou, E.I.; Kolida, I.; et al. Genome Analysis of the Multidrug-Resistant Campylobacter coli BCT3 of the Sequence Type (ST) 872 Isolated from a Pediatric Diarrhea Case. Microorganisms 2025, 13, 1420. https://doi.org/10.3390/microorganisms13061420
Papadimitriou K, Ioannidis A, Slavko A, Chronopoulou G, Marmaras N, Pangalis A, Olntasi E, Vassilaki N, Koufogeorgou EI, Kolida I, et al. Genome Analysis of the Multidrug-Resistant Campylobacter coli BCT3 of the Sequence Type (ST) 872 Isolated from a Pediatric Diarrhea Case. Microorganisms. 2025; 13(6):1420. https://doi.org/10.3390/microorganisms13061420
Chicago/Turabian StylePapadimitriou, Konstantinos, Anastasios Ioannidis, Aleksandra Slavko, Genovefa Chronopoulou, Nektarios Marmaras, Anastasia Pangalis, Elisavet Olntasi, Niki Vassilaki, Efthymia Ioanna Koufogeorgou, Iris Kolida, and et al. 2025. "Genome Analysis of the Multidrug-Resistant Campylobacter coli BCT3 of the Sequence Type (ST) 872 Isolated from a Pediatric Diarrhea Case" Microorganisms 13, no. 6: 1420. https://doi.org/10.3390/microorganisms13061420
APA StylePapadimitriou, K., Ioannidis, A., Slavko, A., Chronopoulou, G., Marmaras, N., Pangalis, A., Olntasi, E., Vassilaki, N., Koufogeorgou, E. I., Kolida, I., Theodoridis, D., & Chatzipanagiotou, S. (2025). Genome Analysis of the Multidrug-Resistant Campylobacter coli BCT3 of the Sequence Type (ST) 872 Isolated from a Pediatric Diarrhea Case. Microorganisms, 13(6), 1420. https://doi.org/10.3390/microorganisms13061420







