Adaptive Laboratory Evolution of a Microbial Consortium Enhancing Non-Protein Nitrogen Assimilation for Feed Protein Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Wheat Straw, and Medium Used in the Experiment
2.2. Adaptive Laboratory Evolution of the Microbial Consortium
2.3. SSF of Wheat Straw by the Microbial Consortium
2.3.1. Optimization of Fermentation Conditions
2.3.2. Pretreatment of Wheat Straw
2.4. Scale-Up Fermentation of the Wheat Straw
2.5. Analysis Methods
2.6. Statistical Analysis
3. Results and Discussion
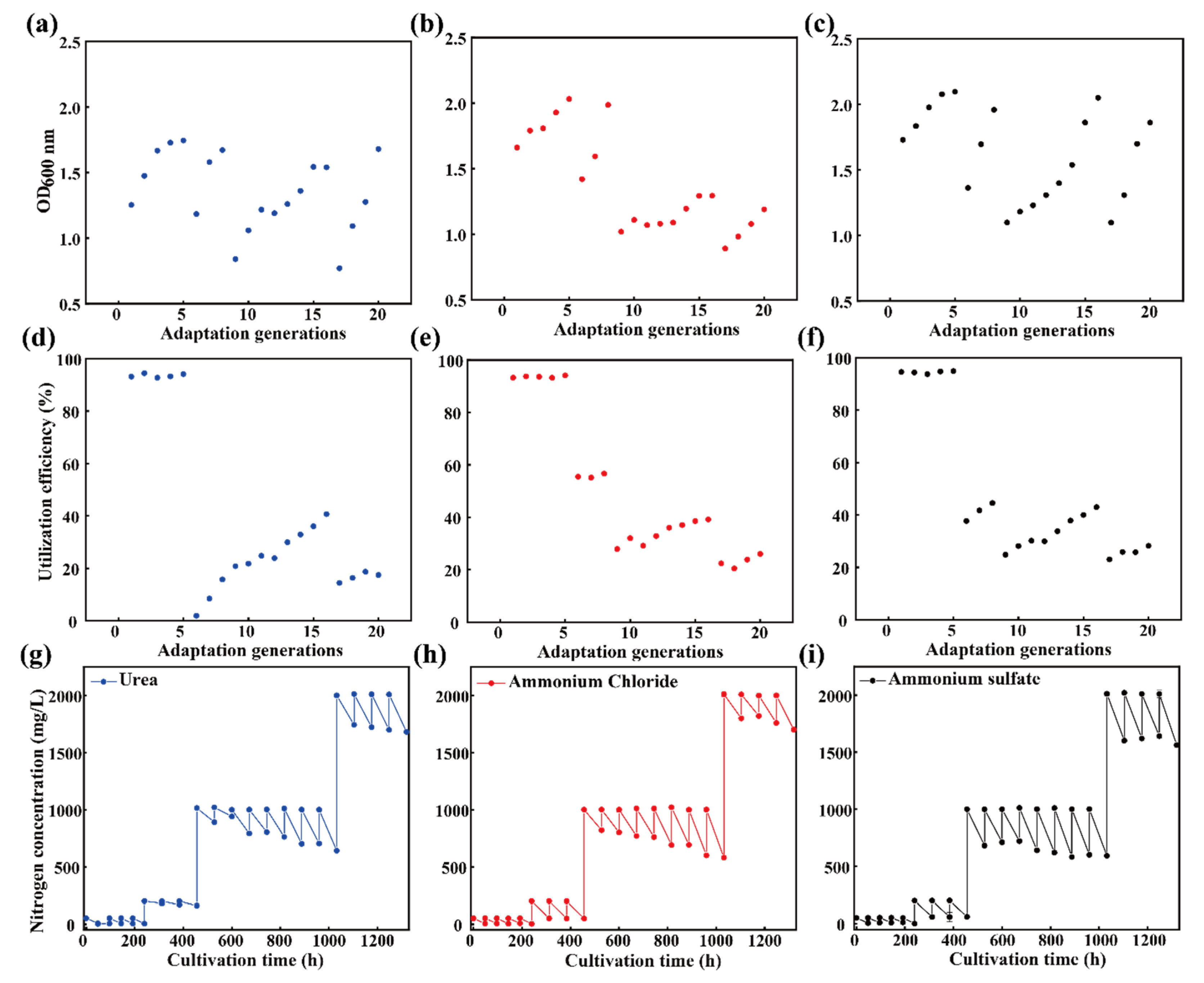
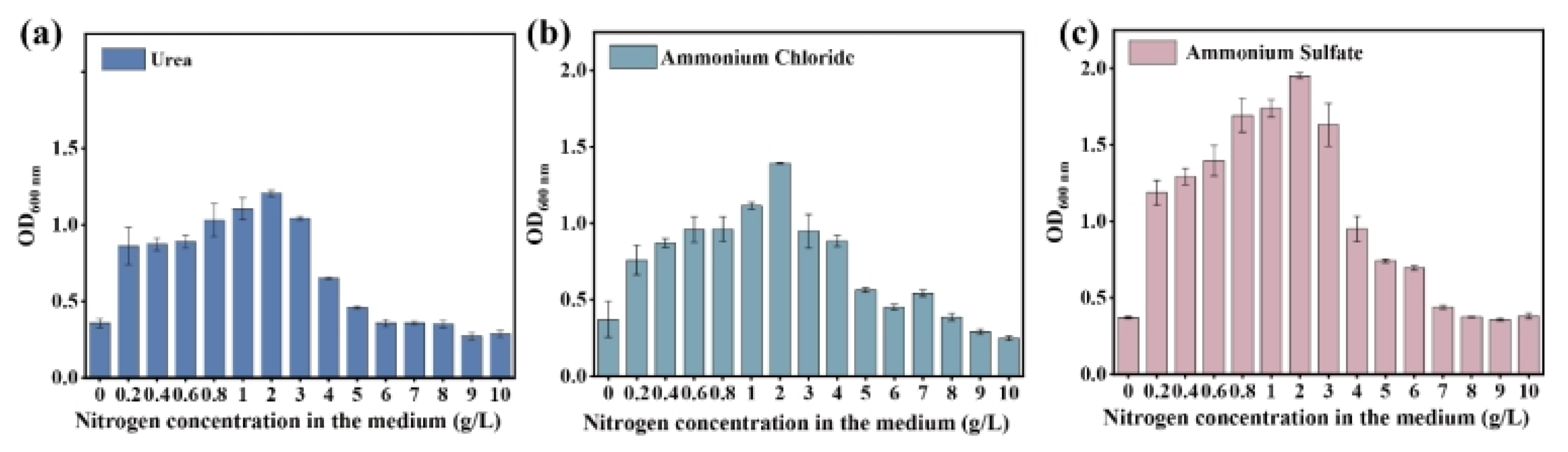
3.1. Adaptive Laboratory Evolution of a Microbial Consortium Enhancing NPN Assimilation
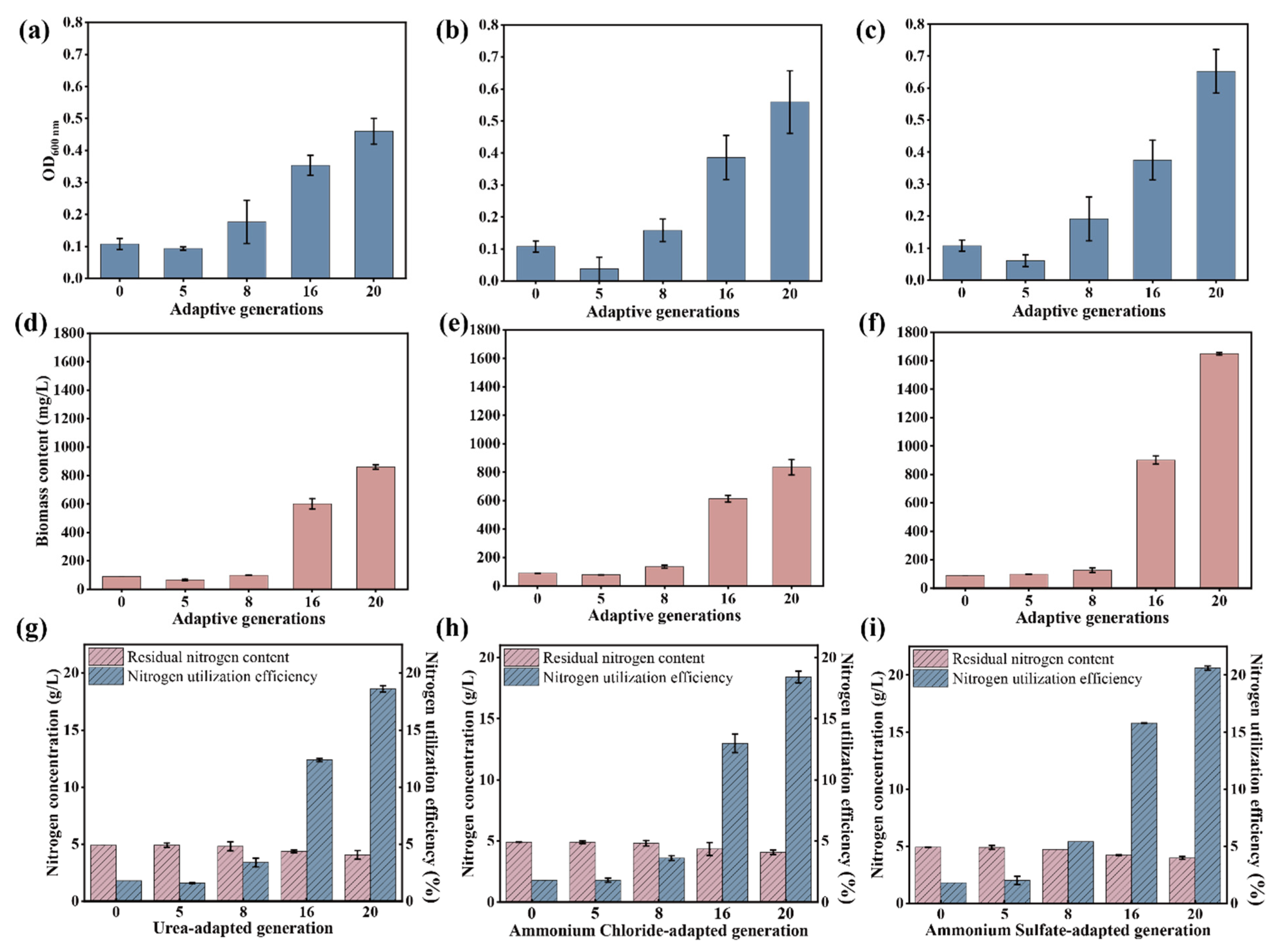
3.2. NPN Assimilation Capacity of the Evolved Microbial Consortium
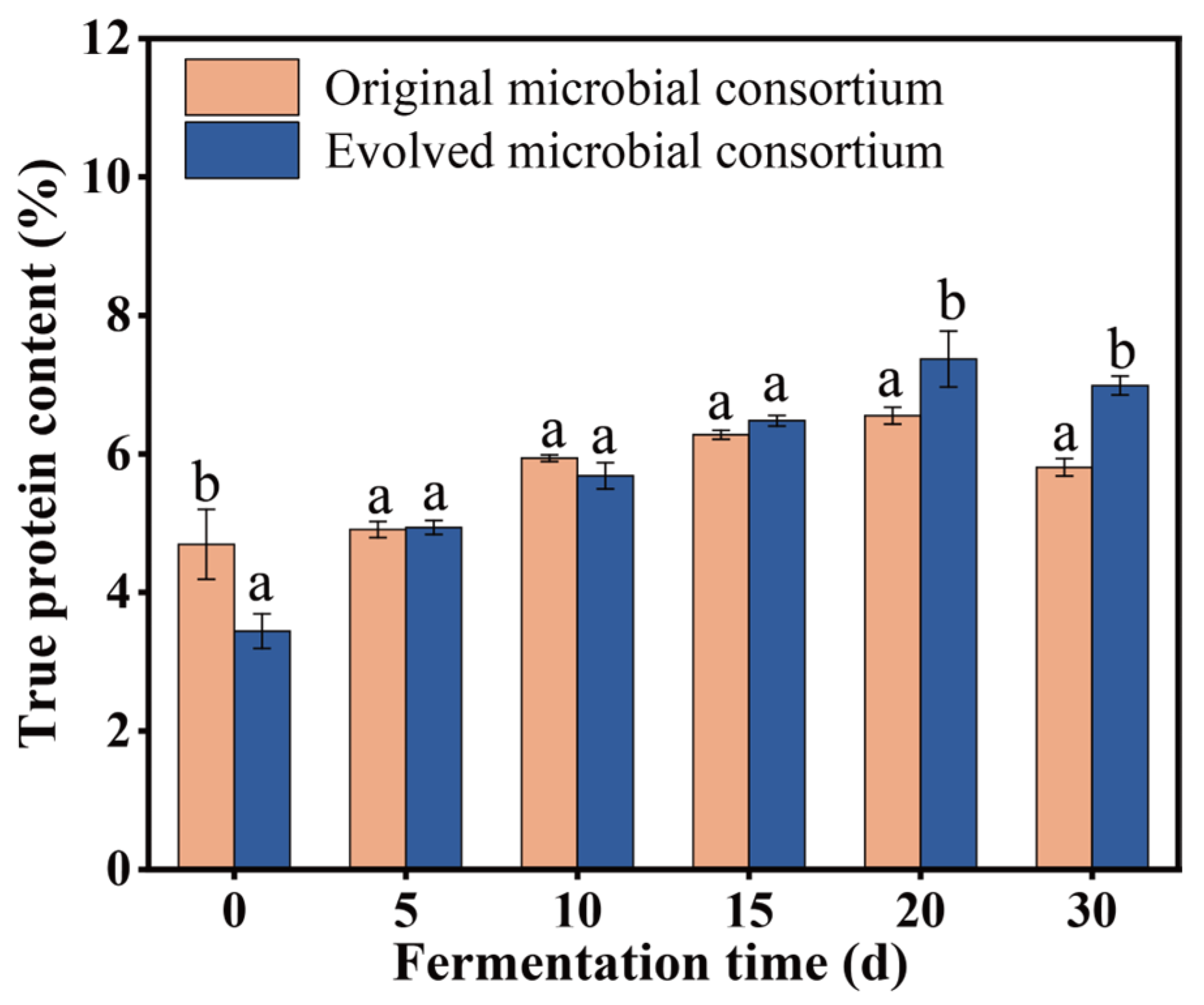
3.3. Production of Feed Protein by Using the Original and the Evolved Microbial Consortium
3.4. Optimization of SSF Conditions to Improve the True Protein Content of the Wheat Straw
3.4.1. The Effect of Alkali Pretreatment
3.4.2. The Effect of Fermentation Time, Inoculation Amount, Nitrogen Source Addition Amount, Initial Moisture Content, Temperature, and Substrate Sterilization
3.5. Scale-Up of the SSF of the Wheat Straw to Produce Feed Protein
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niehues, J.; McElroy, C.; Croon, A.; Jan, P.; Martin, F.; Florian, S. Bacterial Lighthouses—Real-Time Detection of Yersinia enterocolitica by Quorum Sensing. Biosensors 2021, 11, 517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, X.; Gao, L. New strategy for the biosynthesis of alternative feed protein: Single-cell protein production from straw-based biomass. GCB Bioenergy 2024, 16, e13120. [Google Scholar] [CrossRef]
- Wang, R.; Lin, F.; Feng, K. Soybean overweight shock (SOS): The impact of trade liberalization in China on overweight prevalence. China Econ. Rev. 2024, 87, 102224. [Google Scholar] [CrossRef]
- Kong, S.; Wang, S.; He, Y.; Wang, N.; Wang, Z.; Weng, L.; Liu, D.; Zhao, X.; Chen, J.; Xu, J.; et al. Three-stage solid-state fermentation technology for Distillers’ Grain feed protein based on different microorganisms considering oxygen requirements. Fermentation 2024, 10, 550. [Google Scholar] [CrossRef]
- Li, X.; Xu, W.; Yang, J.; Zhao, H.; Pan, C.; Ding, X.; Zhang, Y. Effects of applying lactic acid bacteria to the fermentation on a mixture of corn steep liquor and air-dried rice straw. Anim. Nutr. 2016, 2, 229–233. [Google Scholar] [CrossRef]
- Wadhave, A.A.; Jadhav, A.I.; Arsul, V.A. Plant proteins applications: A review. World J. Pharm. Pharm. Sci. 2014, 3, 702–712. [Google Scholar]
- Roukas, T.; Kotzekidou, P. From food industry wastes to second generation bioethanol: A review. Rev. Environ. Sci. Bio/Technol. 2022, 21, 299–329. [Google Scholar] [CrossRef]
- Mohammadi, M.; Alian, M.; Dale, B.; Ubanwa, B.; Balan, V. Multifaced application of AFEX-pretreated biomass in producing second-generation biofuels, ruminant animal feed, and value-added bioproducts. Biotechnol. Adv. 2024, 72, 108341. [Google Scholar] [CrossRef]
- Khilji, S.A.; Sajid, Z.A.; Fayyaz, S.; Shah, A.A.; Shah, A.N.; Rauf, M.; Arif, M.; Yang, S.H.; Fiaz, S. Fulvic acid alleviates paper sludge toxicity in canola (Brassica napus L.) by reducing Cr, Cd, and Pb uptake. Front. Plant Sci. 2022, 13, 874723. [Google Scholar] [CrossRef]
- Kratky, L.; Jirout, T. Biomass size reduction machines for enhancing biogas production. Chem. Eng. Technol. 2011, 34, 391–399. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, Y.; Luo, L.; Zhang, H.; Liao, Y.; Gou, C. Enhancement of the nutritional value of fermented corn stover as ruminant feed using the fungi Pleurotus spp. Sci. Rep. 2021, 11, 11961. [Google Scholar]
- Wang, Z.; He, X.; Yan, L.; Wang, J.; Hu, X.; Dun, Q.; Zhang, H. Enhancing enzymatic hydrolysis of corn stover by twin-screw extrusion pretreatment. Ind. Crops Prod. 2020, 143, 111960. [Google Scholar] [CrossRef]
- Mancini, G.; Papirio, S.; Lens, P.N.L. Increased biogas production from wheat straw by chemical pretreatments. Renew. Energy 2018, 119, 608–614. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Zhao, J.; Esposito, G. Solid-state fermentation of ammoniated corn straw to produce feed protein and toxicological assessment of the product. Environ. Sci. Pollut. Res. 2020, 27, 13895–13901. [Google Scholar] [CrossRef]
- Kumssa, G.; Eshetu, M. On-farm effect of urea treated straw and urea molasses block on cross-bred dairy cows at Lume District, Ethiopia. Agric. Res. Technol. 2018, 17, 82–85. [Google Scholar] [CrossRef]
- Zhao, L.; Ren, L.; Zhou, Z.; Meng, Q.; Huo, Y.; Wang, F. Improving ruminal degradability and energetic values of bamboo shoot shell using chemical treatments. Anim. Sci. J. 2016, 87, 896–903. [Google Scholar] [CrossRef]
- Shanakhat, H.; Sorrentino, A.; Raiola, A.; Raiola, A.; Romano, A.; Masi, P.; Cavella, S. Current methods for mycotoxins analysis and innovative strategies for their reduction in cereals: An overview. J. Sci. Food Agric. 2018, 98, 4003–4013. [Google Scholar] [CrossRef]
- Xing, D.; Magdouli, S.; Zhang, J.; Koubaa, A. Microbial remediation for the removal of inorganic contaminants from treated wood: Recent trends and challenges. Chemosphere 2020, 258, 127429. [Google Scholar] [CrossRef]
- Li, B.; Zhao, C.; Sun, Q.; Chen, K.; Zhao, X.; Xu, L.; Yang, Z.; Peng, H. Effects of ammonification–steam explosion pretreatment on the production of true protein from rice straw during solid-state fermentation. Sustainability 2023, 15, 5964. [Google Scholar] [CrossRef]
- Ye, Y.; Cai, Y.; Wang, F.; He, Y.; Yang, Y.; Guo, Z.; Liu, M.; Ren, H.; Wang, S.; Liu, D.; et al. Industrial microbial technologies for feed protein production from non-protein nitrogen. Microorganisms 2025, 13, 742. [Google Scholar] [CrossRef]
- Li, X.; Han, C.; Li, W.; Chen, G.; Wang, L. Insights into the cellulose degradation mechanism of the thermophilic fungus Chaetomium thermophilum based on integrated functional omics. Biotechnol. Biofuels 2020, 13, 143. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, K.; Liao, H.; Hector, S.B.; Shi, X.; Li, J.; Liu, B.; Xu, T.; Tong, C.; Liu, X.; et al. Genomic and secretomic insight into lignocellulolytic system of an endophytic bacterium Pantoea ananatis Sd-1. Biotechnol. Biofuels 2016, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Ma, Y.; Chang, H.; Li, B.; Ding, M.; Yuan, Y. Evaluation of PET degradation using artificial microbial consortia. Front. Microbiol. 2021, 12, 778828. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Awasthi, M.K.; Liu, Y.; Cheng, Q.; Qu, J.; Sun, Y. Studies on the degradation of corn straw by combined bacterial cultures. Bioresour. Technol. 2021, 320, 124174. [Google Scholar] [CrossRef]
- Karuppiah, V.; Zhixiang, L.; Liu, H.; Murugappan, V.; Kumaran, S.; Anahas, A.M.P.; Chen, J. Co-cultivation of T. asperellum GDFS1009 and B. amyloliquefaciens 1841: Strategy to regulate the production of ligno-cellulolytic enzymes for the lignocellulose biomass degradation. J. Environ. Manag. 2022, 301, 113833. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Solem, C.; Lu, T.; Jensen, P.R. Harnessing lactic acid bacteria in synthetic microbial consortia. Trends Biotechnol. 2022, 40, 8–11. [Google Scholar] [CrossRef]
- Hou, Y.; Zeng, W.; Hou, M.; Wang, Z.; Luo, Y.; Lei, G.; Zhou, B.; Huang, J. Responses of the soil microbial community to salinity stress in maize fields. Biology 2021, 10, 1114. [Google Scholar] [CrossRef]
- Chen, J.; Cai, Y.; Wang, Z.; Xu, Z.; Zhuang, W.; Liu, D.; Lv, Y.; Wang, S.; Xu, J.; Ying, H. Solid-state fermentation of corn straw using synthetic microbiome to produce fermented feed: The feed quality and conversion mechanism. Sci. Total Environ. 2024, 920, 171034. [Google Scholar] [CrossRef]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome engineering: Synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2021, 39, 244–261. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.K.; Jeon, J.; Harris, A.; Lee, Y. Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome 2020, 8, 20. [Google Scholar] [CrossRef]
- Ruan, Z.; Chen, K.; Cao, W.; Meng, L.; Yang, B.; Xu, M.; Xing, Y.; Li, P.; Freilich, S.; Chen, C. Engineering natural microbiomes toward enhanced bioremediation by microbiome modeling. Nat. Commun. 2024, 15, 4694. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cai, Y.; Wang, Z.; Xu, Z.; Li, J.; Ma, X.; Zhuang, W.; Liu, D.; Wang, S.; Song, A.; et al. Construction of a synthetic microbial community based on multiomics linkage technology and analysis of the mechanism of lignocellulose degradation. Bioresour. Technol. 2023, 389, 129799. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Pal, D.B.; Alkhanani, M.F.; Almalki, A.H.; Areeshi, M.Y.; Haque, S.; Srivastava, N. Prospects of soil microbiome application for lignocellulosic biomass degradation: An overview. Sci. Total Environ. 2022, 838, 155966. [Google Scholar] [CrossRef]
- Wang, G.; Li, Q.; Zhang, Z.; Yin, X.; Wang, B.; Yang, X. Recent progress in adaptive laboratory evolution of industrial microorganisms. J. Ind. Microbiol. Biotechnol. 2023, 50, kuac023. [Google Scholar] [CrossRef]
- Min, B.; Yoo, D.A.; Lee, Y.; Seo, M.; Kim, H. Complete Genomic Analysis of Enterococcus faecium heat-resistant strain developed by two-step adaptation laboratory evolution method. Front. Bioeng. Biotechnol. 2020, 8, 828. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; de Groot, A.; Boeren, S.; Abee, T.; Smid, E. Lactococcus lactis mutants obtained from laboratory evolution showed elevated vitamin K2 content and enhanced resistance to oxidative stress. Front. Microbiol. 2021, 12, 746770. [Google Scholar] [CrossRef]
- Gan, Y.; Bai, M.; Lin, X.; Liu, K.; Huang, B.; Jiang, X.; Liu, Y.; Gao, C. Improvement of macrolactins production by the genetic adaptation of Bacillus siamensis A72 to saline stress via adaptive laboratory evolution. Microb. Cell Factories 2022, 21, 147. [Google Scholar] [CrossRef]
- Lee, S.R.; Kim, P. Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 2020, 30, 793. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Z.; Wang, N.; Wang, S.; Zeng, S.; Xu, Z.; Liu, D.; Zhao, X.; Liu, F.; Xu, J.; et al. Efficient conversion of corn straw to feed protein through solid-state fermentation using a thermophilic microbial consortium. Waste Manag. 2025, 194, 298–308. [Google Scholar] [CrossRef]
- Du, R.; Li, C.; Lin, W.; Lin, C.; Yan, J. Domesticating a bacterial consortium for efficient lignocellulosic biomass conversion. Renew. Energy 2022, 189, 359–368. [Google Scholar] [CrossRef]
- Chen, R.; Huang, J.; Li, X.; Yang, C.; Wu, X. Functional characterization of an efficient ibuprofen-mineralizing bacterial consortium. J. Hazard. Mater. 2023, 447, 130751. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.M.; Barbano, D.M. Kjeldahl nitrogen analysis as a reference method for protein determination in dairy products. J. AOAC Int. 1999, 82, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Hong, C.; Zheng, B.; Jiang, D.; Qin, W. Improving enzymatic digestibility of wheat straw pretreated by a cellulase-free xylanase-secreting Pseudomonas boreopolis G22 with simultaneous production of bioflocculants. Biotechnol. Biofuels 2018, 11, 250. [Google Scholar] [CrossRef]
- Liu, J.; Wang, S.; Wang, Z.; Shen, C.; Liu, D.; Shen, X.; Weng, L.; He, Y.; Wang, S.; Wang, J. Pretreatment of Luzhou distiller’s grains for feed protein production using crude enzymes produced by a synthetic microbial consortium. Bioresour. Technol. 2023, 390, 129852. [Google Scholar] [CrossRef]
- Salvachúa, D.; Martínez, A.T.; Tien, M.; López-Lucendo, M.; García, M.; Ríos, V.; Martínez, M.; Prieto, A. Differential proteomic analysis of the secretome of Irpex lacteus and other white-rot fungi during wheat straw pretreatment. Biotechnol. Biofuels 2013, 6, 115. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, X.; Zahoor Khan, M.; Xiao, J.; Liu, S.; Wang, J.; Maswayi Alugongo, G.; Cao, Z. Biodegradation and hydrolysis of rice straw with corn steep liquor and urea-alkali pretreatment. Front. Nutr. 2022, 9, 989239. [Google Scholar] [CrossRef]
- Dong, M.; Wang, S.; Xu, F.; Wang, J.; Yang, N.; Li, Q.; Chen, J.; Li, W. Pretreatment of sweet sorghum straw and its enzymatic digestion: Insight into the structural changes and visualization of hydrolysis process. Biotechnol. Biofuels 2019, 12, 276. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.Y.; Teo, K.T.K.; Tan, M.K.; Tham, H.J. Fermentation process control and optimization. Chem. Eng. Technol. 2022, 45, 1731–1747. [Google Scholar] [CrossRef]
- Fang, J.; Liu, Y.; Huan, C.C.; Xu, L.; Ji, G.; Yan, Z. Comparison of poly-γ-glutamic acid production between sterilized and non-sterilized solid-state fermentation using agricultural waste as substrates. J. Clean. Prod. 2020, 255, 120248. [Google Scholar] [CrossRef]
- Tian, Y.; Li, J.; Meng, J.; Li, J. High-yield production of single-cell protein from starch processing wastewater using co-cultivation of yeasts. Bioresour. Technol. 2023, 370, 128527. [Google Scholar] [CrossRef]
- Buchinski, M.; Carley, M.; Olufemi, O.; Anna, K.; Daniel, A. PSII-10 ammonium phosphate is an appropriate nitrogen source in nitrogen-limiting diets for growing pigs. J. Anim. Sci. 2023, 101, 285. [Google Scholar] [CrossRef]
- Zhang, Y.; Ye, Q.; Liu, B.; Feng, Z.; Zhang, X.; Liu, M. Fermenting distiller’s grains by the domesticated microbial consortium to release ferulic acid. J. Agric. Food Chem. 2024, 72, 9259–9267. [Google Scholar] [CrossRef] [PubMed]

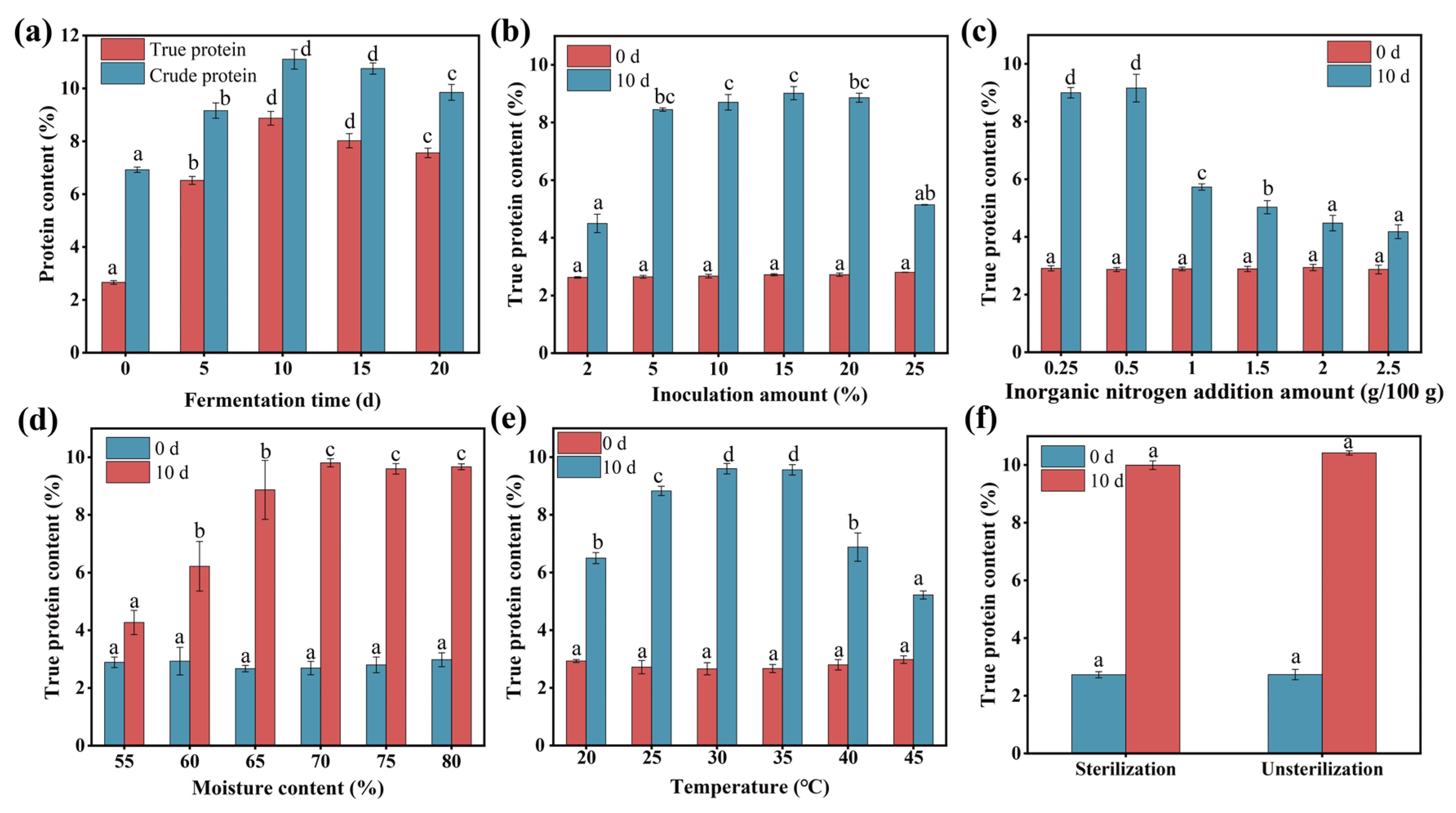
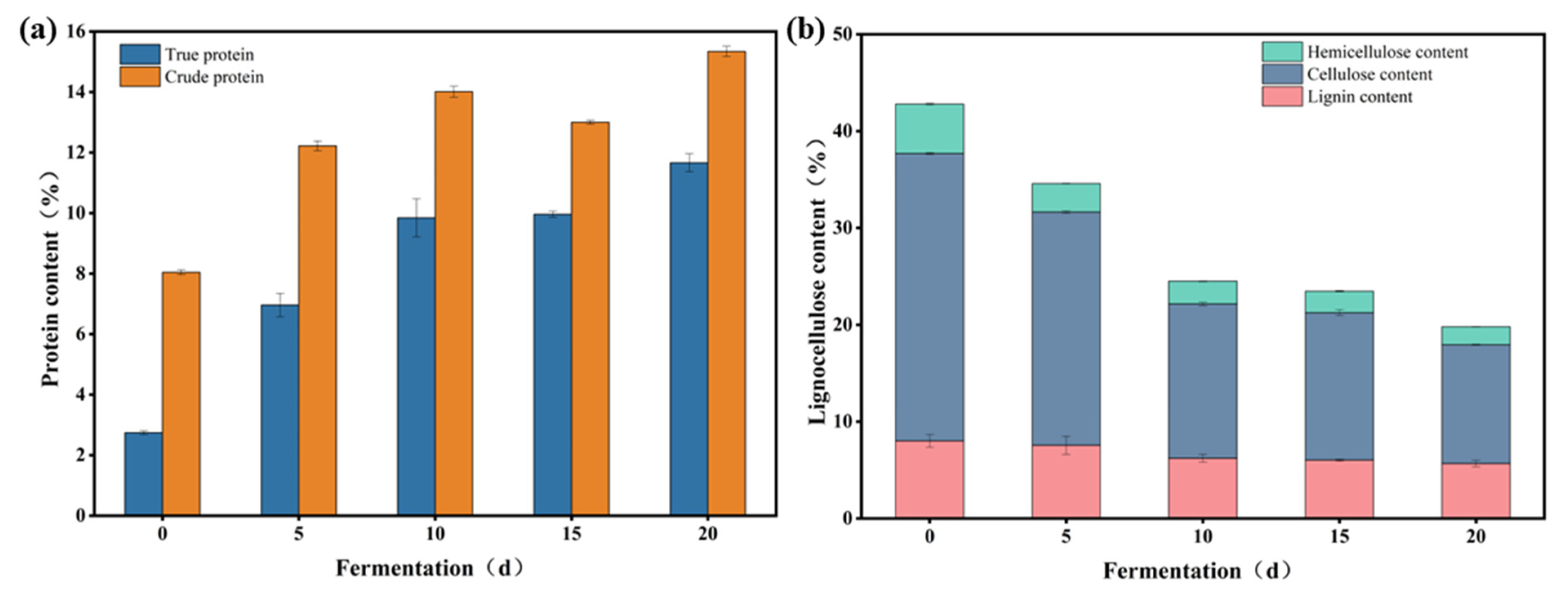
| Conditions | Pretreatment of Wheat Straw | Fermentation Time | Inoculation Amount | Nitrogen Addition Amount | Moisture Content | Temperature | Sterilization |
| Optimal conditions | 15% ammonium hydroxide | 10 days | 15% v/w | 0.5% w/w | 70% w/w | 30 °C | Non-sterilization |
| True protein content (%) | 10.42 ± 0.07 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Wang, S.; Mi, Y.; Liu, M.; Ren, H.; Guo, Z.; Chen, Z.; Cai, Y.; Xu, J.; Liu, D.; et al. Adaptive Laboratory Evolution of a Microbial Consortium Enhancing Non-Protein Nitrogen Assimilation for Feed Protein Production. Microorganisms 2025, 13, 1416. https://doi.org/10.3390/microorganisms13061416
He Y, Wang S, Mi Y, Liu M, Ren H, Guo Z, Chen Z, Cai Y, Xu J, Liu D, et al. Adaptive Laboratory Evolution of a Microbial Consortium Enhancing Non-Protein Nitrogen Assimilation for Feed Protein Production. Microorganisms. 2025; 13(6):1416. https://doi.org/10.3390/microorganisms13061416
Chicago/Turabian StyleHe, Yi, Shilei Wang, Yifan Mi, Mengyu Liu, Huimin Ren, Zhengxiang Guo, Zhen Chen, Yafan Cai, Jingliang Xu, Dong Liu, and et al. 2025. "Adaptive Laboratory Evolution of a Microbial Consortium Enhancing Non-Protein Nitrogen Assimilation for Feed Protein Production" Microorganisms 13, no. 6: 1416. https://doi.org/10.3390/microorganisms13061416
APA StyleHe, Y., Wang, S., Mi, Y., Liu, M., Ren, H., Guo, Z., Chen, Z., Cai, Y., Xu, J., Liu, D., Zhu, C., Wang, Z., & Ying, H. (2025). Adaptive Laboratory Evolution of a Microbial Consortium Enhancing Non-Protein Nitrogen Assimilation for Feed Protein Production. Microorganisms, 13(6), 1416. https://doi.org/10.3390/microorganisms13061416










