The Identification of Novel Mutations in ATP-Dependent Protease ClpC1 Assists in the Molecular Diagnosis of Obscured Pyrazinamide-Resistant Tuberculosis Clinical Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Drug Susceptibility Testing (DST) of Mtb Clinical Isolates
2.2. PZase Activity Assay
2.3. Amplification and Sequencing of PZA Resistance-Associated Genes
2.4. Whole-Genome Sequencing (WGS) and Bioinformatic Analysis
2.5. Validation of Novel Mutations in clpC1
2.6. ClpC1P1P2 Proteolytic Activity Assay
2.7. Detection of Beijing and Non-Beijing Genotypes
2.8. Structural and Statistical Analyses
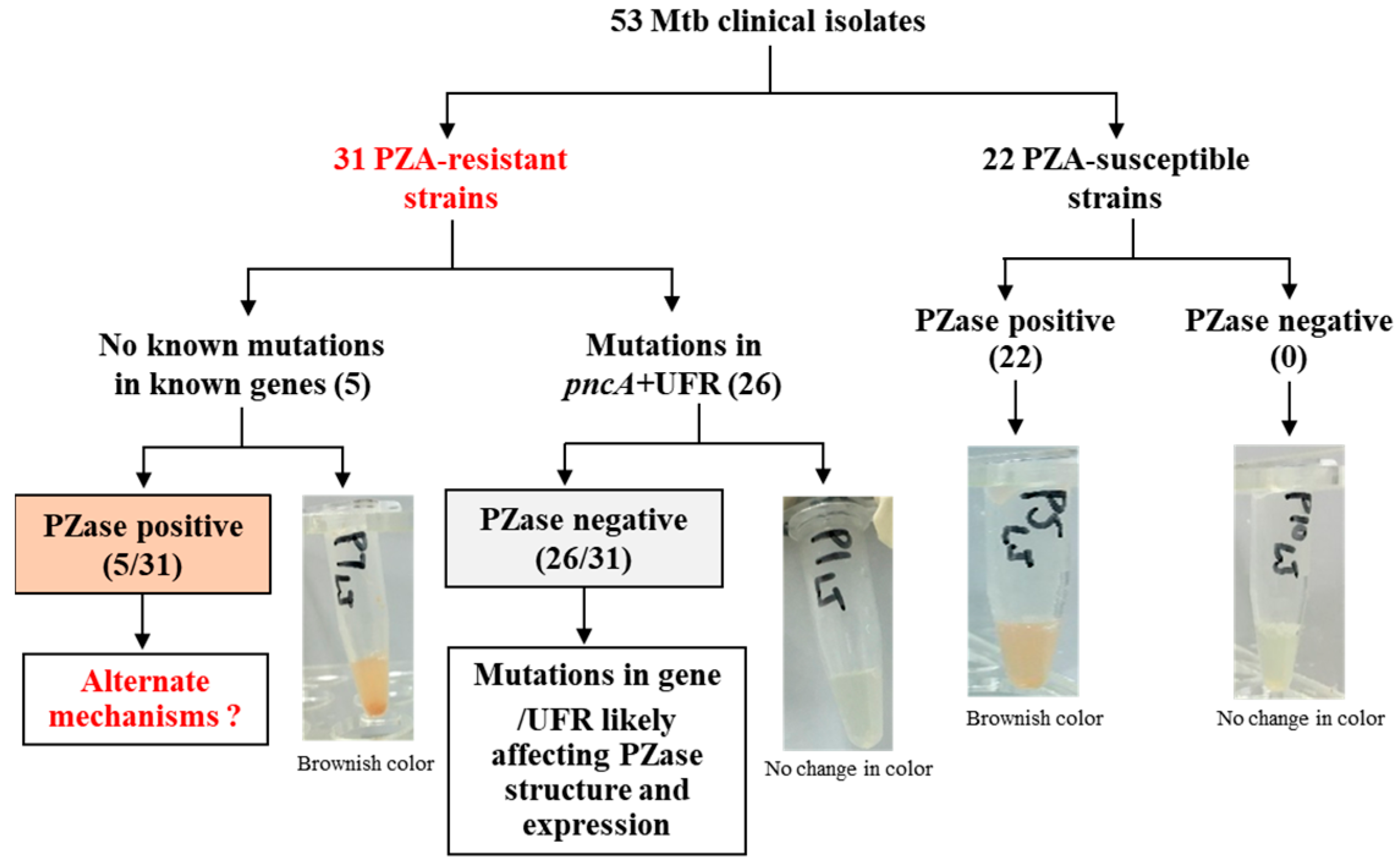
3. Results
3.1. DST Profiles of Mtb Clinical Isolates
3.2. Genotyping of PZA-Resistant Isolates
3.3. Genetic Characterization and Association of Mutations with PZase Activity
3.4. Identification of Novel Mutations in clpC1 of the Sequenced PZAR Strains Using WGS
3.5. Assessment of ClpC1P1P2 Proteolytic Activity
3.6. Comparative Assessment of Molecular Versus Phenotypic Diagnostic Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Operational Handbook on Tuberculosis. Module 4: Treatment-Drug-Resistant Tuberculosis Treatment, 2022 Update; World Health Organization: Geneva, Switzerland, 2022.
- Gopal, P.; Grüber, G.; Dartois, V.; Dick, T. Pharmacological and molecular mechanisms behind the sterilizing activity of pyrazinamide. Trends Pharmacol. Sci. 2019, 40, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Ju, Y.; Han, X.; Tian, X.; Ding, J.; Wang, S.; Hameed, H.M.A.; Gao, Y.; Li, L.; Li, Y.; et al. Bactericidal and sterilizing activity of sudapyridine-clofazimine-TB47 combined with linezolid or pyrazinamide in a murine model of tuberculosis. Antimicrob. Agents Chemother. 2024, 68, e00124-24. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Busby, S.; Rodwell, T.; Fink, L.; Catanzaro, D.; Jackson, R.; Pettigrove, M.; Catanzaro, A.; Valafar, F. A multinational analysis of mutations and heterogeneity in PZase, RpsA, and PanD associated with pyrazinamide resistance in M/XDR Mycobacterium tuberculosis. Sci. Rep. 2017, 7, 3790. [Google Scholar] [CrossRef] [PubMed]
- Dalton, T.; Cegielski, P.; Kurbatova, E.; Ershova, J.; Caoili, J.C. Resistance to second-line drugs in multidrug-resistant tuberculosis—Authors’ reply. Lancet 2013, 381, 626. [Google Scholar] [CrossRef]
- Kendall, E.A.; Fofana, M.O.; Dowdy, D.W. Burden of transmitted multidrug resistance in epidemics of tuberculosis: A transmission modelling analysis. Lancet. Respir. Med. 2015, 3, 963–972. [Google Scholar] [CrossRef]
- Lange, C.; Dheda, K.; Chesov, D.; Mandalakas, A.M.; Udwadia, Z.; Horsburgh, C.R., Jr. Management of drug-resistant tuberculosis. Lancet 2019, 394, 953–966. [Google Scholar] [CrossRef]
- Lawn, S.D.; Nicol, M.P. Xpert® MTB/RIF assay: Development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance. Future Microbiol. 2011, 6, 1067–1082. [Google Scholar] [CrossRef]
- Omoteso, O.A.; Fadaka, A.O.; Walker, R.B.; Khamanga, S.M. Innovative Strategies for Combating Multidrug-Resistant Tuberculosis: Advances in Drug Delivery Systems and Treatment. Microorganisms 2025, 13, 722. [Google Scholar] [CrossRef]
- Zhang, Y.; Yew, W.W. Mechanisms of drug resistance in Mycobacterium tuberculosis: Update 2015. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Against Tuberc. Lung Dis. 2015, 19, 1276–1289. [Google Scholar] [CrossRef]
- Goletti, D.; Meintjes, G.; Andrade, B.B.; Zumla, A.; Lee, S.S. Insights from the 2024 WHO Global Tuberculosis Report–More Comprehensive Action, Innovation, and Investments required for achieving WHO End TB goals. Int. J. Infect. Dis. 2025, 150, 107325. [Google Scholar] [CrossRef]
- Hameed, H.A.; Tan, Y.; Islam, M.M.; Lu, Z.; Chhotaray, C.; Wang, S.; Liu, Z.; Fang, C.; Tan, S.; Yew, W.W. Detection of novel gene mutations associated with pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates in Southern China. Infect. Drug Resist. 2020, 13, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Hameed, H.A.; Islam, M.M.; Chhotaray, C.; Wang, C.; Liu, Y.; Tan, Y.; Li, X.; Tan, S.; Delorme, V.; Yew, W.W. Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-Mycobacterium tuberculosis strains. Front. Cell. Infect. Microbiol. 2018, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Zhang, X.; Jiang, X.; Yuan, H.; Lee, J.S.; Barry, C.E., 3rd; Wang, H.; Zhang, W.; Zhang, Y. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 2011, 333, 1630–1632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, J.; Shi, W.; Liu, W.; Zhang, W.; Zhang, Y. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg. Microbes Infect. 2013, 2, 1–5. [Google Scholar] [CrossRef]
- Njire, M.; Tan, Y.; Mugweru, J.; Wang, C.; Guo, J.; Yew, W.; Tan, S.; Zhang, T. Pyrazinamide resistance in Mycobacterium tuberculosis: Review and update. Adv. Med. Sci. 2016, 61, 63–71. [Google Scholar] [CrossRef]
- Gopal, P.; Sarathy, J.P.; Yee, M.; Ragunathan, P.; Shin, J.; Bhushan, S.; Zhu, J.; Akopian, T.; Kandror, O.; Lim, T.K. Pyrazinamide triggers degradation of its target aspartate decarboxylase. Nat. Commun. 2020, 11, 1661. [Google Scholar] [CrossRef]
- Liu, Q.; Ma, A.; Wei, L.; Pang, Y.; Wu, B.; Luo, T.; Zhou, Y.; Zheng, H.X.; Jiang, Q.; Gan, M.; et al. China’s tuberculosis epidemic stems from historical expansion of four strains of Mycobacterium tuberculosis. Nat. Ecol. Evol. 2018, 2, 1982–1992. [Google Scholar] [CrossRef]
- Merker, M.; Blin, C.; Mona, S.; Duforet-Frebourg, N.; Lecher, S.; Willery, E.; Blum, M.G.B.; Rüsch-Gerdes, S.; Mokrousov, I.; Aleksic, E.; et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat. Genet. 2015, 47, 242–249. [Google Scholar] [CrossRef]
- Shitikov, E.; Kolchenko, S.; Mokrousov, I.; Bespyatykh, J.; Ischenko, D.; Ilina, E.; Govorun, V. Evolutionary pathway analysis and unified classification of East Asian lineage of Mycobacterium tuberculosis. Sci. Rep. 2017, 7, 9227. [Google Scholar] [CrossRef]
- Lan, N.T.; Lien, H.T.; Tung, L.B.; Borgdorff, M.W.; Kremer, K.; van Soolingen, D. Mycobacterium tuberculosis Beijing genotype and risk for treatment failure and relapse, Vietnam. Emerg. Infect. Dis. 2003, 9, 1633–1635. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, D.; Martinez, L.; Lu, P.; Zhu, L.; Lu, W.; Wang, J. Mycobacterium tuberculosis Beijing genotype strains and unfavourable treatment outcomes: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2020, 26, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, D.; Zhao, L.; Fleming, J.; Lin, N.; Wang, T.; Liu, Z.; Li, C.; Galwey, N.; Deng, J.; et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 2013, 45, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, M.G.; Soeters, H.M.; Warren, R.M.; York, T.; Sampson, S.L.; Streicher, E.M.; Van Helden, P.D.; Van Rie, A. A global perspective on pyrazinamide resistance: Systematic review and meta-analysis. PLoS ONE 2015, 10, e0133869. [Google Scholar] [CrossRef]
- Pang, Y.; Dong, H.; Tan, Y.; Deng, Y.; Cai, X.; Jing, H.; Xia, H.; Li, Q.; Ou, X.; Su, B.; et al. Rapid diagnosis of MDR and XDR tuberculosis with the MeltPro TB assay in China. Sci. Rep. 2016, 6, 25330. [Google Scholar] [CrossRef]
- World Health Organization. Technical Report on Critical Concentrations for Drug Susceptibility Testing of Medicines Used in the Treatment of Drug-Resistant Tuberculosis; World Health Organization: Geneva, Switzerland, 2018.
- Zhou, B.; Gao, Y.; Zhao, H.; Liu, B.; Zhang, H.; Fang, C.; Yuan, H.; Wang, J.; Li, Z.; Zhao, Y. Structural Insights into Bortezomib-Induced Activation of the Caseinolytic Chaperone-Protease System in Mycobacterium tuberculosis. Nat. Commun. 2025, 16, 3466. [Google Scholar] [CrossRef]
- Modlin, S.J.; Mansjö, M.; Werngren, J.; Ejike, C.M.; Hoffner, S.E.; Valafar, F. Pyrazinamide-resistant Tuberculosis Obscured From Common Targeted Molecular Diagnostics. Drug Resist. Updates 2023, 68, 100959. [Google Scholar] [CrossRef]
- Piersimoni, C.; Mustazzolu, A.; Giannoni, F.; Bornigia, S.; Gherardi, G.; Fattorini, L. Prevention of false resistance results obtained in testing the susceptibility of Mycobacterium tuberculosis to pyrazinamide with the Bactec MGIT 960 system using a reduced inoculum. J. Clin. Microbiol. 2013, 51, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Mok, S.; Roycroft, E.; Flanagan, P.R.; Montgomery, L.; Borroni, E.; Rogers, T.R.; Fitzgibbon, M.M. Overcoming the Challenges of Pyrazinamide Susceptibility Testing in Clinical Mycobacterium tuberculosis Isolates. Antimicrob. Agents Chemother. 2021, 65, 8. [Google Scholar] [CrossRef]
- Miotto, P.; Tessema, B.; Tagliani, E.; Chindelevitch, L.; Starks, A.M.; Emerson, C.; Hanna, D.; Kim, P.S.; Liwski, R.; Zignol, M.; et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur. Respir. J. 2017, 50, 1701354. [Google Scholar] [CrossRef]
- Liu, B.; Su, P.; Hu, P.; Yan, M.; Li, W.; Yi, S.; Chen, Z.; Zhang, X.; Guo, J.; Wan, X. Prevalence, transmission and genetic diversity of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Hunan, China. Infect. Drug Resist. 2024, 17, 403–416. [Google Scholar] [CrossRef]
- Che, Y.; Bo, D.; Lin, X.; Chen, T.; He, T.; Lin, Y. Phenotypic and molecular characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Ningbo, China. BMC Infect. Dis. 2021, 21, 605. [Google Scholar] [CrossRef] [PubMed]
- Ei, P.W.; Mon, A.S.; Htwe, M.M.; Win, S.M.; Aye, K.T.; San, L.L.; Zaw, N.N.; Nyunt, W.W.; Myint, Z.; Lee, J.S. Pyrazinamide resistance and pncA mutations in drug resistant Mycobacterium tuberculosis clinical isolates from Myanmar. Tuberculosis 2020, 125, 102013. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jo, K.; Shim, T. Treatment outcomes in multidrug-resistant tuberculosis according to pyrazinamide susceptibility. Int. J. Tuberc. Lung Dis. 2020, 24, 233–239. [Google Scholar] [CrossRef]
- Wang, G.; Jiang, G.; Jing, W.; Zong, Z.; Yu, X.; Chen, S.; Li, W.; Huang, H. Prevalence and molecular characterizations of seven additional drug resistance among multidrug-resistant tuberculosis in China: A subsequent study of a national survey. J. Infect. 2021, 82, 371–377. [Google Scholar] [CrossRef]
- Calderón, R.I.; Velásquez, G.E.; Becerra, M.C.; Zhang, Z.; Contreras, C.C.; Yataco, R.M.; Galea, J.T.; Lecca, L.W.; Kritski, A.L.; Murray, M.B. Prevalence of pyrazinamide resistance and Wayne assay performance analysis in a tuberculosis cohort in Lima, Peru. Int. J. Tuberc. Lung Dis. 2017, 21, 894–901. [Google Scholar] [CrossRef]
- Peterson, N.D.; Rosen, B.C.; Dillon, N.A.; Baughn, A.D. Uncoupling environmental pH and intrabacterial acidification from pyrazinamide susceptibility in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2015, 59, 7320–7326. [Google Scholar] [CrossRef]
- Shrestha, D.; Maharjan, B.; Thapa, J.; Akapelwa, M.L.; Bwalya, P.; Chizimu, J.Y.; Nakajima, C.; Suzuki, Y. Detection of mutations in pncA in Mycobacterium tuberculosis clinical isolates from Nepal in association with pyrazinamide resistance. Curr. Issues Mol. Biol. 2022, 44, 4132–4141. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Wei, S.; Zhao, Y.; Dai, Z.; Lin, J.; Pang, Y. Genetic diversity and drug susceptibility profiles of multidrug-resistant tuberculosis strains in Southeast China. Infect. Drug Resist. 2021, 14, 3979–3989. [Google Scholar] [CrossRef]
- Daum, L.; Konstantynovska, O.; Solodiankin, O.; Poteiko, P.; Bolotin, V.; Rodriguez, J.; Gerilovych, A.; Chambers, J.; Fischer, G. Characterization of novel Mycobacterium tuberculosis pncA gene mutations in clinical isolates from the Ukraine. Diagn. Microbiol. Infect. Dis. 2019, 93, 334–338. [Google Scholar] [CrossRef]
- Pang, Y.; Zhu, D.; Zheng, H.; Shen, J.; Hu, Y.; Liu, J.; Zhao, Y. Prevalence and molecular characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. BMC Infect. Dis. 2017, 17, 1–8. [Google Scholar] [CrossRef]
- Li, K.; Yang, Z.; Gu, J.; Luo, M.; Deng, J.; Chen, Y. Characterization of pncA Mutations and Prediction of PZA Resistance in Mycobacterium tuberculosis Clinical Isolates From Chongqing, China. Front. Microbiol. 2021, 11-2020, 594171. [Google Scholar] [CrossRef]
- Sodja, E.; Koren, S.; Toplak, N.; Truden, S.; Žolnir-Dovč, M. Next-generation sequencing to characterise pyrazinamide resistance in Mycobacterium tuberculosis isolates from two Balkan countries. J. Glob. Antimicrob. Resist. 2022, 29, 507–512. [Google Scholar] [CrossRef]
- Shi, D.; Zhou, Q.; Xu, S.; Zhu, Y.; Li, H.; Xu, Y. Pyrazinamide resistance and pncA mutation profiles in multidrug resistant Mycobacterium tuberculosis. Infect. Drug Resist. 2022, 15, 4985–4994. [Google Scholar] [CrossRef] [PubMed]
- Weinhäupl, K.; Gragera, M.; Bueno-Carrasco, M.T.; Arranz, R.; Krandor, O.; Akopian, T.; Soares, R.; Rubin, E.; Felix, J.; Fraga, H. Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action. J. Biol. Chem. 2022, 298, 102553. [Google Scholar] [CrossRef]
- Bajaj, D.; Batra, J.K. The C-terminus of ClpC1 of Mycobacterium tuberculosis is crucial for its oligomerization and function. PLoS ONE 2012, 7, e51261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, J.; Shi, W.; Cui, P.; Zhang, J.; Cho, S.; Zhang, W.; Zhang, Y. Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg. Microbes Infect. 2017, 6, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Gavrish, E.; Sit, C.S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef]
- Choules, M.P.; Wolf, N.M.; Lee, H.; Anderson, J.R.; Grzelak, E.M.; Wang, Y.; Ma, R.; Gao, W.; McAlpine, J.B.; Jin, Y.-Y. Rufomycin targets ClpC1 proteolysis in Mycobacterium tuberculosis and M. abscessus. Antimicrob. Agents Chemother. 2019, 63, 5806–5816. [Google Scholar] [CrossRef]
- Gao, W.; Kim, J.-Y.; Anderson, J.R.; Akopian, T.; Hong, S.; Jin, Y.-Y.; Kandror, O.; Kim, J.-W.; Lee, I.-A.; Lee, S.-Y. The cyclic peptide ecumicin targeting ClpC1 is active against Mycobacterium tuberculosis in vivo. Antimicrob. Agents Chemother. 2015, 59, 880–889. [Google Scholar] [CrossRef]
- Dougan, D.A.; Mogk, A.; Zeth, K.; Turgay, K.; Bukau, B. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 2002, 529, 6–10. [Google Scholar] [CrossRef]
- Schmitt, E.K.; Riwanto, M.; Sambandamurthy, V.; Roggo, S.; Miault, C.; Zwingelstein, C.; Krastel, P.; Noble, C.; Beer, D.; Rao, S.P. The natural product cyclomarin kills Mycobacterium tuberculosis by targeting the ClpC1 subunit of the caseinolytic protease. Angew. Chem. Int. Ed. 2011, 50, 5889–5891. [Google Scholar] [CrossRef]
- Gao, Y.; Fang, C.; Zhou, B.; Hameed, H.A.; Sun, C.; Tian, X.; He, J.; Han, X.; Zhang, H.; Ju, J. Ilamycins are dual inhibitors of ClpX and ClpC1 involved in proteostasis in Mycobacteria. bioRxiv, 2024; preprint. [Google Scholar] [CrossRef]
- Reinhardt, L.; Thomy, D.; Lakemeyer, M.; Westermann, L.M.; Ortega, J.; Sieber, S.A.; Sass, P.; Brötz-Oesterhelt, H. Antibiotic acyldepsipeptides stimulate the Streptomyces Clp-ATPase/ClpP complex for accelerated proteolysis. Mbio 2022, 13, e01413-22. [Google Scholar] [CrossRef]
- Wolf, N.M.; Lee, H.; Choules, M.P.; Pauli, G.F.; Phansalkar, R.; Anderson, J.R.; Gao, W.; Ren, J.; Santarsiero, B.D.; Lee, H. High-resolution structure of ClpC1-rufomycin and ligand binding studies provide a framework to design and optimize anti-tuberculosis leads. ACS Infect. Dis. 2019, 5, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Valafar, S.J. Systematic review of mutations associated with isoniazid resistance points to continuing evolution and subsequent evasion of molecular detection, and potential for emergence of multidrug resistance in clinical strains of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2021, 65, 5806–5816. [Google Scholar] [CrossRef]
- World Health Organization. High Priority Target Product Profiles for New Tuberculosis Diagnostics: Report of a Consensus Meeting, 28–29 April 2014, Geneva, Switzerland; World Health Organization: Geneva, Switzerland, 2014.
- Liu, W.; Chen, J.; Shen, Y.; Jin, J.; Wu, J.; Sun, F.; Wu, Y.; Xie, L.; Zhang, Y.; Zhang, W. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis clinical isolates in Hangzhou, China. Clin. Microbiol. Infect. 2018, 24, 1016.e1–1016.e5. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Su, R.; Zheng, D.; Zhu, Y.; Ma, X.; Wang, S.; Li, H.; Sun, D. Pyrazinamide resistance and mutation patterns among multidrug-resistant Mycobacterium tuberculosis from Henan Province. Infect. Drug Resist. 2020, 13, 2929–2941. [Google Scholar] [CrossRef]
- Whitfield, M.G.; Engelthaler, D.M.; Allender, C.; Folkerts, M.; Heupink, T.H.; Limberis, J.; Warren, R.M.; Van Rie, A.; Metcalfe, J.Z. Comparative performance of genomic methods for the detection of pyrazinamide resistance and heteroresistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 2022, 60, e01907–e01921. [Google Scholar] [CrossRef]
- Modlin, S.J.; Marbach, T.; Werngren, J.; Mansjö, M.; Hoffner, S.E.; Valafar, F. Atypical genetic basis of pyrazinamide resistance in monoresistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2021, 65, 5806–5816. [Google Scholar] [CrossRef]



| Gene | Protein | Functional Activity | Primer Name | Oligonucleotide Sequence (5′→3′) | Product Size ~ (−200 to +200) |
|---|---|---|---|---|---|
| pncA (Rv2043c) | Pyrazinamidase/nicotinamidase (PZase) | Convert PZA into POA | pncA-F | TCGCTCACTACATCACCGGC | 892 bp |
| pncA-R | TCGTAGAAGCGGCCGATGGC | ||||
| * Rv2044c + pncA + Rv2042c | Conserved hypothetical protein- PZase-Conserved protein | pncAf-F | GTGCCGCATCGAGTTCGATCCGCA | 2070 bp | |
| pncAf-R | GATATCGGGATAGCGCCGCTGGA | ||||
| rpsA (Rv1630) | 30S ribosomal protein S1 (RpsA) | Trans-translation | rpsA-F | ACTGAGTGCCGAGCGTGCATC | 1800 bp |
| rpsA-R | ACCGAACGCGTCGACCAGCG | ||||
| panD (Rv3601c) | Aspartate alpha-decarboxylase (PanD) | Pantothenate biosynthesis | panD-F | TCGACTACCTGGAGCTGCGC | 755 bp |
| panD-R | TCGATCGTCAGTGCCAGTTC | ||||
| Rv2783c | Bifunctional protein polyribonucleotide Nucleotidyltransferase (GpsI, Pnpase) and synthesize and hydrolyze (p)ppGpp | Synthesis/ degradation of ssDNA/ssRNA and (p)ppGpp | gpsI-F | ATTCAGACCTTTTCTCCTGGG | 2547 bp |
| gpsI-R | GTCGACTTGAACAGCAAATG | ||||
| clpC1 (Rv3596c) | ATP-dependent protease ATP-binding subunit (ClpC1) | Hydrolyses proteins in the presence of ATP | clpC1-F | ACGCTTGGGTGGTTTTCTCGTT | 2816 bp |
| clpC1-R | ACAAACCGACGTCAGCAGAGT | ||||
| Locus | Nucleotide Change | Codon Change | Amino Acid Change | PZase Activity | No. of Isolates |
|---|---|---|---|---|---|
| UFR | A-11C | - | - | N | 1 |
| A-11G | - | - | N | 2 | |
| T-122 | deletion | Frameshift | N | 1 | |
| C-114, A-11G | deletion + substitution | Frameshift + substitution | N | 1 | |
| pncA + UFR | G-115C, T476G | CTG → CGG | Leu159Arg | N | 1 |
| C-110G, T355G | TGG → GGG | Trp119 Gly | N | 1 | |
| A403C | ACC → CCC | Thr135 Pro | N | 1 | |
| A422C | CAG → CCG | Gln141 Pro | N | 2 | |
| A-144ins, T416C | GTG → GCG | Val139Ala | N | 1 | |
| C28T | CAG → TAG | Gln10Stop | N | 1 | |
| T80G | CTG → CGG | Leu27Arg | N | 1 | |
| pncAc | A128 | deletion | Frameshift | N | 1 |
| G133T | GTG → TTG | Val45Leu | N | 1 | |
| A142T | AAG → TAG | Lys48Stop | N | 1 | |
| C161A | CCG → CAG | Pro54 Gln | N | 1 | |
| C185T | CCG → CTG | Pro62Leu | N | 1 | |
| C189A | GAC → GAA | Asp63Glu | N | 1 | |
| G233A | GGC → GAC | Gly78Asp | N | 2 | |
| C282G | TTC → TTG | Phe94Leu | N | 1 | |
| A286G | AAG → GAG | Lys96Glu | N | 2 | |
| C299T | ACC → ATC | Thr100Ile | N | 1 | |
| G311T | AGC → ATC | Ser104Ile | N | 1 |
| Genes | PZAR Isolates n = 31 | PZAS Isolates n = 22 | Sensitivity% (95% CI) | Specificity% (95% CI) | Accuracy% (95% CI) | ||
|---|---|---|---|---|---|---|---|
| Non-Synonymous Mutations (%) | WT or Synonymous Mutations (%) | Non-Synonymous Mutations (%) | WT or Synonymous Mutations (%) | ||||
| pncAc | 15 (48.3) | 16 (51.6) | 0 (0.0) | 22 (100) | 48.3 (30.1–66.9) | 100 (84.5–100) | 69.8 (55.6–81.6) |
| UFR | 5 (16.1) | 26 (83.8) | 0 (0.0) | 22 (100) | 16.1 (5.45–33.7) | 100 (84.5–100) | 50.9 (36.8–64.9) |
| pncA + UFR* | 6 (19.3) | 25 (80.6) | 0 (0.0) | 22 (100) | 19.3 (7.45–37.4) | 100 (84.5–100) | 52.8 (38.6–66.7) |
| pncA + FR# | 26 (83.8) | 5 (16.1) | 0 (0.0) | 22 (100) | 83.8 (66.2–94.5) | 100 (84.5–100) | 90.5 (79.3–96.8) |
| clpC1 | 5 (16.1) | 26 (83.8) | 0 (0.0) | 22 (100) | 16.1 (5.45–33.7) | 100 (84.5–100) | 50.9 (36.8–64.9) |
| Total | 31 (100) | 0 (0.0) | 0 (0.0) | 22 (100) | 100 (88.7–100) | 100 (84.5–100) | 100 (93.2–100) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hameed, H.M.A.; Fang, C.; Liu, Z.; Gao, Y.; Wang, S.; Chen, X.; Zhong, N.; Aung, H.L.; Hu, J.; Zhang, T. The Identification of Novel Mutations in ATP-Dependent Protease ClpC1 Assists in the Molecular Diagnosis of Obscured Pyrazinamide-Resistant Tuberculosis Clinical Isolates. Microorganisms 2025, 13, 1401. https://doi.org/10.3390/microorganisms13061401
Hameed HMA, Fang C, Liu Z, Gao Y, Wang S, Chen X, Zhong N, Aung HL, Hu J, Zhang T. The Identification of Novel Mutations in ATP-Dependent Protease ClpC1 Assists in the Molecular Diagnosis of Obscured Pyrazinamide-Resistant Tuberculosis Clinical Isolates. Microorganisms. 2025; 13(6):1401. https://doi.org/10.3390/microorganisms13061401
Chicago/Turabian StyleHameed, H. M. Adnan, Cuiting Fang, Zhiyong Liu, Yamin Gao, Shuai Wang, Xinwen Chen, Nanshan Zhong, Htin Lin Aung, Jinxing Hu, and Tianyu Zhang. 2025. "The Identification of Novel Mutations in ATP-Dependent Protease ClpC1 Assists in the Molecular Diagnosis of Obscured Pyrazinamide-Resistant Tuberculosis Clinical Isolates" Microorganisms 13, no. 6: 1401. https://doi.org/10.3390/microorganisms13061401
APA StyleHameed, H. M. A., Fang, C., Liu, Z., Gao, Y., Wang, S., Chen, X., Zhong, N., Aung, H. L., Hu, J., & Zhang, T. (2025). The Identification of Novel Mutations in ATP-Dependent Protease ClpC1 Assists in the Molecular Diagnosis of Obscured Pyrazinamide-Resistant Tuberculosis Clinical Isolates. Microorganisms, 13(6), 1401. https://doi.org/10.3390/microorganisms13061401







