ESBL-Producing Escherichia coli and Klebsiella pneumoniae Exhibit Divergent Paths During In-Human Evolution Towards Carbapenem Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Isolates
2.2. DNA Extraction
2.3. Rapid Amplification of Polymorphic DNA (RAPD) Assay
2.4. Whole Genome Sequencing and Analysis
2.5. Statistical Analysis
3. Results
3.1. ESBL Clinical Strains from Diverse Genetic Backgrounds Evolved Different Carbapenem-Resistant Phenotypes
3.2. Strains with Absent or Progressive Loss of CRISPR-Cas Systems Support Plasmid-Bearing blaKPC
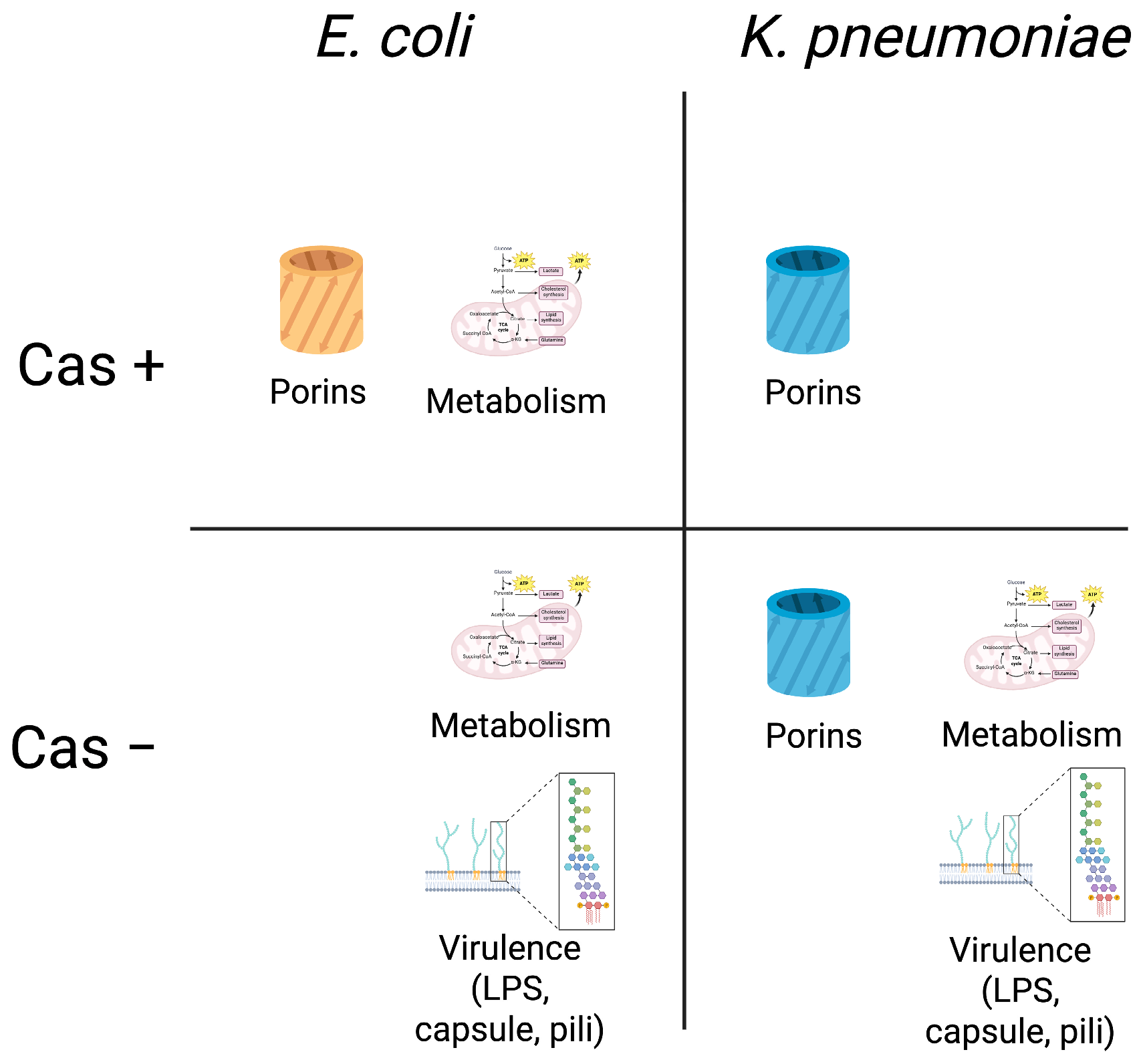
3.3. Acquisition of Porin Mutations Differs Between ESBL EC and KP Strains by CRISPR-Cas Background
3.4. Acquisition of Virulence Gene Mutations Favored CRISPR-Cas Negative EC and KP Strains
3.5. Acquisition of Mutations in Metabolism-Related Genes Favors CRISPR-Cas Negative KP but Not EC Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EC | Escherichia coli |
| KP | Klebsiella pneumoniae |
| ESBL | extended-spectrum beta-lactamase |
| cHR | carbapenem heteroresistance |
| blaKPC | gene encoding K. pneumoniae carbapenemase enzyme |
| CRKP | Carbapenem-resistant K. pneumoniae |
References
- Centers for Disease Control and Prevention (USA). Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention (USA): Atlanta, GA, USA, 2019. Available online: https://stacks.cdc.gov/view/cdc/82532 (accessed on 4 October 2024). [CrossRef]
- Centers for Disease Control and Prevention (USA). COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; Centers for Disease Control and Prevention (USA): Atlanta, GA, USA, 2022. Available online: https://stacks.cdc.gov/view/cdc/117915 (accessed on 4 October 2024). [CrossRef]
- Tan, K.; Kelsom, C.; Chron, A.; Nieberg, P.; Huse, H.; Wong-Beringer, A. Risk factors and outcome associated with infection or colonization due to carbapenem-heteroresistant Escherichia coli. JAC-Antimicrob. Resist. 2021, 3, dlab036. [Google Scholar] [CrossRef] [PubMed]
- Ny, P.; Nieberg, P.; Wong-Beringer, A. Impact of carbapenem resistance on epidemiology and outcomes of nonbacteremic Klebsiella pneumoniae infections. Am. J. Infect. Control 2015, 43, 1076–1080. [Google Scholar] [CrossRef]
- Tamma, P.D.; Heil, E.L.; Justo, J.A.; Mathers, A.J.; Satlin, M.J.; Bonomo, R.A. Infectious Diseases Society of America 2024 Guidance on the Treatment of Antimicrobial-Resistant Gram-Negative Infections. Clin. Infect. Dis. 2024, ciae403. [Google Scholar] [CrossRef]
- Karaiskos, I.; Giamarellou, H. Carbapenem-Sparing Strategies for ESBL Producers: When and How. Antibiotics 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; He, L.L.; Pironti, A.; Laibinis, H.H.; Ernst, C.M.; Manson, A.L.; Bhattacharyya, R.P.; Earl, A.M.; Livny, J.; Hung, D.T. Genetic determinants facilitating the evolution of resistance to carbapenem antibiotics. eLife 2021, 10, e67310. [Google Scholar] [CrossRef]
- Kalu, M.; Tan, K.; Algorri, M.; Jorth, P.; Wong-Beringer, A. In-Human Multiyear Evolution of Carbapenem-Resistant Klebsiella pneumoniae Causing Chronic Colonization and Intermittent Urinary Tract Infections: A Case Study. mSphere 2022, 7, e0019022. [Google Scholar] [CrossRef]
- Adams-Sapper, S.; Nolen, S.; Donzelli, G.F.; Lal, M.; Chen, K.; da Silva, L.H.J.; Moreira, B.M.; Riley, L.W. Rapid induction of high-level carbapenem resistance in heteroresistant KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2015, 59, 3281–3289. [Google Scholar] [CrossRef]
- Nicoloff, H.; Hjort, K.; Levin, B.R.; Andersson, D.I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 2019, 4, 504–514. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Sun, J.D.; Huang, S.F.; Yang, S.S.; Pu, S.L.; Zhang, C.M.; Zhang, L.P. Impact of carbapenem heteroresistance among clinical isolates of invasive Escherichia coli in Chongqing, southwestern China. Clin. Microbiol. Infect. 2015, 21, 469.e1–469.e10. [Google Scholar] [CrossRef]
- Palmer, K.L.; Gilmore, M.S. Multidrug-Resistant Enterococci Lack CRISPR-cas. mBio 2010, 1, e00227-10. [Google Scholar] [CrossRef] [PubMed]
- Westra, E.R.; Swarts, D.C.; Staals, R.H.J.; Jore, M.M.; Brouns, S.J.J.; van der Oost, J. The CRISPRs, They Are A-Changin’: How Prokaryotes Generate Adaptive Immunity. Annu. Rev. Genet. 2012, 46, 311–339. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-Y.; Teng, C.-H.; Lin, W.-H.; Lin, W.-H.; Yan, J.-J.; Wang, M.-C.; Teng, C.-H.; Tseng, C.-C.; Wu, J.-J. Characterization of CRISPR-Cas Systems in Clinical Klebsiella pneumoniae Isolates Uncovers Its Potential Association with Antibiotic Susceptibility. Front. Microbiol. 2018, 9, 1595. [Google Scholar] [CrossRef]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchêne, S.; Jenney, A.; Holt, K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLOS Genet. 2019, 15, e1008114. [Google Scholar] [CrossRef] [PubMed]
- Mackow, N.A.; Shen, J.; Adnan, M.; Khan, A.S.; Fries, B.C.; Diago-Navarro, E. CRISPR-Cas influences the acquisition of antibiotic resistance in Klebsiella pneumoniae. PLoS ONE 2019, 14, e0225131. [Google Scholar] [CrossRef]
- Lin, T.L.; Pan, Y.J.; Hsieh, P.F.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci. Rep. 2016, 6, 31644. [Google Scholar] [CrossRef]
- Tan, K.; Nguyen, J.; Nguyen, K.; Huse, H.K.; Nieberg, P.H.; Wong-Beringer, A. Prevalence of the carbapenem-heteroresistant phenotype among ESBL-producing Escherichia coli and Klebsiella pneumoniae clinical isolates. J. Antimicrob. Chemother. 2020, 75, 1506–1512. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Ninth Edition; CLSI document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; Volume 32. [Google Scholar]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of Mutations in Laboratory-Evolved Microbes from Next-Generation Sequencing Data Using Breseq. In Engineering and Analyzing Multicellular Systems; Sun, L., Shou, W., Eds.; Springer: New York, NY, USA, 2014; Volume 1151, pp. 165–188. [Google Scholar]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Shi, Q.; Wang, S.; Shi, Y.; Sun, D.; Yu, Y. The Characterization of OXA-232 Carbapenemase-Producing ST437 Klebsiella pneumoniae in China. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 5626503. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Li, C.; Ge, H.; Qiao, J.; Fang, L.; Liu, C.; Gou, J.; Guo, X. Difference analysis and characteristics of incompatibility group plasmid replicons in gram-negative bacteria with different antimicrobial phenotypes in Henan, China. BMC Microbiol. 2024, 24, 64. [Google Scholar] [CrossRef] [PubMed]
- Zavan, L.; Hor, L.; Johnston, E.L.; Paxman, J.; Heras, B.; Kaparakis, M. Antigen 43 associated with Escherichia coli membrane vesicles contributes to bacterial cell association and biofilm formation. Microbiol. Spectr. 2025, 13, e01890-24. [Google Scholar] [CrossRef]
- Klein, R.D.; Hultgren, S.J. Urinary tract infections: Microbial pathogenesis, host–pathogen interactions and new treatment strategies. Nat. Rev. Microbiol. 2020, 18, 211–226. [Google Scholar] [CrossRef]
- Ernst, C.M.; Braxton, J.R.; Rodriguez-Osorio, C.A.; Zagieboylo, A.P.; Li, L.; Pironti, A.; Manson, A.L.; Nair, A.V.; Benson, M.; Cummins, K.; et al. Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat. Med. 2020, 26, 705–711. [Google Scholar] [CrossRef]
- Chen, L.; Mathema, B.; Pitout, J.D.D.; DeLeo, F.R.; Kreiswirth, B.N. Epidemic Klebsiella pneumoniae ST258 is a Hybrid Strain. mBio 2014, 5, e01355-14. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, Y.; Fu, P.; Tian, D.; Yu, L.; Huang, Y.; Li, G.; Li, M.; Wang, Y.; Yang, Z.; et al. The type I-E CRISPR-Cas system influences the acquisition of blaKPC-IncF plasmid in Klebsiella pneumonia. Emerg. Microbes Infect. 2020, 9, 1011–1022. [Google Scholar] [CrossRef]
- Zakrzewska, M.; Burmistrz, M. Mechanisms regulating the CRISPR-Cas systems. Front. Microbiol. 2023, 14, 1060337. [Google Scholar] [CrossRef]
- Louwen, R.; Staals, R.H.J.; Endtz, H.P.; Van Baarlen, P.; Van Der Oost, J. The Role of CRISPR-Cas Systems in Virulence of Pathogenic Bacteria. Microbiol. Mol. Biol. Rev. 2014, 78, 74–88. [Google Scholar] [CrossRef]
- Chu, W.H.W.; Tan, Y.H.; Tan, S.Y.; Chen, Y.; Yong, M.; Lye, D.C.; Kalimuddin, S.; Archuleta, S.; Gan, Y.-H. Acquisition of regulator on virulence plasmid of hypervirulent Klebsiella allows bacterial lifestyle switch in response to iron. mBio 2023, 14, e01297-23. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Liu, C.; Fan, S.; Baker, S.; Guo, J. The Role of Plasmid and Resistance Gene Acquisition in the Emergence of ST23 Multi-Drug Resistant, Hypervirulent Klebsiella pneumoniae. Microbiol. Spectr. 2022, 10, e01929-21. [Google Scholar] [CrossRef] [PubMed]
- Ikhimiukor, O.O.; Zac Soligno, N.I.; Akintayo, I.J.; Marcovici, M.M.; Souza, S.S.; Workman, A.; Martin, I.W.; Andam, C.P. Clonal background and routes of plasmid transmission underlie antimicrobial resistance features of bloodstream Klebsiella pneumoniae. Nat. Commun. 2024, 15, 6969. [Google Scholar] [CrossRef]
- Samson, J.E.; Magadan, A.H.; Moineau, S. The CRISPR-Cas Immune System and Genetic Transfers: Reaching an Equilibrium. Microbiol. Spectr. 2015, 3, PLAS-0034-2014. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, R.M.; MacLean, R.C. CRISPR-Cas systems restrict horizontal gene transfer in Pseudomonas aeruginosa. ISME J. 2021, 15, 1420–1433. [Google Scholar] [CrossRef]
- Hall, R.J.; Snaith, A.E.; Thomas, M.J.N.; Brockhurst, M.A.; McNally, A. Multidrug resistance plasmids commonly reprogram the expression of metabolic genes in Escherichia coli. mSystems 2024, 9, e01193-23. [Google Scholar] [CrossRef]
- Samson, J.E.; Magadan, A.H.; Moineau, S. Fine-tuning carbapenem resistance by reducing porin permeability of bacteria activated in the selection process of conjugation. Sci. Rep. 2018, 8, 15248. [Google Scholar]
- Lopatkin, A.J.; Bening, S.C.; Manson, A.L.; Stokes, J.M.; Kohanski, M.A.; Badran, A.H.; Earl, A.M.; Cheney, N.J.; Yang, J.H.; Collins, J.J. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 2021, 371, eaba0862. [Google Scholar] [CrossRef]
- Palomino, A.; Gewurz, D.; DeVine, L.; Zajmi, U.; Moralez, J.; Abu-Rumman, F.; Smith, R.P.; Lopatkin, A.J. Metabolic genes on conjugative plasmids are highly prevalent in Escherichia coli and can protect against antibiotic treatment. ISME J. 2023, 17, 151–162. [Google Scholar] [CrossRef]
- Hurst, M.N.; Beebout, C.J.; Hollingsworth, A.; Guckes, K.R.; Purcell, A.; Bermudez, T.A.; Williams, D.; Reasoner, S.A.; Trent, M.S.; Hadjifrangiskou, M. The QseB response regulator imparts tolerance to positively charged antibiotics by controlling metabolism and minor changes to LPS. mSphere 2023, 8, e00059-23. [Google Scholar] [CrossRef]
- Pearcy, N.; Hu, Y.; Baker, M.; Maciel-Guerra, A.; Xue, N.; Wang, W.; Kaler, J.; Peng, Z.; Li, F.; Dottorini, T. Genome-Scale Metabolic Models and Machine Learning Reveal Genetic Determinants of Antibiotic Resistance in Escherichia coli and Unravel the Underlying Metabolic Adaptation Mechanisms. mSystems 2021, 6, e0091320. [Google Scholar] [CrossRef]
- Masi, M.; Pinet, E.; Pagès, J.-M. Complex Response of the CpxAR Two-Component System to β-Lactams on Antibiotic Resistance and Envelope Homeostasis in Enterobacteriaceae. Antimicrob. Agents Chemother. 2020, 64, e00291-20. [Google Scholar] [CrossRef]
- Liu, Z.; Guan, J.; Chen, Z.; Tai, C.; Deng, Z.; Chao, Y.; Ou, H.Y. CpxR promotes the carbapenem antibiotic resistance of Klebsiella pneumoniae by directly regulating the expression and the dissemination of blaKPC on the IncFII conjugative plasmid. Emerg. Microbes Infect. 2023, 12, 2256427. [Google Scholar] [CrossRef]
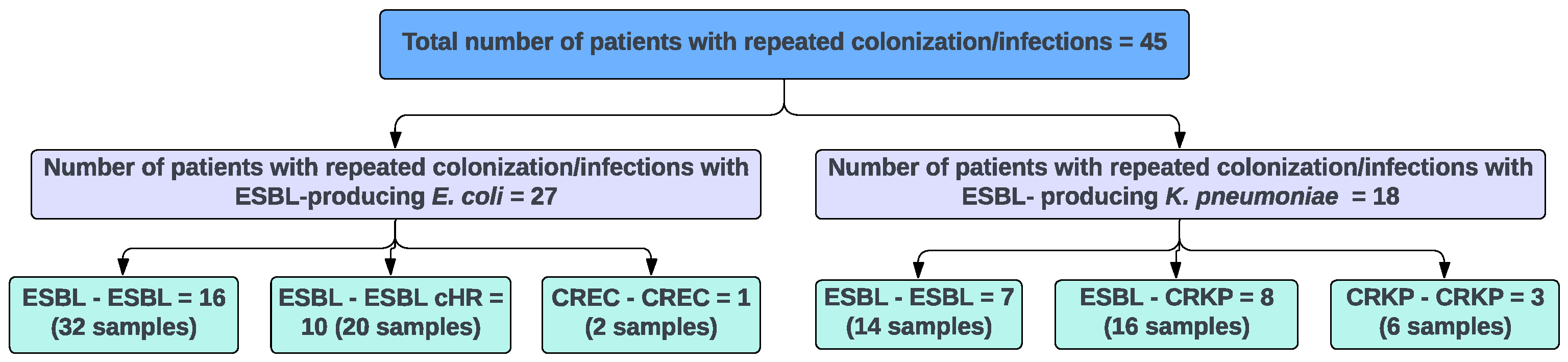
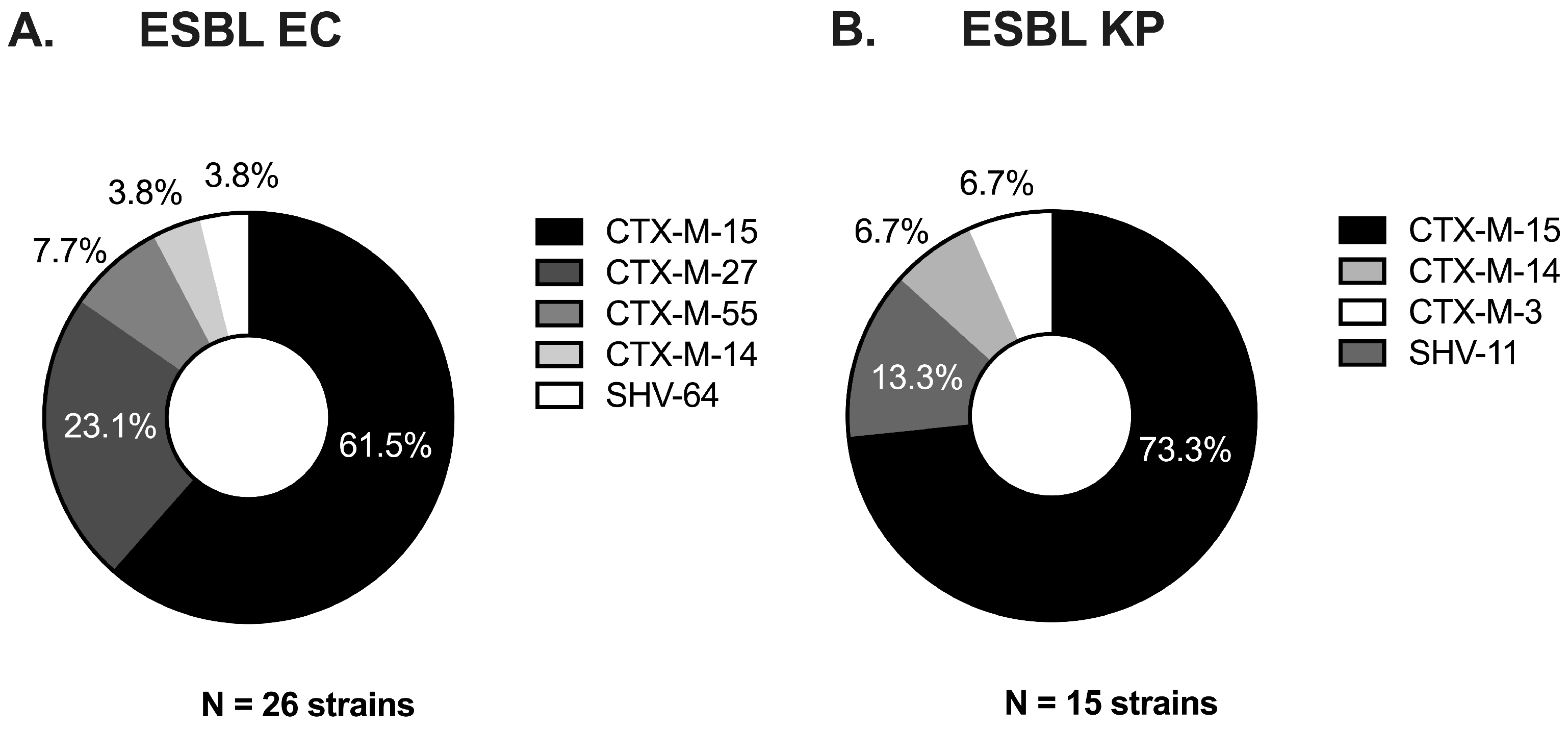
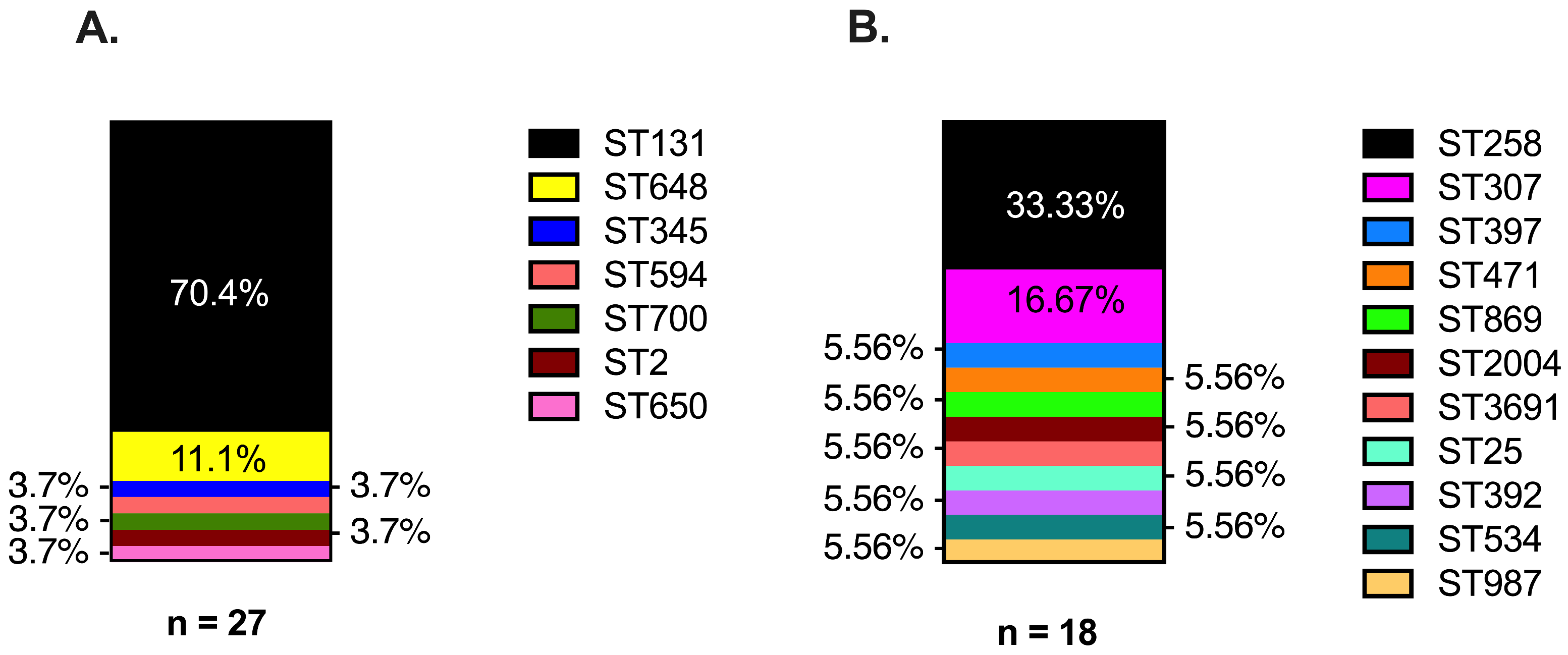
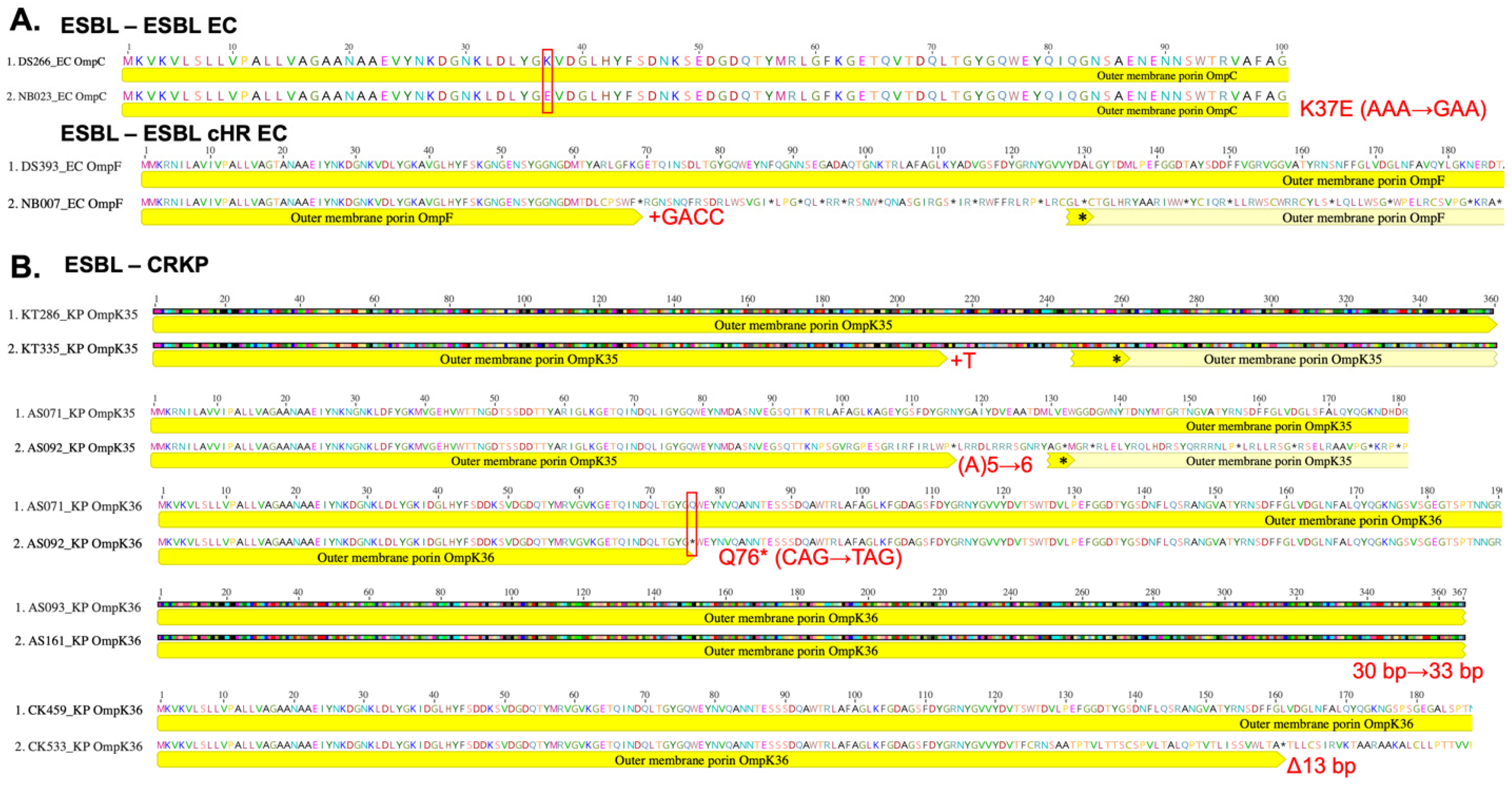
| Index Strain a,b | Evolved Strain a,b | ||||||
|---|---|---|---|---|---|---|---|
| Isolate ID | AMR c | Cas | CRISPR Spacers 5′-3′ | Isolate ID | AMR | Cas | CRISPR Spacers 5′-3′ |
| CK063 | ESBL EC | Cas3 | CGTTTTTAGCCTACCTATAAGGAATTGAAAC | CK178 | ESBL EC | none | GCCGGATGCGGCGTGAACGCCTTATCCGGCCTACAAAAGAAATGCAG |
| CCACCTTTTTTACCTGCTTCAGATGC | TTTTTGATAGTTGGAGTCGCTTTGTCTT | ||||||
| ATCTGCCTGTACGGCAGTGAACT | TCTACAAGGACACAGACACACTTC | ||||||
| ATCTGCCTGTACGGCAGTGAACT | |||||||
| PN312 | ESBL EC | Cas3 | GCCGGATGCGGCGTGAACGCCTTATCCGGCCTACAAAAGAAATGCAG | DS181 | ESBL cHR EC | none | GCCGGATGCGGCGTGAACGCCTTATCCGGCCTACAAAAGAAATGCAG |
| CGACCCCCACCATGTCAAGGTGGTGCTCTAACCAACTGAGCTA | CCAGAGAAGCCGCCAAAGCCGCTTCCGCC | ||||||
| GTTTTTAGCCTACCTATAAGGAATTGAAACAGGT | TTTTTGATAGTTGGAGTCGCTTTGTCTT | ||||||
| GTTTTTAGCCTACCTATAAGGAATTGAAAC | AGTTCACTGCCGTACAGGCAGCT | ||||||
| CCACCTTTTTTACCTGCTTCAGATGC | |||||||
| ATCTGCCTGTACGGCAGTGAACT | |||||||
| KT168 | ESBL KP | Cas5 | TTGTGCCAACAGAATGCCAACAAAGTGCCA | KT212 | CRKP | none | TTGTGCCAACAGAATGCCAACAAAGTGCCA |
| AATAAAAACCATAAAAACCACAGT | AATAAAAACCATAAAAACCACAGT | ||||||
| GTTTTTAGCCTACCTATAAGGAATTGAAAC | |||||||
| AAGGCGTCAGCCGCCGCCCGGCA | |||||||
| Genes Mutated | Gene Function | Total N = 27 | CRISPR-Cas Positive n = 6 | CRISPR-Cas Negative n = 21 | ESBL-ESBL n = 16 | ESBL-cHR n = 10 | CREC-CREC n = 1 |
|---|---|---|---|---|---|---|---|
| Resistance | |||||||
| ompC | porin OmpC | 1 (3.70%) | 1 (16.67%) | 1 (6.25%) | |||
| ompF | porin OmpF | 1 (3.70%) | 1 (16.67%) | 1 (1.00%) | |||
| ompR | Two component system response regulator OmpR | 1 (3.70%) | 1 (16.67%) | 1 (1.00%) | |||
| Virulence | |||||||
| rfaQ | lipopolysaccharide core heptosyltransferase RfaQ | 1 (3.70%) | 1 (16.67%) | 1 (1.00%) | |||
| lptF | LPS export permease LptF | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| wzzB | LPS O-antigen chain length determinant protein WzzB | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| arnD | 4-deoxy-4-formamido-L-arabinose- phosphoundecaprenol deformylase | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| wcaM | colanic acid biosynthesis protein WcaM | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| waaU | glycosyltransferase family 9 protein | 1 (3.70%) | 1 (4.76%) | 1 (100%) | |||
| fyuA | siderophore yersiniabactin receptor FyuA | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| entF | enterobactin synthetase EntF | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| ycgR | flagellar brake protein | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| fecR | ferric citrate uptake regulator FecR | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| agn43 | autotransporter adhesin Ag43 | 2 (7.40%) | 2 (9.52%) | 1 (6.25%) | 1 (1.00%) | ||
| iutA | ferric aerobactin receptor IutA | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| yggR | type IV pilus twitching motility protein PilT | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| papX | transcriptional regulator PapX | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| cheY | chemotaxis response regulator CheY | 1 (3.70%) | 1 (4.76%) | 1 (1.00%) | |||
| Metabolism | |||||||
| cydA | cytochrome ubiquinol oxidase subunit I | 1 (3.70%) | 1 (16.67%) | 1 (1.00%) | |||
| lacY | lactose permease | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| dtpA | dipeptide/tripeptide permease DtpA | 1 (3.70%) | 1 (16.67%) | 1 (1.00%) | |||
| dtpC | dipeptide/tripeptide permease DtpC | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| hycE | formate hydrogenlyase subunit HycE | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| fdrA | acyl-CoA synthetase FdrA | 1 (3.70%) | 1 (4.76%) | 1 (6.25%) | |||
| aaeB | p-hydroxybenzoic acid efflux subunit AaeB | 1 (3.70%) | 1 (16.67%) | 1 (1.00%) |
| Genes Mutated | Gene Function | Total N = 18 | CRISPR-Cas Positive n = 5 | CRISPR-Cas Negative n = 13 | ESBL-ESBL n = 7 | ESBL-CRKP n = 8 | CRKP-CRKP n = 3 |
|---|---|---|---|---|---|---|---|
| Resistance | |||||||
| ompK36 | porin OmpK36 | 2 (11.11%) | 1 (20.00%) | 1 (7.69%) | 2 (25.00%) | ||
| ompK35 | Porin OmpK35 | 2 (11.11%) | 2 (15.38%) | 2 (25.00%) | |||
| Virulence | |||||||
| ecpD | fimbrial adhesin EcpD | 1 (5.56%) | 1 (7.69%) | 1 (12.50%) | |||
| lptB | Lipopolysaccharide export system ATP-binding protein LptB | 1 (5.56%) | 1 (7.69%) | 1 (12.50%) | |||
| wzi | capsule assembly Wzi family protein | 1 (5.56%) | 1 (7.69%) | 1 (12.50%) | |||
| wbgU | UDP-N-acetylglucosamine 4-epimerase | 1 (5.56%) | 1 (7.69%) | 1 (33.33%) | |||
| Metabolism | |||||||
| rsxC | electron transport complex subunit RsxC | 1 (5.56%) | 1 (20.00%) | 1 (14.29%) | |||
| uhpT | hexose-6-phosphate:phosphate antiporter | 1 (5.56%) | 1 (20.00%) | 1 (14.29%) | |||
| kbl | glycine C-acetyltransferase | 1 (5.56%) | 1 (20.00%) | 1 (14.29%) | |||
| gyrA | DNA topoisomerase subunit A | 1 (5.56%) | 1 (20.00%) | 1 (12.50%) | |||
| metF | methylenetetrahydrofolate reductase | 1 (5.56%) | 1 (20.00%) | 1 (12.50%) | |||
| cpxA | Sensor histidine kinase CpxA | 1 (5.56%) | 1 (7.69%) | 1 (12.50%) | |||
| sodA | superoxide dismutase | 1 (5.56%) | 1 (7.69%) | 1 (12.50%) | |||
| cydB | cytochrome d ubiquinol oxidase subunit II | 1 (5.56%) | 1 (7.69%) | 1 (12.50%) | |||
| cyoA | cytochrome o ubiquinol oxidase subunit II | 1 (5.56%) | 1 (7.69%) | 1 (12.50%) | |||
| phoQ | Sensor protein PhoQ | 1 (5.56%) | 1 (7.69%) | 1 (33.33%) | |||
| narL | Nitrate/nitrite response regulator protein NarL | 1 (5.56%) | 1 (7.69%) | 1 (33.33%) |
| Isolate ID | MLST | AMR a | Presence of CRISPR | KPC Gene | Porin Gene Mutated | Mutation | Meropenem MIC (μg/mL) |
|---|---|---|---|---|---|---|---|
| DS393 | 10 | ESBL EC | Yes | ≤0.015625 | |||
| NB007 | ESBL cHR EC | Yes | ompF | Addition; +GACC | 0.03125 | ||
| KT286 | 25 | ESBL KP | No | 0.0625 | |||
| KT335 | CRKP | No | No | ompK35 | Addition; +T | 8 | |
| AS071 | 307 | ESBL KP | No | 2 | |||
| AS092 | CRKP | No | No | ompK35 ompK36 | Addition; A(5)->(6) SNP; Q76 * | 16 | |
| CK459 | 534 | ESBL KP | Yes | 0.03125 | |||
| CK533 | CRKP | Yes | No | ompK36 | Deletion; Δ13 bp | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalu, M.C.; Acharya, A.; Jorth, P.; Wong-Beringer, A. ESBL-Producing Escherichia coli and Klebsiella pneumoniae Exhibit Divergent Paths During In-Human Evolution Towards Carbapenem Resistance. Microorganisms 2025, 13, 1387. https://doi.org/10.3390/microorganisms13061387
Kalu MC, Acharya A, Jorth P, Wong-Beringer A. ESBL-Producing Escherichia coli and Klebsiella pneumoniae Exhibit Divergent Paths During In-Human Evolution Towards Carbapenem Resistance. Microorganisms. 2025; 13(6):1387. https://doi.org/10.3390/microorganisms13061387
Chicago/Turabian StyleKalu, Michelle Chioma, Akanksha Acharya, Peter Jorth, and Annie Wong-Beringer. 2025. "ESBL-Producing Escherichia coli and Klebsiella pneumoniae Exhibit Divergent Paths During In-Human Evolution Towards Carbapenem Resistance" Microorganisms 13, no. 6: 1387. https://doi.org/10.3390/microorganisms13061387
APA StyleKalu, M. C., Acharya, A., Jorth, P., & Wong-Beringer, A. (2025). ESBL-Producing Escherichia coli and Klebsiella pneumoniae Exhibit Divergent Paths During In-Human Evolution Towards Carbapenem Resistance. Microorganisms, 13(6), 1387. https://doi.org/10.3390/microorganisms13061387







