Virulence and Antibiotic Resistance of Pathogenic Aeromonas caviae from Diseased Macrobrachium rosenbergii
Abstract
1. Introduction
2. Materials and Methods
2.1. Prawn Disease Description and Pathogen Examination
2.2. Morphological Observation of Aeromonas caviae GMRS4
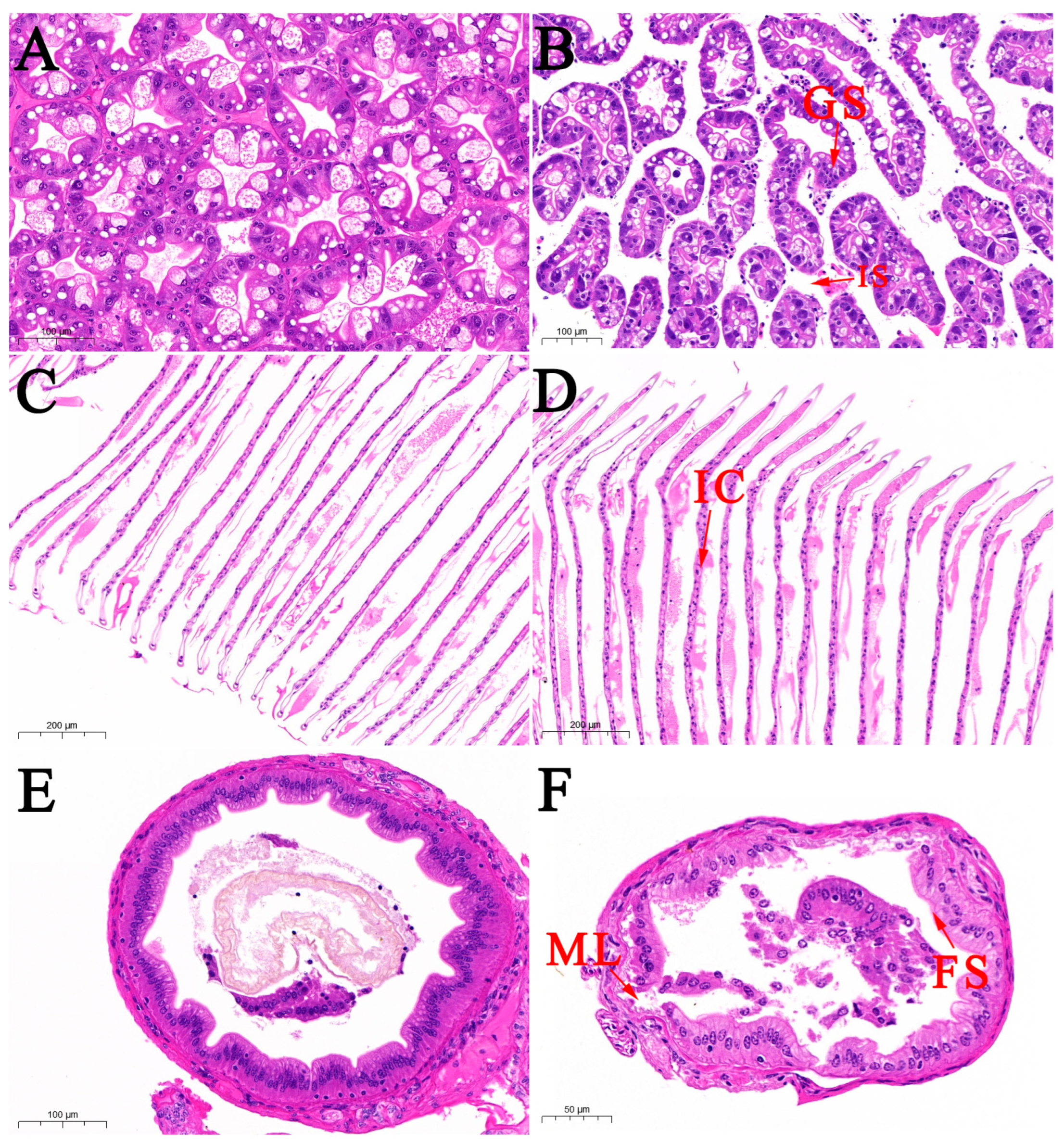
2.3. Histopathology
2.4. Identification of Bacterial Isolates
2.5. Bacterial Virulence Assay
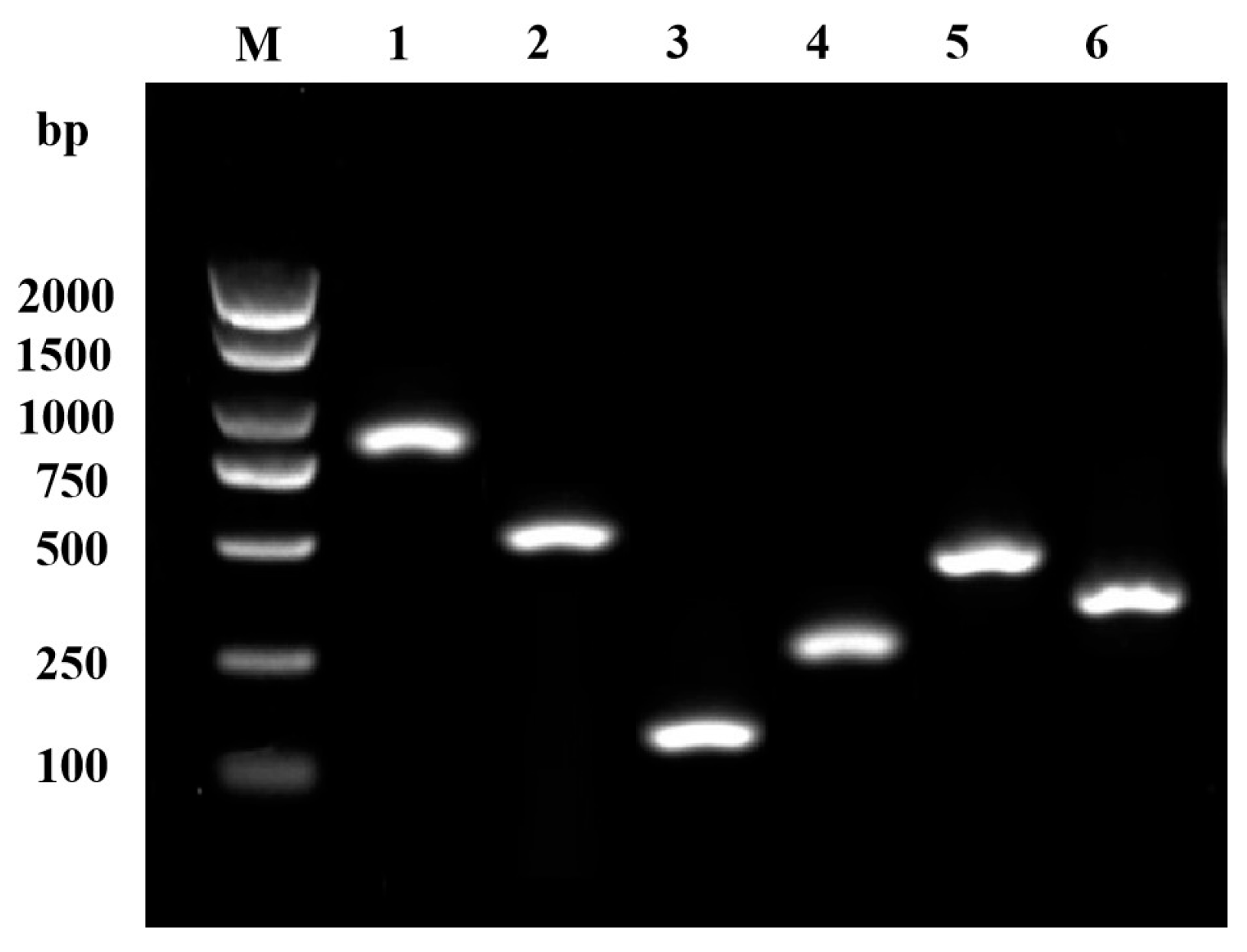
2.6. Determination of Virulence-Related Factors
2.7. Antibiotic Sensitivity Test
2.8. Whole-Genome Sequencing and Assembly
2.9. Gene Prediction and Database Annotation
3. Results
3.1. Electron Microscopic Observation of the Isolate GMRS4
3.2. Histological Observation
3.3. Physiological and Biochemical Characterization
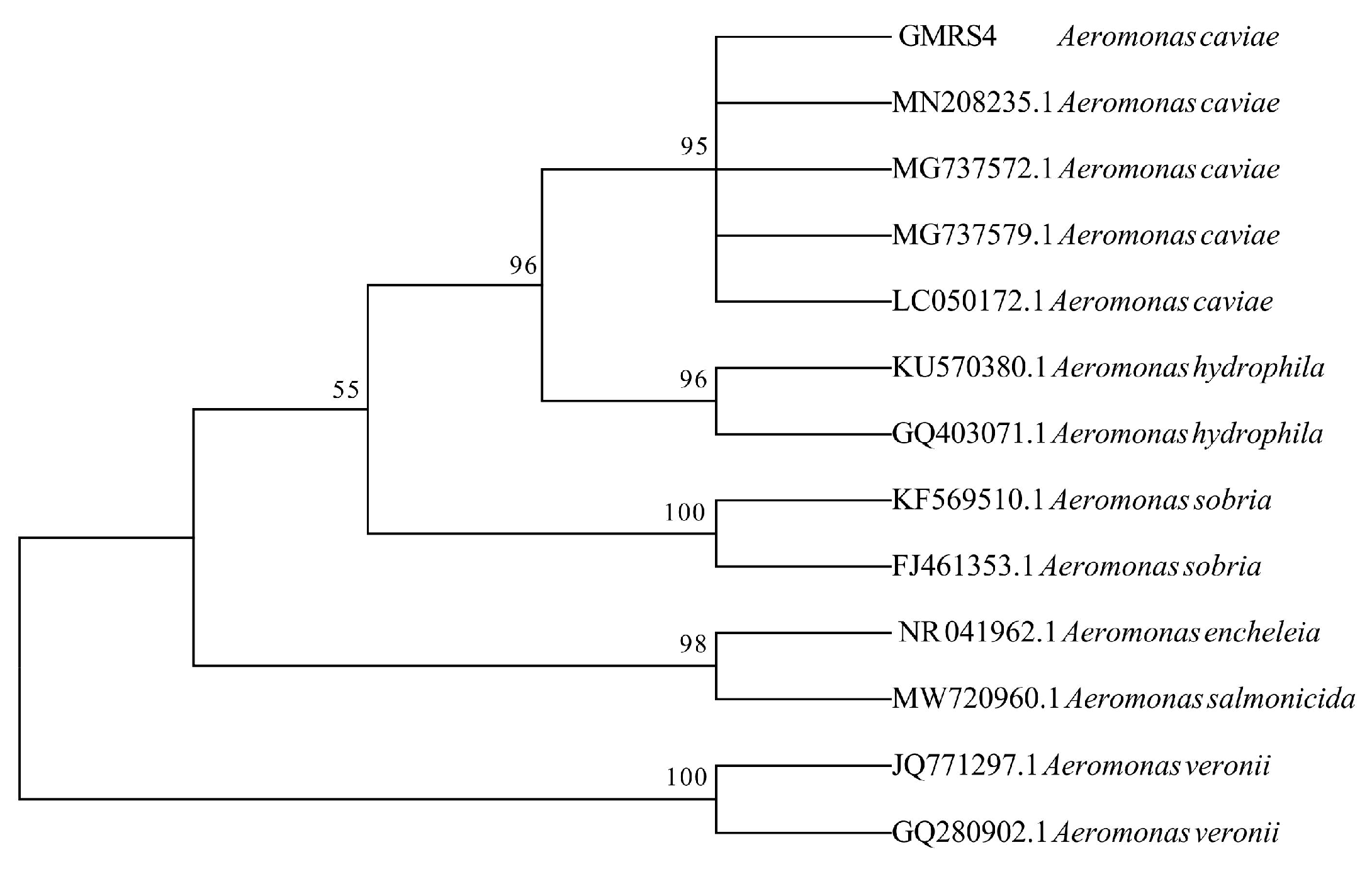
3.4. Molecular Identification
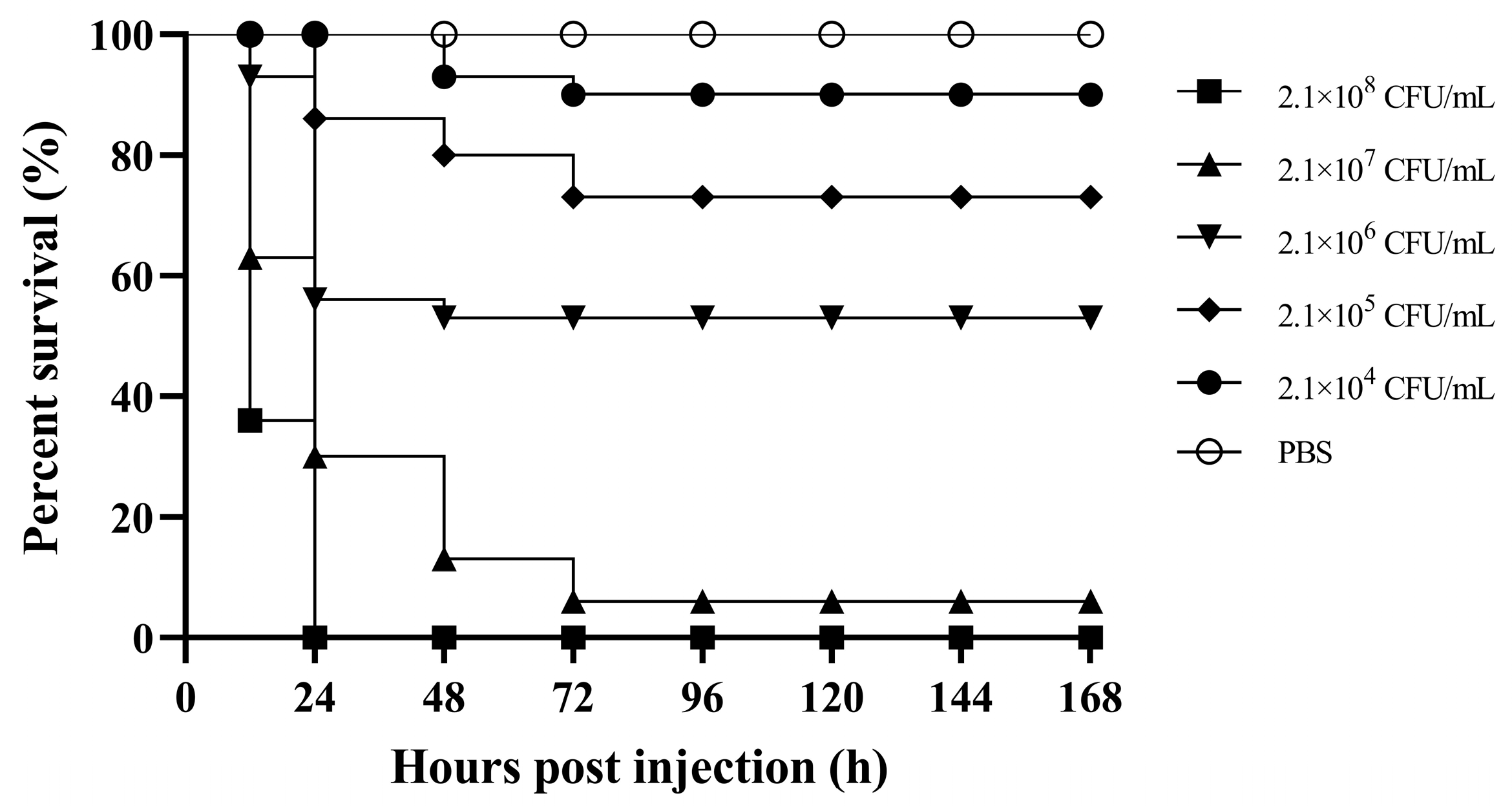
3.5. Virulence of A. caviae GMRS4
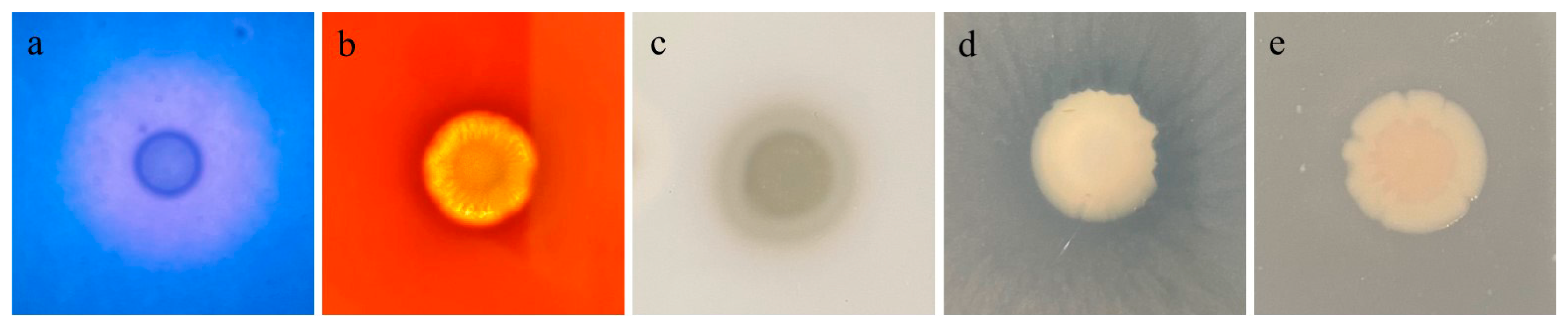
3.6. Virulence Factors of GMRS4
3.7. Antibiotic Sensitivity of GMRS4
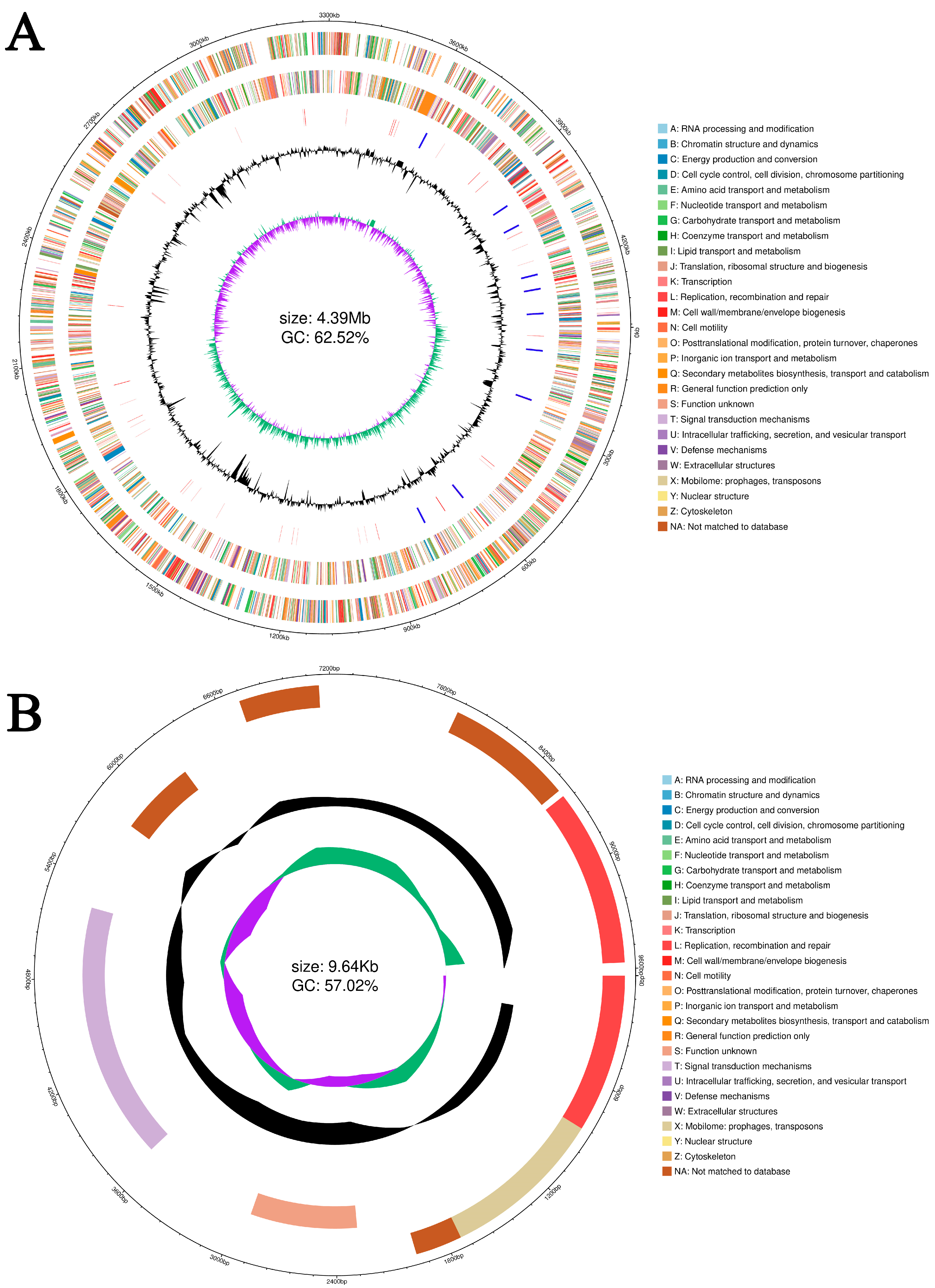
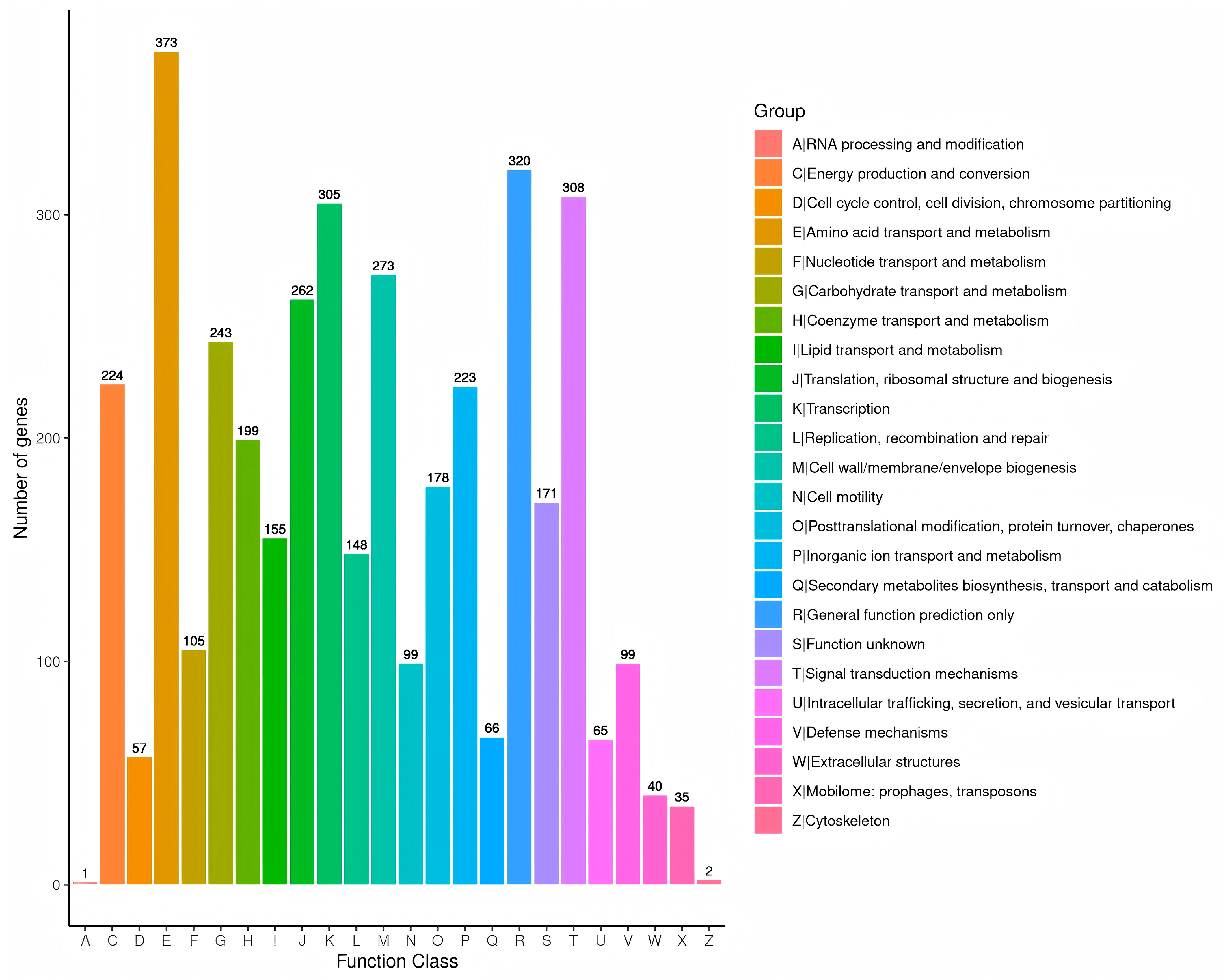
3.8. Genome Structure and General Features of A. caviae GMRS4 Genome
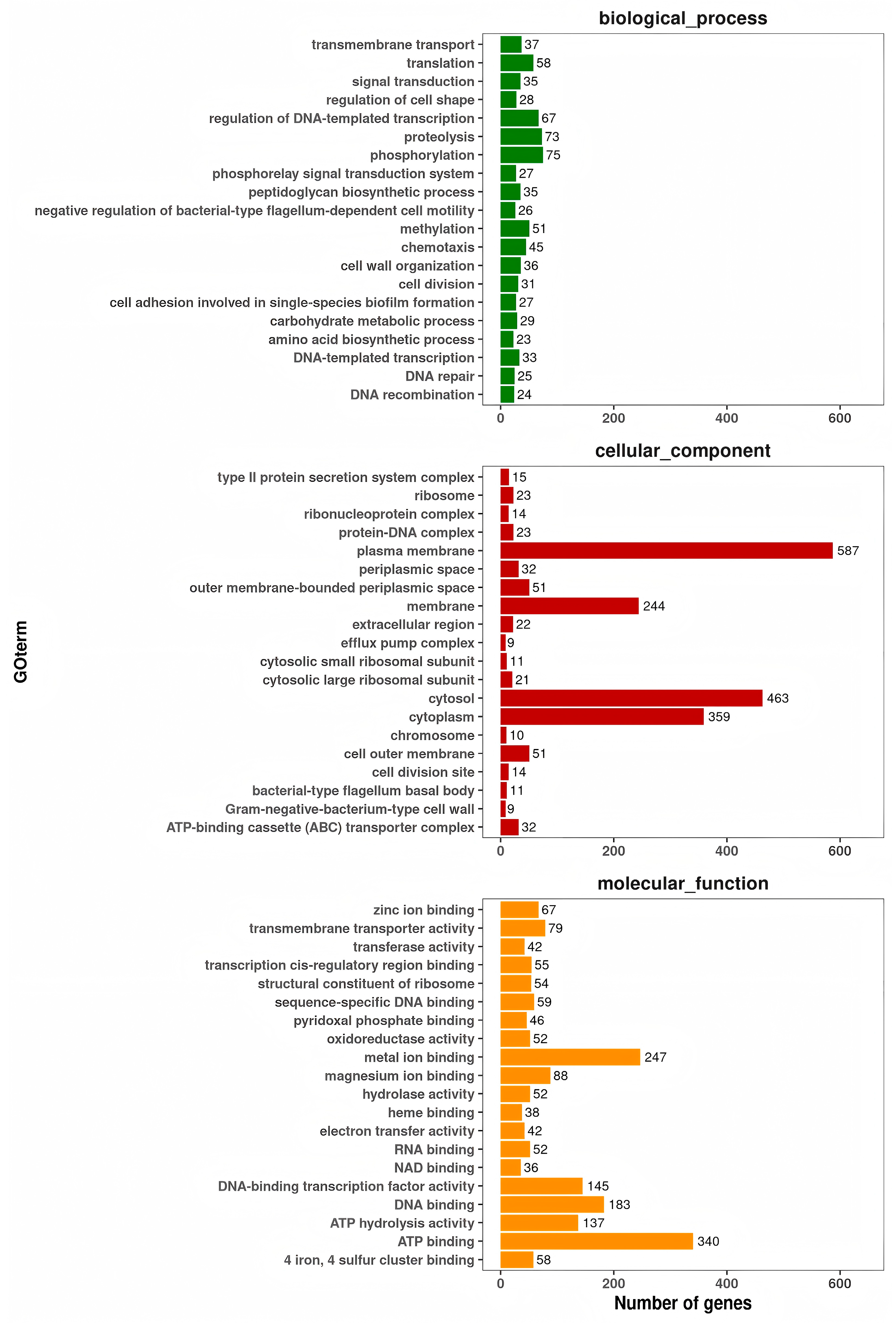
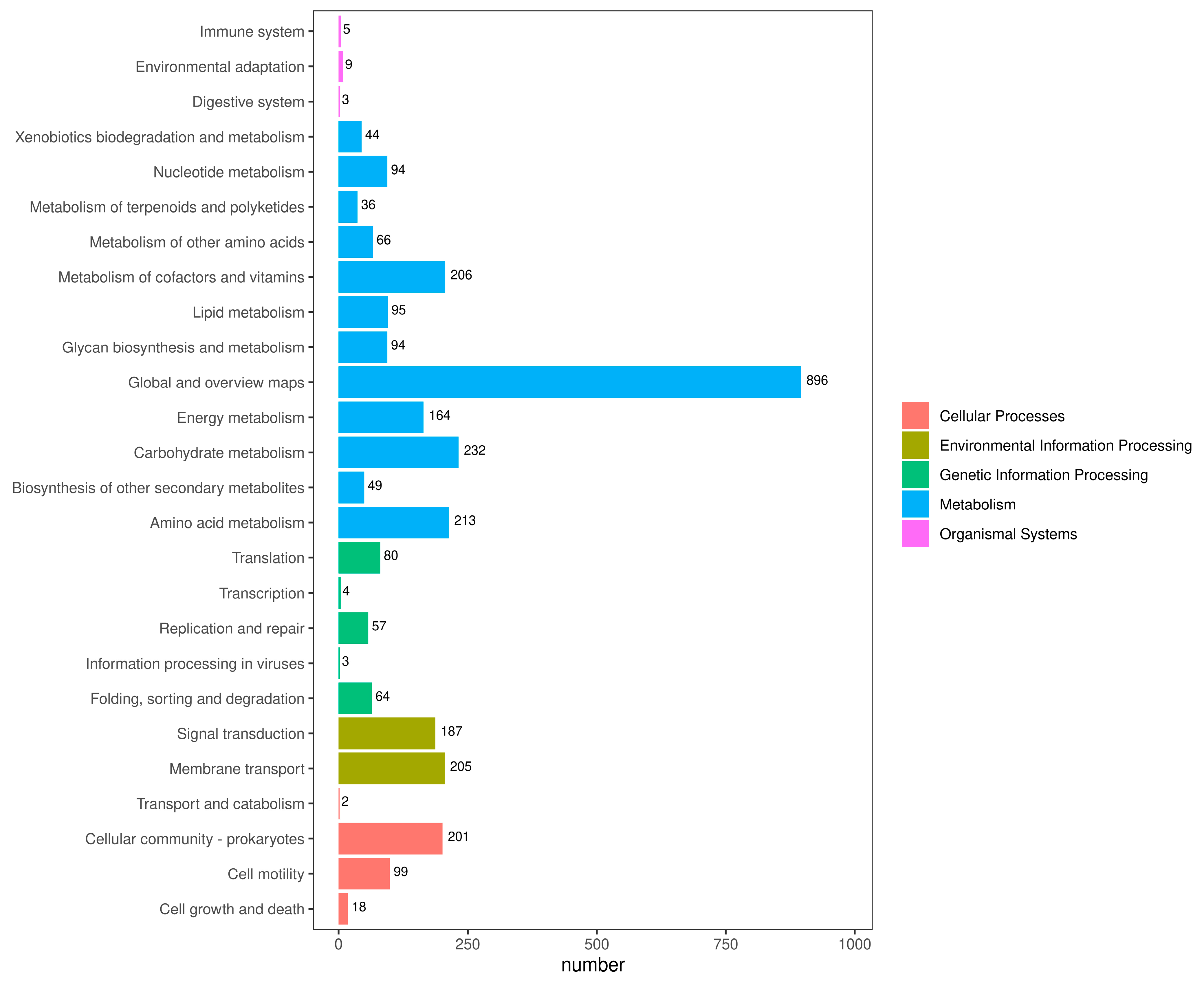
3.9. Prediction of Virulence Genes of A. caviae GMRS4
3.10. Prediction of Resistance Genes of A. caviae GMRS4
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chavanne, H.; Janssen, K.; Hofherr, J.; Contini, F.; Haffray, P.; Consortium, A.; Komen, H.; Nielsen, E.E.; Bargelloni, L. A comprehensive survey on selective breeding programs and seed market in the European aquaculture fish industry. Aquac. Int. 2016, 24, 1287–1307. [Google Scholar] [CrossRef]
- Regan, T.; Bean, T.P.; Ellis, T.; Davie, A.; Carboni, S.; Migaud, H.; Houston, R.D. Genetic improvement technologies to support the sustainable growth of UK aquaculture. Rev. Aquac. 2021, 13, 1958–1985. [Google Scholar] [CrossRef]
- Li, J.; Xu, S.; Cai, Y.; Jiang, Y.; Chen, H.; Lin, L.; Lv, X. The effects of net-chasing training on survival and growth are related to stocking density in the freshwater prawn Macrobrachium rosenbergii. Aquaculture 2022, 561, 738621. [Google Scholar] [CrossRef]
- Tan, K.; Wang, W. The early life culture and gonadal development of giant freshwater prawn, Macrobrachium rosenbergii: A review. Aquaculture 2022, 559, 738357. [Google Scholar] [CrossRef]
- Gao, X.; Miao, Z.; Li, X.; Chen, N.; Gu, W.; Liu, X.; Yang, H.; Wei, W.; Zhang, X. Pathogenicity of non-O1/O139 Vibrio cholerae and its induced immune response in Macrobrachium rosenbergii. Fish. Shellfish. Immun. 2019, 92, 300–307. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Y.; Jiang, Q.; Yang, H.; Pi, D.; Liu, X.; Gao, X.; Chen, N.; Zhang, X. Virulence properties of Vibrio vulnificus isolated from diseased zoea of freshness shrimp Macrobrachium rosenbergii. Microb. Pathog. 2019, 127, 166–171. [Google Scholar] [CrossRef]
- Jayaprakash, N.; Pai, S.; Philip, R.; Singh, I. Isolation of a pathogenic strain of Vibrio alginolyticus from necrotic larvae of Macrobrachium rosenbergii (de Man). J. Fish. Dis. 2006, 29, 187–191. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, H.; Jiang, Q.; Chen, N.; Li, X.; Liu, X.; Yang, H.; Wei, W.; Zhang, X. Enterobacter cloacae associated with mass mortality in zoea of giant freshwater prawns Macrobrachium rosenbergii and control with specific chicken egg yolk immunoglobulins (IgY). Aquaculture 2019, 501, 331–337. [Google Scholar] [CrossRef]
- Gao, X.; Chen, Z.; Zhang, Z.; Qian, Q.; Chen, A.; Qin, L.; Tang, X.; Jiang, Q.; Zhang, X. Pathogenicity of Aeromonas veronii Isolated from Diseased Macrobrachium rosenbergii and Host Immune-Related Gene Expression Profiles. Microorganisms 2024, 12, 694. [Google Scholar] [CrossRef]
- Sung, H.H.; Hwang, S.F.; Tasi, F.M. Responses of giant freshwater prawn (Macrobrachium rosenbergii) to challenge by two strains of Aeromonas spp. J. Invertebr. Pathol. 2000, 76, 278–284. [Google Scholar] [CrossRef]
- Xu, W.; Zhao, C.; Pan, X.; He, X.; Liu, X.; Hou, J.; Pang, J. Transmission Routes and a New Freshwater Crustacean Host for Infectious Precocity Virus (IPV). Transbound. Emerg. Dis. 2023, 1, 3950107. [Google Scholar] [CrossRef]
- Low, C.F.; Yusoff, M.R.; Kuppusamy, G.; Ahmad, N.F. Molecular biology of Macrobrachium rosenbergii nodavirus infection in giant freshwater prawn. J. Fish. Dis. 2018, 41, 1771–1781. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.; Debnath, P.P.; Stentiford, G.D.; Bateman, K.S.; Salin, K.R.; Bass, D. Diseases of the giant river prawn Macrobrachium rosenbergii: A review for a growing industry. Rev. Aquac. 2023, 15, 738–758. [Google Scholar] [CrossRef]
- Qian, Q.; Zhou, Y.; Chen, Z.; Zhu, Y.; Xu, J.; Gao, X.; Jiang, Q.; Wang, J.; Zhang, X. Pathogenesis and complete genome sequence of Decapod iridescent virus 1 (DIV1) associated with mass mortality in Macrobrachium rosenbergii. Aquaculture 2023, 566, 739220. [Google Scholar] [CrossRef]
- Jones, B.L.; Wilcox, M.H. Aeromonas infections and their treatment. J. Antimicrob. Chemoth. 1995, 35, 453–461. [Google Scholar] [CrossRef]
- De Schryver, P.; Vadstein, O. Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J. 2014, 8, 2360–2368. [Google Scholar] [CrossRef] [PubMed]
- Cholewińska, P.; Moniuszko, H.; Wojnarowski, K.; Pokorny, P.; Szeligowska, N.; Dobicki, W.; Polechoński, R.; Górniak, W. The occurrence of microplastics and the formation of biofilms by pathogenic and opportunistic bacteria as threats in aquaculture. Int. J. Environ. Res. Public Health 2022, 19, 8137. [Google Scholar] [CrossRef] [PubMed]
- Mulia, D.S.; Isnansetyo, A.; Pratiwi, R.; Asmara, W. Molecular characterizations of Aeromonas caviae isolated from catfish (Clarias sp.). AACL Bioflux 2020, 13, 2717–2732. [Google Scholar]
- Agarwal, P.; Kayala, P.; Chandrasekaran, N.; Mukherjee, A.; Shah, S.; Thomas, J. Antioxidant and antibacterial activity of Gelidium pusillum (Stackhouse) against Aeromonas caviae and its applications in aquaculture. Aquac. Int. 2021, 29, 845–858. [Google Scholar] [CrossRef]
- Thomas, J.; Madan, N.; Nambi, K.S.N.; Majeed, S.A.; Basha, A.N.; Hameed, A.S. Studies on ulcerative disease caused by Aeromonas caviae-like bacterium in Indian catfish, Clarias batrachus (Linn). Aquaculture 2013, 376, 146–150. [Google Scholar] [CrossRef]
- Roy, A.; Singha, J.; Abraham, T.J. Histopathology of Aeromonas caviae infection in challenged Nile tilapia Oreochromis niloticus (Linnaeus, 1758). Int. J. Aquac. 2018, 8, 151. [Google Scholar] [CrossRef]
- Xue, M.; Xiao, Z.; Li, Y.; Jiang, N.; Liu, W.; Meng, Y.; Fan, Y.D.; Zeng, L.B.; Zhou, Y. Isolation, identification and characteristics of Aeromonas caviae from diseased largemouth bass (Micropterus salmoides). Fishes 2022, 7, 119. [Google Scholar] [CrossRef]
- Dai, Y.; Gao, J.; Jiang, M. Case Report: A rare infection of multidrug-resistant Aeromonas caviae in a pediatric case with acute lymphoblastic leukemia and review of the literature. Front. Pediatr. 2024, 12, 1233600. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Sharma, C.; Srivastava, A.; Kumar, B.N.; Pathak, D.; Rai, S. Isolation, characterization and complete genome sequencing of fish pathogenic Aeromonas veronii from diseased Labeo rohita. Aquaculture 2022, 553, 738085. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.A. Host-pathogen interactions: Redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 1999, 67, 3703–3713. [Google Scholar] [CrossRef]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence factors of Pseudomonas aeruginosa and antivirulence strategies to combat its drug resistance. Front. Cell. Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, F.; Jia, L.; Yan, H.; Gao, L.; Tian, Y.; Su, X.; Zhang, X.; Zhang, X.; Lv, C.; et al. Edwardsiella piscicida infection reshapes the intestinal microbiome and metabolome of big-belly seahorses: Mechanistic insights of synergistic actions of virulence factors. Front. Immunol. 2023, 14, 1135588. [Google Scholar] [CrossRef]
- Haslinger-Löffler, B.; Kahl, B.C.; Grundmeier, M.; Strangfeld, K.; Wagner, B.; Fischer, U.; Cheung, A.L.; Peters, G.; Schulze, O.K.; Sinha, B. Multiple virulence factors are required for Staphylococcus aureus-induced apoptosis in endothelial cells. Cell. Microbiol. 2005, 7, 1087–1097. [Google Scholar] [CrossRef]
- Nguyen, Q.; Nguyen, T.T.N.; Pham, P.; Chau, V.; Nguyen, L.P.H.; Nguyen, T.D.; Ha, T.T.; Le, N.T.Q.; Vu, D.T.; Baker, S.; et al. Genomic insights into the circulation of pandemic fluoroquinolone-resistant extra-intestinal pathogenic Escherichia coli ST1193 in Vietnam. Microb. Genom. 2021, 7, 000733. [Google Scholar] [CrossRef]
- Zhu, L.; Lian, Y.; Lin, D.; Huang, D.; Yao, Y.; Ju, F.; Wang, M. Insights into microbial contamination in multi-type manure-amended soils: The profile of human bacterial pathogens, virulence factor genes and antibiotic resistance genes. J. Hazard. Mater. 2022, 437, 129356. [Google Scholar] [CrossRef]
- Gao, X.; Zhou, Y.; Zhu, X.; Tang, H.; Li, X.; Jiang, Q.; Wei, W.; Zhang, X. Enterobacter cloacae: A probable etiological agent associated with slow growth in the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 2021, 530, 735826. [Google Scholar] [CrossRef]
- Brenner, D.J.; Krieg, N.R.; Staley, J.T. Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: Greer, SC, USA, 2021; Volume 2, Part B, pp. 404–406. [Google Scholar]
- Zhang, X.J.; Bai, X.S.; Yan, B.L.; Bi, K.R.; Qin, L. Vibrio harveyi as a causative agent of mass mortalities of megalopa in the seed production of swimming crab Portunus trituberculatus. Aquac. Int. 2014, 22, 661–672. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Behreans, A.; Karber, L. Determination of LD50. In Screening in Pharmacology; Academic Press: New York, NY, USA, 1953. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics 2016, 32, 2103–2110. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Bertelli, C.; Brinkman, F.S. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 2018, 34, 2161–2167. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32 (Suppl. S1), D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32 (Suppl. S1), D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.D.; White, O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003, 31, 371–373. [Google Scholar] [CrossRef]
- Kimura, M.; Araoka, H.; Yoneyama, A. Aeromonas caviae is the most frequent pathogen amongst cases of Aeromonas bacteremia in Japan. Scand. J. Infect. Dis. 2013, 45, 304–309. [Google Scholar] [CrossRef]
- Wu, R.; Shen, J.; Tian, D.; Yu, J.; He, T.; Yi, J.; Li, Y. A potential alternative to traditional antibiotics in aquaculture: Yeast glycoprotein exhibits antimicrobial effect in vivo and in vitro on Aeromonas caviae isolated from Carassius auratus gibelio. Vet. Med. Sci. 2020, 6, 639–648. [Google Scholar] [CrossRef]
- Zhou, L.; Jang, Z.; Qian, Q.; Chen, Z.; Gu, S.; Li, J.; Ji, P.; Gao, X.; Jang, O.; Zhang, X. Identification and pathogenicity of pathogenic Aeromonas caviae isolated from Eriocheir sinensis. Freshw. Fish. 2022, 52, 58–65. [Google Scholar]
- Zeng, X.; Cao, H.; Mu, C. Isolation and antibiotic susceptibility ofan Aeromonas caviae pathogen from yellow legdisease-infected Penaeus vannamei. J. Aquac. 2020, 41, 28–33. [Google Scholar]
- Roberts, A.J.; Wiedmann, M. Pathogen, host and environmental factors contributing to the pathogenesis of listeriosis. Cell. Mol. Life Sci. 2003, 60, 904–918. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Figueras, M.J. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 2013, 36, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, J.M.; Kidd, S.P.; Schmidt, R. Secreted enzymes of Aeromonas. FEMS Microbiol. Lett. 1997, 152, 1–10. [Google Scholar] [CrossRef]
- Wang, G.; Clark, C.G.; Liu, C.; Pucknell, C.; Munro, C.K.; Kruk, T.M.; Caldeira, R.; Woodward, D.; Rodgers, F.G. Detection and characterization of the hemolysin genes in Aeromonas hydrophila and Aeromonas sobria by multiplex PCR. J. Clin. Microbiol. 2003, 41, 1048–1054. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Gryllos, I.; Tomás, J.M.; Shaw, J.G. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect. Immun. 2001, 69, 4257–4267. [Google Scholar] [CrossRef] [PubMed]
- Hanchanachai, N.; Chumnanpuen, P.; E-kobon, T. Effect of conditioned media from Aeromonas caviae on the transcriptomic changes of the porcine isolates of Pasteurella multocida. BMC Microbiol. 2022, 22, 272. [Google Scholar] [CrossRef] [PubMed]
- Pugazhendhi, A.S.; Wei, F.; Hughes, M.; Coathup, M. Bacterial adhesion, virulence, and biofilm formation. In Musculoskeletal Infection; Springer International Publishing: Cham, Switzerland, 2022; pp. 19–64. [Google Scholar]
- Ghssein, G.; Ezzeddine, Z. A review of Pseudomonas aeruginosa metallophores: Pyoverdine, pyochelin and pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Cornelis, P.; Tahrioui, A.; Lesouhaitier, O.; Bouffartigues, E.; Feuilloley, M.; Baysse, C.; Chevalier, S. High affinity iron uptake by pyoverdine in Pseudomonas aeruginosa involves multiple regulators besides Fur, PvdS, and FpvI. Biometals 2023, 36, 255–261. [Google Scholar] [CrossRef]
- Cherian, T.; Ragavendran, C.; Vijayan, S.; Kurien, S.; Peijnenburg, W.J. A review on the fate, human health and environmental impacts, as well as regulation of antibiotics used in aquaculture. Environ. Adv. 2023, 13, 100411. [Google Scholar] [CrossRef]
- Inuwa, A.B.; Pervez, A.; Nazir, R. Microalgae-based wastewater treatment system: Current state, antibiotic resistant bacteria and antibiotic resistance genes reduction potentials. Int. J. Environ. Sci. Technol. 2023, 20, 14053–14072. [Google Scholar] [CrossRef]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and bacterial resistance—A short story of an endless arms race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef]
- Nijland, M.; Lefebvre, S.N.; Thangaratnarajah, C.; Slotboom, D.J. Bidirectional ATP-driven transport of cobalamin by the mycobacterial ABC transporter BacA. Nat. Commun. 2024, 15, 2626. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cao, Z.; Cui, L.; Hu, T.; Guo, K.; Zhang, F.; Wang, X.; Peng, Z.; Liu, Q.; Dai, M. Enrofloxacin promotes plasmid-mediated conjugation transfer of fluoroquinolone-resistance gene qnrS. Front. Microbiol. 2022, 12, 773664. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qin, L.; Zhu, Y.; Qian, Q.; Gao, X.; Jiang, Q.; Wang, J.; Liu, G.; Zhang, X. Characteristics and complete genome analysis of a pathogenic Aeromonas Veronii SJ4 from diseased Siniperca Chuatsi. Mar. Biotechnol. 2023, 25, 966–982. [Google Scholar] [CrossRef] [PubMed]

| Sequence ID | Temperature/°C; | Product Length (bp) | Primer Sequences (5′-3′) | Gene |
|---|---|---|---|---|
| ctg_01684 | 55 | 911 | ATTGGATCCCTGCCTA | ahp |
| GCTAAGCTTGCATCCG | ||||
| ctg_00796 | 55 | 513 | ACACGGTCAAGGAGATCAAC | ahyB |
| ACACGGTCAAGGAGATCAAC | ||||
| ctg_01472 | 63 | 194 | GCTACTGTCAAGCTGGACTC | flgM |
| AGATTGGCCTCGAAACTG | ||||
| ctg_02189 | 63 | 289 | CGGACGATTATCAGGATGG | hly |
| CAAGAACGAGTTTCAGTGGC | ||||
| ctg_03161 | 63 | 442 | TGACCCAGTCCTGG | alt |
| GGTGATCGATCACC | ||||
| ctg_04074 | 63 | 301 | AACCGAACTCTCCAT | aer |
| CGCCTTGTCCTTGTA |
| Aeromonas caviae * | GMRS4 | Characteristics |
|---|---|---|
| + | + | Cellobiose |
| + | + | Xylose |
| + | + | Sucrose |
| + | + | Rhamnose |
| + | + | Mannose |
| − | − | Lactose |
| − | − | Arabinose |
| + | + | D-Trehalose anhydrous |
| + | + | Escin |
| − | − | Citrate |
| − | − | Inositol |
| − | − | Dacron |
| − | − | Raffinose |
| − | − | Tartrate |
| + | + | Salicin |
| − | − | Lysine |
| − | − | Hydrogen sulfide |
| + | + | Ammonium Gluconate |
| + | + | Arginine dihydrolase |
| + | + | α-Methyl-d-glucose ammonium |
| − | − | Peptone |
| Sensitivity | Diameter of Inhibition Zone/mm | Drug Content of Paper Disc/µg/disc | Drug | Drug Classification |
|---|---|---|---|---|
| R | 8 | 30 | Cefazolin | Cephalosporins |
| R | 8 | 30 | Ceftriaxone | |
| I | 16 | 30 | Cefuroxime | |
| S | 22 | 75 | Cefoperazone | |
| S | 30 | 30 | Cefotaxime | |
| S | 23 | 30 | Ceftriaxone | |
| S | 19 | 30 | Cefepime | |
| S | 20.5 | 30 | Ceftazidime | |
| S | 8 | 30 | Cefoxitin | |
| S | 19 | 30 | Amikacin | Aminoglycosides |
| S | 16 | 10 | Streptomycin | |
| I | 13.5 | 10 | Tobramycin | |
| S | 18 | 100 | Spectinomycin | |
| I | 17 | 30 | Kanamycin | |
| S | 18 | 10 | Gentamicin | |
| I | 13.5 | 30 | Neomycin | |
| R | 8 | 1 | Oxacillin | Penicillin |
| R | 8 | 10 | Penicillin G | |
| R | 8 | 10 | Ampicillin | |
| R | 14 | 100 | Piperacillin | |
| I | 14 | 15 | Erythromycin | Macrolides |
| I | 16 | 15 | Clarithromycin | |
| R | 8 | 2 | Clindamycin | |
| S | 19 | 30 | Tetracycline | Tetracyclines |
| S | 20 | 30 | Minocycline | |
| S | 26 | 10 | Norfloxacin | Quinolones |
| S | 24 | 5 | Ciprofloxacin | |
| S | 23 | 5 | Levofloxacin | |
| S | 25 | 5 | Ofloxacin | |
| S | 26 | 10 | Norfloxacin | Polypeptide |
| S | 24 | 5 | Ciprofloxacin | Furan |
| S | 23 | 5 | Levofloxacin | Amphenicols |
| S | 25 | 5 | Ofloxacin | Monobactams |
| I | 11.5 | 30 | Polymyxin B | Amides |
| R | 8 | 23.75 | Trimethoprim/sulfamethoxazole | Sulfonamides |
| Number | Related Gene | VF Name | VF Category |
|---|---|---|---|
| 65 | bcs1, kpsD, cpsA/uppS, lipA, kpsF, etc. | Capsule | Immune modulation |
| 39 | lpxC, lpxH, galE, licA, lpxB, msbB, waaQ, etc. | LOS | |
| 11 | sadH, fadD13, adhD, etc. | MymA operon | |
| 11 | ddrA, ppsC, etc. | PDIM | |
| 27 | lpxA/glmU, flmF2, etc. | LPS | |
| 14 | YPO_RS16495, YPTB_RS05510, etc. | O-antigen | |
| 15 | fbpB, fbpC, etc. | FbpABC | |
| 22 | hitC, hitB, hitA, etc. | HitABC | |
| 16 | sugC, sugA, sugB, etc. | Trehalose-recycling ABC transporter | |
| 14 | hemX, hemD, hemL, hemG, hemN, hemB, etc. | Heme biosynthesis | |
| 11 | allB, allS, allR, etc. | Allantion utilization | |
| 47 | pvdQ, PA2383, pvdN, pvdE, pvdI, ptxR, etc. | Pyoverdine | |
| 17 | pdxJ, CFF8240_RS05385, fleQ, flgP, fliL, etc. | Flagella | Motility |
| 75 | flgB, flrC, fliE, flaA, fliK, flgM, flgA, cheY, etc. | Polar flagella | |
| 17 | tapV, tapP, ASA_RS18105, tapU, tppF, etc. | Tap type IV pili | Adherence |
| 16 | mshN, mshC, mshJ, mshP, mshI1, etc. | MSHA type IV pili | |
| 20 | frpC, etc. | RTX protein | |
| 25 | rpoN, pilS, pilH, etc. | Type IV pili | |
| 14 | vpdC, lirB, LPG_RS00105, lidL, etc. | Dot/Icm T4SS secreted effectors | |
| 14 | exeM, exeN, exeK, exeH, exeA, exeB, exeL, etc. | Exe T2SS | |
| 10 | ETAE_RS04220, MAFF_RS25805, etc. | T3SS | |
| 12 | CBUK_RS04750, coxDFB4, rimP, etc. | T4SS secreted effectors | |
| 14 | mucD, algW, mucC, algQ, mucP, algU, etc. | Alginate | |
| 10 | bopD, etc. | BopD | |
| 34 | papR, etc. | PlcR-PapR quorum sensing | |
| 17 | rtxA, etc. | RtxA | Exotoxin |
| 23 | acfB, etc. | ACF |
| Number | SeqList | Resistance Profile | Description | Resistance Type |
|---|---|---|---|---|
| 1 | ctg_03842 | bacitracin | Undecaprenyl pyrophosphate phosphatase, which consists in the sequestration of Undecaprenyl pyrophosphate. | baca |
| 1 | ctg_04073 | fluoroquinolone | Pentapeptide repeat family, which protects DNA gyrase from the inhibition of quinolones. | qnrs |
| AMR Gene Family | Resistance Mechanism | Drug Class | ARO | Best_Hit_ARO | ORF_ID |
|---|---|---|---|---|---|
| resistance-nodulation-cell division (RND) antibiotic efflux pump | antibiotic efflux | fluoroquinolone antibiotic; tetracycline antibiotic | 3000777 | adeF | ctg_01323 |
| resistance-nodulation-cell division (RND) antibiotic efflux pump | antibiotic efflux | fluoroquinolone antibiotic; tetracycline antibiotic | 3000777 | adeF | ctg_01375 |
| TRU beta-lactamase | antibiotic inactivation | cephalosporin; penam | 3004450 | TRU-1 | ctg_02948 |
| glycopeptide resistance gene cluster; vanT | antibiotic target alteration | glycopeptide antibiotic | 3002972 | vanT gene in vanG cluster | ctg_02455 |
| resistance-nodulation-cell division (RND) antibiotic efflux pump | antibiotic efflux | fluoroquinolone antibiotic; diaminopyrimidine antibiotic; phenicol antibiotic | 3005069 | rsmA | ctg_00475 |
| elfamycin resistant EF-Tu | antibiotic target alteration | elfamycin antibiotic | 3003369 | Escherichia coli EF-Tu mutants conferring resistance to Pulvomycin | ctg_03757 |
| elfamycin resistant EF-Tu | antibiotic target alteration | elfamycin antibiotic | 3003369 | Escherichia coli EF-Tu mutants conferring resistance to Pulvomycin | ctg_03776 |
| quinolone resistance protein (qnr) | antibiotic target protection | fluoroquinolone antibiotic | 3002791 | QnrS2 | ctg_04073 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Qian, Q.; Chen, A.; Zhou, L.; Zhang, Y.; Gao, X.; Jiang, Q.; Zhang, X. Virulence and Antibiotic Resistance of Pathogenic Aeromonas caviae from Diseased Macrobrachium rosenbergii. Microorganisms 2025, 13, 1343. https://doi.org/10.3390/microorganisms13061343
Zhu X, Qian Q, Chen A, Zhou L, Zhang Y, Gao X, Jiang Q, Zhang X. Virulence and Antibiotic Resistance of Pathogenic Aeromonas caviae from Diseased Macrobrachium rosenbergii. Microorganisms. 2025; 13(6):1343. https://doi.org/10.3390/microorganisms13061343
Chicago/Turabian StyleZhu, Xinhai, Qieqi Qian, Anting Chen, Liying Zhou, Yao Zhang, Xiaojian Gao, Qun Jiang, and Xiaojun Zhang. 2025. "Virulence and Antibiotic Resistance of Pathogenic Aeromonas caviae from Diseased Macrobrachium rosenbergii" Microorganisms 13, no. 6: 1343. https://doi.org/10.3390/microorganisms13061343
APA StyleZhu, X., Qian, Q., Chen, A., Zhou, L., Zhang, Y., Gao, X., Jiang, Q., & Zhang, X. (2025). Virulence and Antibiotic Resistance of Pathogenic Aeromonas caviae from Diseased Macrobrachium rosenbergii. Microorganisms, 13(6), 1343. https://doi.org/10.3390/microorganisms13061343





