Occurrence and Genetic Diversity of Cryptosporidium spp. and Giardia intestinalis from Yaks (Bos grunniens) in Ganzi, Sichuan Province, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Genomic DNA Extraction
2.3. PCR Amplification
2.4. Sequence Analysis
2.5. Statistical Analysis
2.6. Nucleotide Sequence Accession Numbers
3. Results
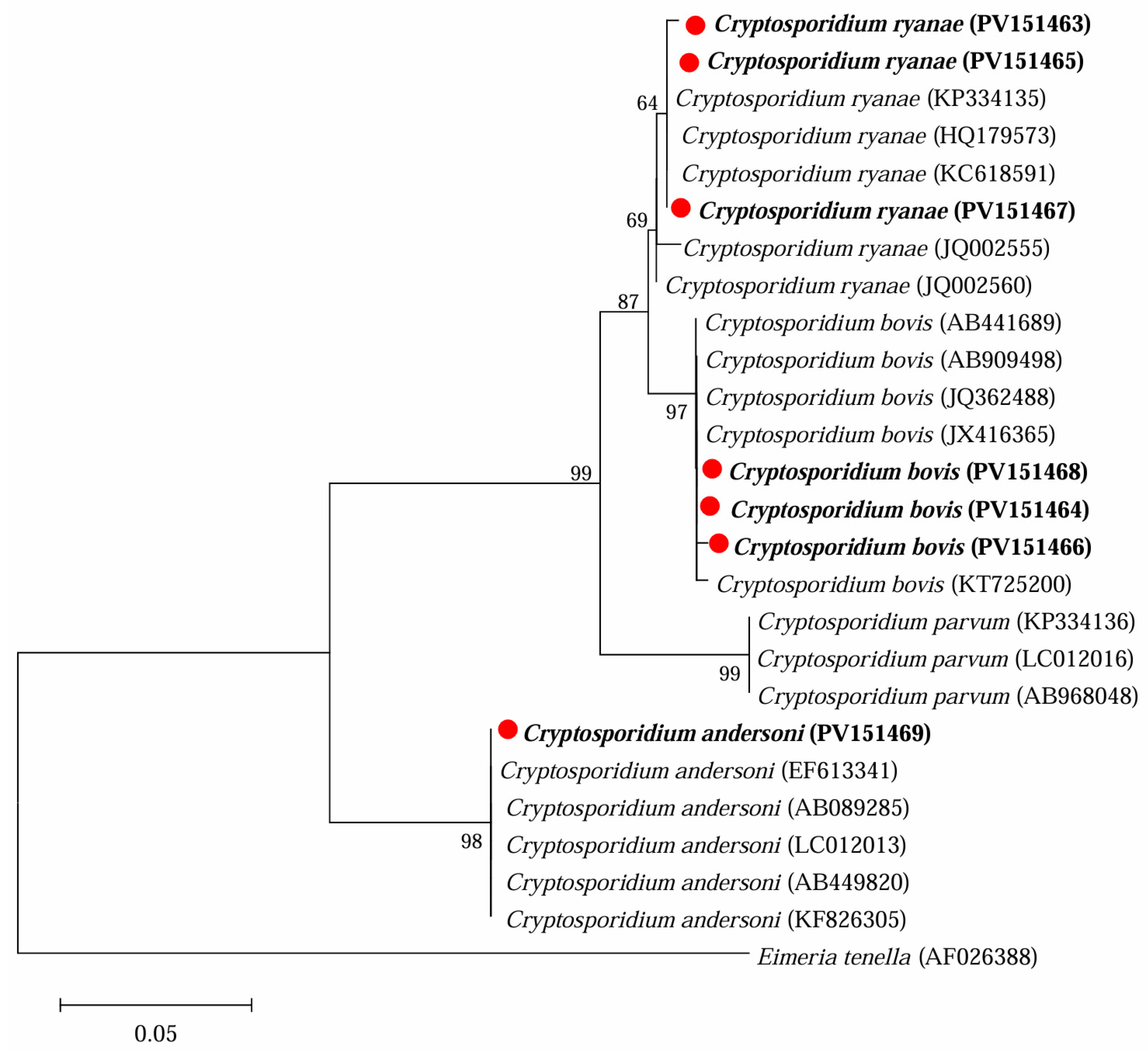
3.1. Occurrence of Cryptosporidium spp. in Yaks in Ganzi
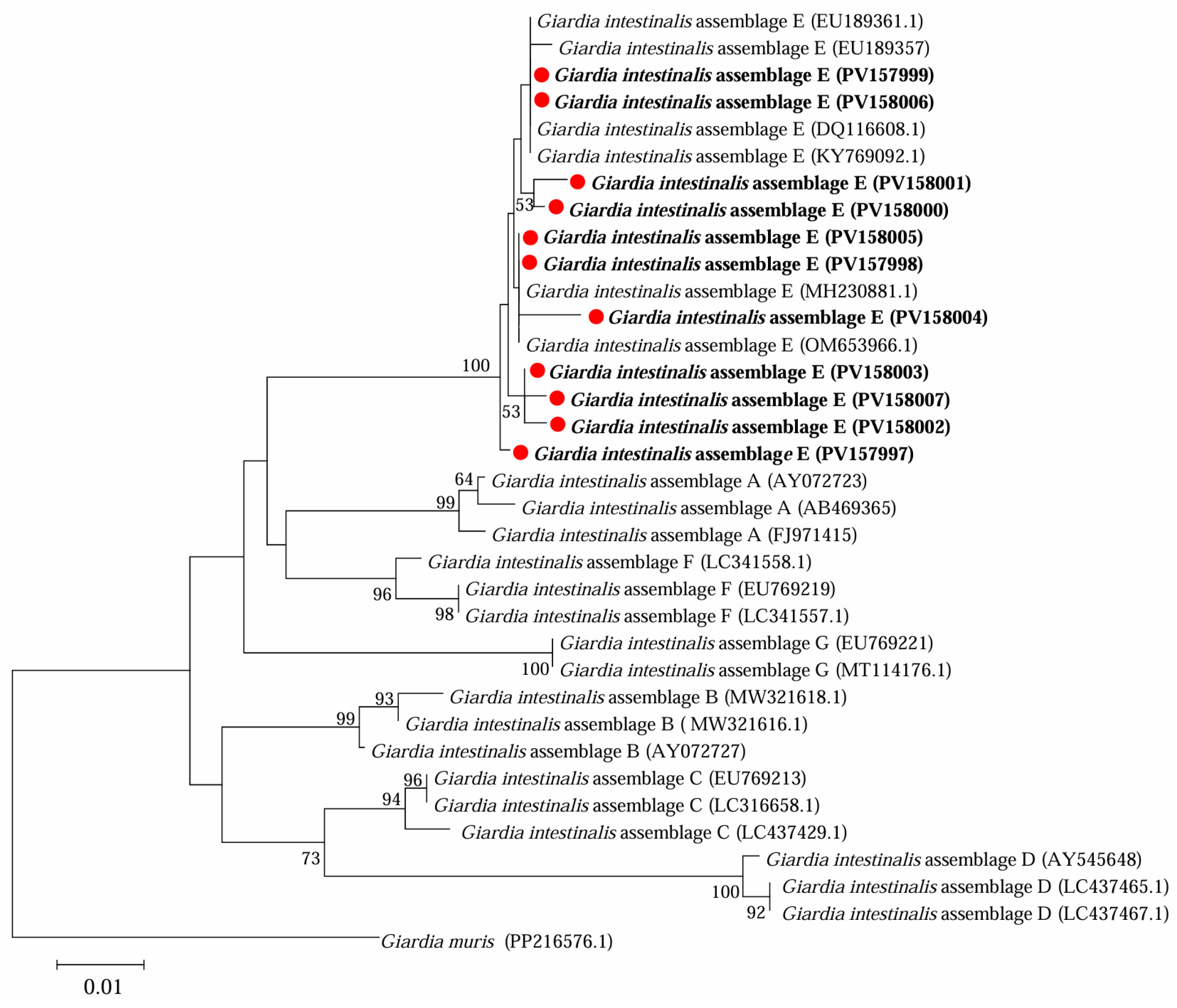
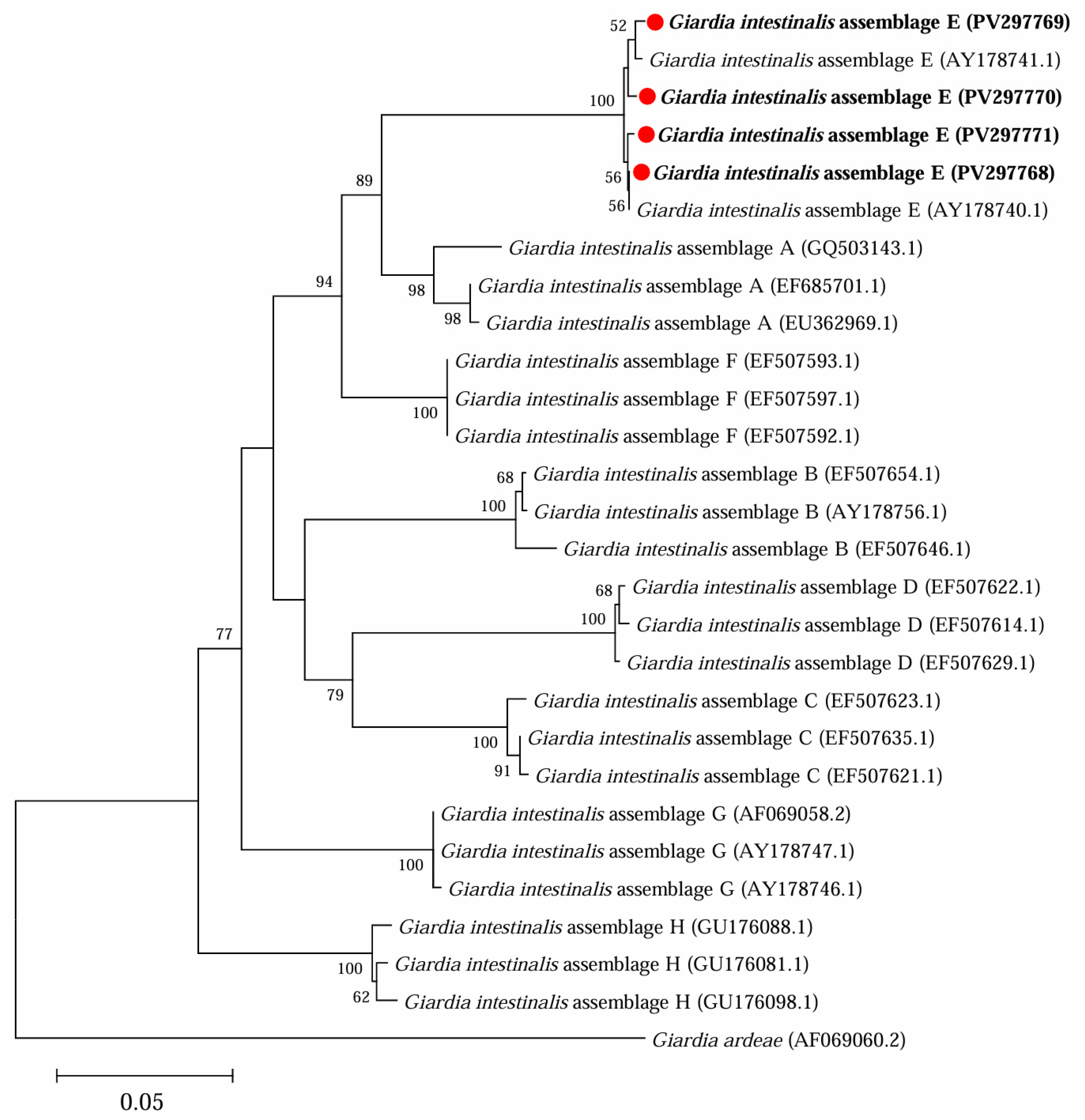
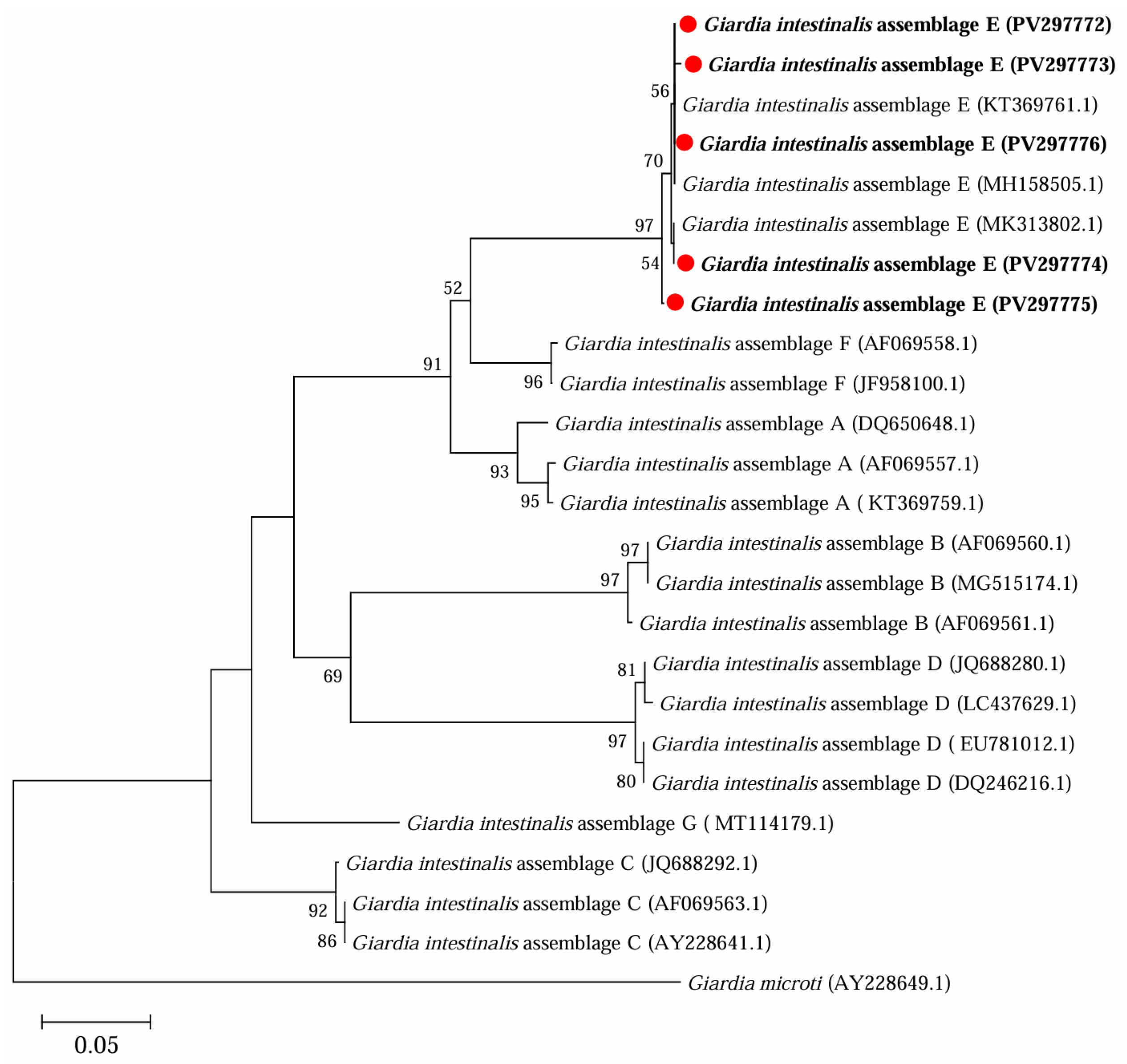
3.2. Occurrence of G. intestinalis in Yaks in Ganzi
3.3. Co-Infection of Cryptosporidium spp. and G. intestinalis in Yaks in Ganzi
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xiao, L.; Feng, Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017, 8–9, 14–32. [Google Scholar] [CrossRef] [PubMed]
- Hijjawi, N.; Zahedi, A.; Al-Falah, M.; Ryan, U. A review of the molecular epidemiology of Cryptosporidium spp. and Giardia duodenalis in the Middle East and North Africa (MENA) region. Infect. Genet. Evol. 2022, 98, 105212. [Google Scholar] [CrossRef]
- Ali, M.; Ji, Y.; Xu, C.; Hina, Q.; Javed, U.; Li, K. Food and waterborne cryptosporidiosis from a one health perspective: A comprehensive review. Animals 2024, 14, 3287. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Z.; Kang, C.; Wei, J.; Ma, H.; Liu, G.; Zhao, J.P.; Zhang, H.S.; Yang, X.B.; Wang, X.Y.; Yang, L.H.; et al. Meta-analysis of the prevalence of Giardia duodenalis in cattle in China. Foodborne Pathog. Dis. 2023, 20, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.M.; Feng, Y.; Fayer, R.; Xiao, L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia—A 50 year perspective (1971–2021). Int. J. Parasitol. 2021, 51, 1099–1119. [Google Scholar] [CrossRef]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011, 24, 110. [Google Scholar] [CrossRef]
- Cai, W.; Ryan, U.; Xiao, L.; Feng, Y. Zoonotic giardiasis: An update. Parasitol. Res. 2021, 120, 4199–4218. [Google Scholar] [CrossRef]
- Li, P.; Cai, J.; Cai, M.; Wu, W.; Li, C.; Lei, M.; Xu, H.; Feng, L.; Ma, J.; Feng, Y.; et al. Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet. Parasitol. 2016, 215, 58–62. [Google Scholar] [CrossRef]
- Wu, Y.; Chang, Y.; Zhang, X.; Chen, Y.; Li, D.; Wang, L.; Zheng, S.; Wang, R.; Zhang, S.; Jian, F.; et al. Molecular characterization and distribution of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi from yaks in Tibet, China. BMC Vet. Res. 2019, 15, 417. [Google Scholar] [CrossRef]
- Ma, J.; Cai, J.; Ma, J.; Feng, Y.; Xiao, L. Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet. Parasitol. 2014, 202, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Cai, J.; Wang, R.; Li, J.; Jian, F.; Huang, J.; Zhou, H.; Zhang, L. Molecular characterization of Cryptosporidium spp. and Giardia duodenalis from yaks in the central western region of China. BMC Microbiol. 2015, 15, 108. [Google Scholar] [CrossRef] [PubMed]
- Song, G.Y.; Qin, S.Y.; Zhao, G.H.; Zhu, X.Q.; Zhou, D.H.; Song, M.X. Molecular characterization of Giardia duodenalis from white yaks in China. Acta Parasitol. 2016, 61, 397–400. [Google Scholar] [CrossRef]
- Wang, G.; Wang, G.; Li, X.; Zhang, X.; Karanis, G.; Jian, Y.; Ma, L.; Karanis, P. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in 1–2-month-old highland yaks in Qinghai Province, China. Parasitol. Res. 2018, 117, 1793–1800. [Google Scholar] [CrossRef]
- Geng, H.L.; Ni, H.B.; Li, J.H.; Jiang, J.; Wang, W.; Wei, X.Y.; Zhang, Y.; Sun, H.T. Prevalence of Cryptosporidium spp. in yaks (Bos grunniens) in China: A systematic review and meta-analysis. Front. Cell. Infect. Microbiol. 2021, 11, 770612. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, Y.; Chang, Y.; Zhang, X.; Li, D.; Wang, L.; Zheng, S.; Wang, R.; Zhang, S.; Li, J.; et al. Genotyping and identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from free-range Tibetan yellow cattle and cattle-yak in Tibet, China. Acta Trop. 2020, 212, 105671. [Google Scholar] [CrossRef]
- Xiao, L.; Escalante, L.; Yang, C.; Sulaiman, I.; Escalante, A.A.; Montali, R.J.; Fayer, R.; Lal, A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microb. 1999, 65, 1578–1583. [Google Scholar] [CrossRef]
- Lalle, M.; Pozio, E.; Capelli, G.; Bruschi, F.; Crotti, D.; Cacciò, S.M. Genetic heterogeneity at the β-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 2005, 35, 207–213. [Google Scholar] [CrossRef]
- Cacciò, S.M.; Beck, R.; Lalle, M.; Marinculic, A.; Pozio, E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 2008, 38, 1523–1531. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Fayer, R.; Bern, C.; Gilman, R.H.; Trout, J.M.; Schantz, P.M.; Das, P.; Lal, A.A.; Xiao, L. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 2003, 9, 1444–1452. [Google Scholar] [CrossRef]
- Qin, S.Y.; Zhang, X.X.; Zhao, G.H.; Zhou, D.H.; Yin, M.Y.; Zhao, Q.; Zhu, X.Q. First report of Cryptosporidium spp. in white yaks in China. Parasite Vector 2014, 7, 230. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, J.; Chang, Y.; Yu, F.; Zhang, S.; Wang, R.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite 2020, 27, 62. [Google Scholar] [CrossRef] [PubMed]
- Elmahallawy, E.K.; Köster, P.C.; Dashti, A.; Alghamdi, S.Q.; Saleh, A.; Gareh, A.; Alrashdi, B.M.; Hernández-Castro, C.; Bailo, B.; Lokman, M.S.; et al. Molecular detection and characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi infections in dromedary camels (Camelus dromedaries) in Egypt. Front. Vet. Sci. 2023, 10, 1139388. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, L.; Liu, L.; Yu, F.; Jian, Y.; Wang, R.; Zhang, S.; Zhang, L.; Ning, C.; Jian, F. Genotyping of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from sheep and goats in China. BMC Vet. Res. 2022, 18, 361. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Foronda, P.; González, J.F.; Guedes, A.; Abreu-Acosta, N.; Molina, J.M.; Valladares, B. Occurrence and genotype characterization of Giardia duodenalis in goat kids from the Canary Islands, Spain. Vet. Parasitol. 2008, 154, 137–141. [Google Scholar] [CrossRef]
- Zhao, J.Q.; Fan, Y.Y.; Lei, Y.D.; Liu, D.; Wang, J.W.; Yang, X.; Song, J.K.; Zhao, G.H. Molecular characterization of common zoonotic protozoan parasites and bacteria causing diarrhea in dairy calves in Ningxia Hui Autonomous Region, China. Parasite 2024, 31, 60. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zan, X.Q.; Liu, J.B.; Xu, L.H.; Zhao, H.X. Molecular detection and characterization of Cryptosporidium spp. and Giardia duodenalis in dairy calves from Ningxia, China. Acta Parasitol. 2024, 69, 1876–1885. [Google Scholar] [CrossRef]
- Qi, M.Z.; Fang, Y.Q.; Wang, X.T.; Zhang, L.X.; Wang, R.J.; Du, S.Z.; Guo, Y.X.; Jia, Y.Q.; Yao, L.; Liu, Q.D.; et al. Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, north-western China. J. Med. Microbiol. 2015, 64, 111–116. [Google Scholar] [CrossRef]
- Zhang, S.X.; Zhou, Y.M.; Xu, W.; Tian, L.G.; Chen, J.X.; Chen, S.H.; Dang, Z.S.; Gu, W.P.; Yin, J.W.; Serrano, E.; et al. Impact of co-infections with enteric pathogens on children suffering from acute diarrhea in southwest China. Infect. Dis. Poverty 2016, 5, 64. [Google Scholar] [CrossRef]

| Targets | Primer Names | Sequence (5′-3′) | Annealing Temperature |
|---|---|---|---|
| 18S rRNA [17] | 18S-F1 | TTCTAGAGCTAATACATGCG | 55 °C |
| 18S-R1 | CCCATTTCCTTCGAAACAGGA | ||
| 18S-F2 | GGAAGGGTTGTATTTATTAGATAAAG | 55 °C | |
| 18S-R2 | CTCATAAGGTGCTGAAGGAGTA | ||
| bg [18] | bg-F1 | AAGCCCGACGACCTCACCCGCAGTGC | 65 °C |
| bg-R1 | GAGGCCGCCCTGGATCTTCGAGACGAC | ||
| bg-F2 | GAACGAGATCGAGGTCCG | 55 °C | |
| bg-R2 | CTCGACGAGCTTCGTGTT | ||
| gdh [19] | gdh-F1 | TTCCGTGTCCAGTACAACTC | 50 °C |
| gdh-R1 | GCCAGCTTCTCCTCGTTGAA | ||
| gdh-F2 | CGCTTCCACCCCTCTGTCAAT | 50 °C | |
| gdh-R2 | TGTTGTCCTTGCACATCTC | ||
| tpi [20] | tpi-F1 | AATAAATIATGCCTGCTCGTCG | 54 °C |
| tpi-R1 | ATGGACITCCTCTGCCTGCTC | ||
| tpi-F2 | CCCTTCATCGGIGGTAACTTCAA | 58 °C | |
| tpi-R2 | GTGGCCACCACICCCGTGCC |
| Factors | No. Tested | No. Positive (%, 95% CI) | RR (95% CI) | OR (95% CI) | p | Species (n) |
|---|---|---|---|---|---|---|
| Location | ||||||
| Yajiang | 27 | 10 (37.0, 0.2153–0.5577) | 0.0831 (0.0281–0.2454) | 0.054 (0.0152–0.1913) | <0.0001 | C. bovis (7), C. ryanae (3) |
| Seda | 66 | 2 (3.0, 0.0083–0.1039) | 1.0154 (0.1909–5.4012) | 1.0159 (0.1812–5.6951) | C. bovis (1), C. andersoni (1) | |
| Litang | 130 | 4 (3.1, 0.012–0.0765) | Reference | Reference | C. ryanae (3), C. bovis (1) | |
| Age (Months) | ||||||
| <6 | 39 | 8 (20.5, 0.1078–0.3553) | 0.2167 (0.0693–0.6774) | 0.1802 (0.0507–0.6408) | 0.002 | C. bovis (5), C. ryanae (3) |
| 6–12 | 51 | 4 (7.8, 0.0309–0.185) | 0.5667 (0.148–2.1699) | 0.5465 (0.1307–2.2855) | C. bovis (3), C. ryanae (1) | |
| 12–24 | 43 | 0 (0, 0–0.082) | - | - | - | |
| >24 | 90 | 4 (4.4, 0.0174–0.1087) | Reference | Reference | C. ryanae (2), C. bovis (1), C. andersoni (1) | |
| Gender | ||||||
| Male | 62 | 3 (4.8, 0.0166–0.1329) | 0.3444 (0.0368–3.22) | 0.3333 (0.0337–3.2976) | 0.3253 | C. ryanae (2), C. bovis (1) |
| Female | 60 | 1 (1.7, 0.003–0.0886) | Reference | Reference | C. ryanae (1) | |
| NA | 101 | 12 (11.9, 0.0693–0.1963) | C. bovis (8), C. ryanae (3), C. andersoni (1) | |||
| Diarrhea | ||||||
| Yes | 40 | 3 (5.0, 0.0258–0.1986) | 1.0558 (0.3155–3.5331) | 1.0603 (0.2876–3.9092) | 0.9299 | C. bovis (1), C. ryanae (1), C. andersoni (1) |
| No | 183 | 13 (7.1, 0.042–0.1177) | Reference | Reference | C. bovis (8), C. ryanae (5) | |
| Total | 223 | 16 (7.2, 0.0446–0.1133) | - | - | C. bovis (9), C. ryanae (6), C. andersoni (1) |
| Factors | No. Tested | No. Positive (%, 95% CI) | RR (95% CI) | OR (95% CI) | p | Genotype (n) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| gdh | tpi | bg | gdh | tpi | bg | |||||
| Location | ||||||||||
| Yajiang | 27 | 11 (40.7, 0.2451–0.5927) | 4 (14.8, 0.0591–0.3247) | 10 (37, 0.2153–0.5577) | 0.4343 (0.2415–0.781) | 0.3127 (0.1284–0.7614) | <0.0001 | E (11) | E (4) | E (10) |
| Seda | 66 | 1 (1.5, 0.0027–0.081) | 0 (0, 0–0.055) | 0 (0, 0–0.055) | 11.6769 (1.612–84.583) | 13.972 (1.8428–105.9317) | E (1) | - | - | |
| Litang | 130 | 23 (17.7, 0.1209–0.2515) | 12 (9.2, 0.0536–0.1544) | 21 (16.2, 0.1081–0.2343) | Reference | Reference | E (23) | E (12) | E (21) | |
| Age (Months) | ||||||||||
| <6 | 39 | 14 (35.9, 0.2274–0.5158) | 8 (20.5, 0.1078–0.3553) | 13 (33.3, 0.2063–0.4902) | 0.1238 (0.0435–0.3523) | 0.0831 (0.0251–0.275) | <0.0001 | E (14) | E (8) | E (13) |
| 6–12 | 51 | 12 (23.5, 0.1401–0.3676) | 5 (9.8, 0.0426–0.2097) | 12 (23.5, 0.1401–0.3676) | 0.1889 (0.0643–0.5552) | 0.1512 (0.0458–0.4985) | E (12) | E (5) | E (12) | |
| 12–24 | 43 | 5 (11.6, 0.0507–0.2448) | 2 (4.7, 0.0128–0.1545) | 4 (9.3, 0.0368–0.216) | 0.3822 (0.108–1.3524) | 0.3535 (0.0899–1.3899) | E (5) | E (2) | E (4) | |
| >24 | 90 | 4 (4.4, 0.0174–0.1087) | 1 (1.1, 0.002–0.0603) | 2 (2.2, 0.0061–0.0774) | Reference | Reference | E (4) | E (1) | E (2) | |
| Gender | ||||||||||
| Male | 62 | 10 (16.1, 0.09–0.2721) | 4 (6.5, 0.0254–0.1545) | 10 (16.1, 0.09–0.2721) | 1.1367 (0.5214–2.478) | 1.1673 (0.4555–2.9917) | 0.7471 | E (10) | E (4) | E (10) |
| Female | 60 | 11 (18.3, 0.1056–0.2992) | 3 (5, 0.0171–0.137) | 9 (15, 0.081–0.2611) | Reference | Reference | E (11) | E (3) | E (9) | |
| NA | 101 | 14 (13.9, 0.0844–0.2193) | 9 (8.9, 0.0476–0.1607) | 12 (11.9, 0.0693–0.1963) | - | - | E (14) | E (9) | E (12) | |
| Diarrhea | ||||||||||
| Yes | 40 | 6 (15.0, 0.0706–0.2907) | 2 (5, 0.0138–0.165) | 2 (5, 0.0138–0.165) | 1.0565 (0.4701–2.3743) | 1.0671 (0.4109–2.7711) | 0.8939 | E (6) | E (2) | E (2) |
| No | 183 | 29 (15.9, 0.1127–0.2184) | 14 (7.7, 0.0461–0.1243) | 29 (15.9, 0.1127–0.2184) | Reference | Reference | E (29) | E (14) | E (29) | |
| Total | 223 | 35 (15.7, 0.1151–0.2105) | 16 (7.2, 0.0446–0.1133) | 31 (13.9, 0.0997–0.1905) | - | - | E (35) | E (16) | E (31) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Hu, G.; Yang, D.; Hou, X.; Zhang, M.; Niu, Y.; Wang, Z.; Yang, X. Occurrence and Genetic Diversity of Cryptosporidium spp. and Giardia intestinalis from Yaks (Bos grunniens) in Ganzi, Sichuan Province, China. Microorganisms 2025, 13, 1261. https://doi.org/10.3390/microorganisms13061261
Fan Y, Hu G, Yang D, Hou X, Zhang M, Niu Y, Wang Z, Yang X. Occurrence and Genetic Diversity of Cryptosporidium spp. and Giardia intestinalis from Yaks (Bos grunniens) in Ganzi, Sichuan Province, China. Microorganisms. 2025; 13(6):1261. https://doi.org/10.3390/microorganisms13061261
Chicago/Turabian StyleFan, Yingying, Guirong Hu, Danjiao Yang, Xinrui Hou, Mingyi Zhang, Yufeng Niu, Zijie Wang, and Xin Yang. 2025. "Occurrence and Genetic Diversity of Cryptosporidium spp. and Giardia intestinalis from Yaks (Bos grunniens) in Ganzi, Sichuan Province, China" Microorganisms 13, no. 6: 1261. https://doi.org/10.3390/microorganisms13061261
APA StyleFan, Y., Hu, G., Yang, D., Hou, X., Zhang, M., Niu, Y., Wang, Z., & Yang, X. (2025). Occurrence and Genetic Diversity of Cryptosporidium spp. and Giardia intestinalis from Yaks (Bos grunniens) in Ganzi, Sichuan Province, China. Microorganisms, 13(6), 1261. https://doi.org/10.3390/microorganisms13061261






