Molecular Tactics of Biocontrol Fungi to Hack Plant Immunity for Successful Host Colonization—A Focus on Trichoderma Fungi
Abstract
1. Introduction
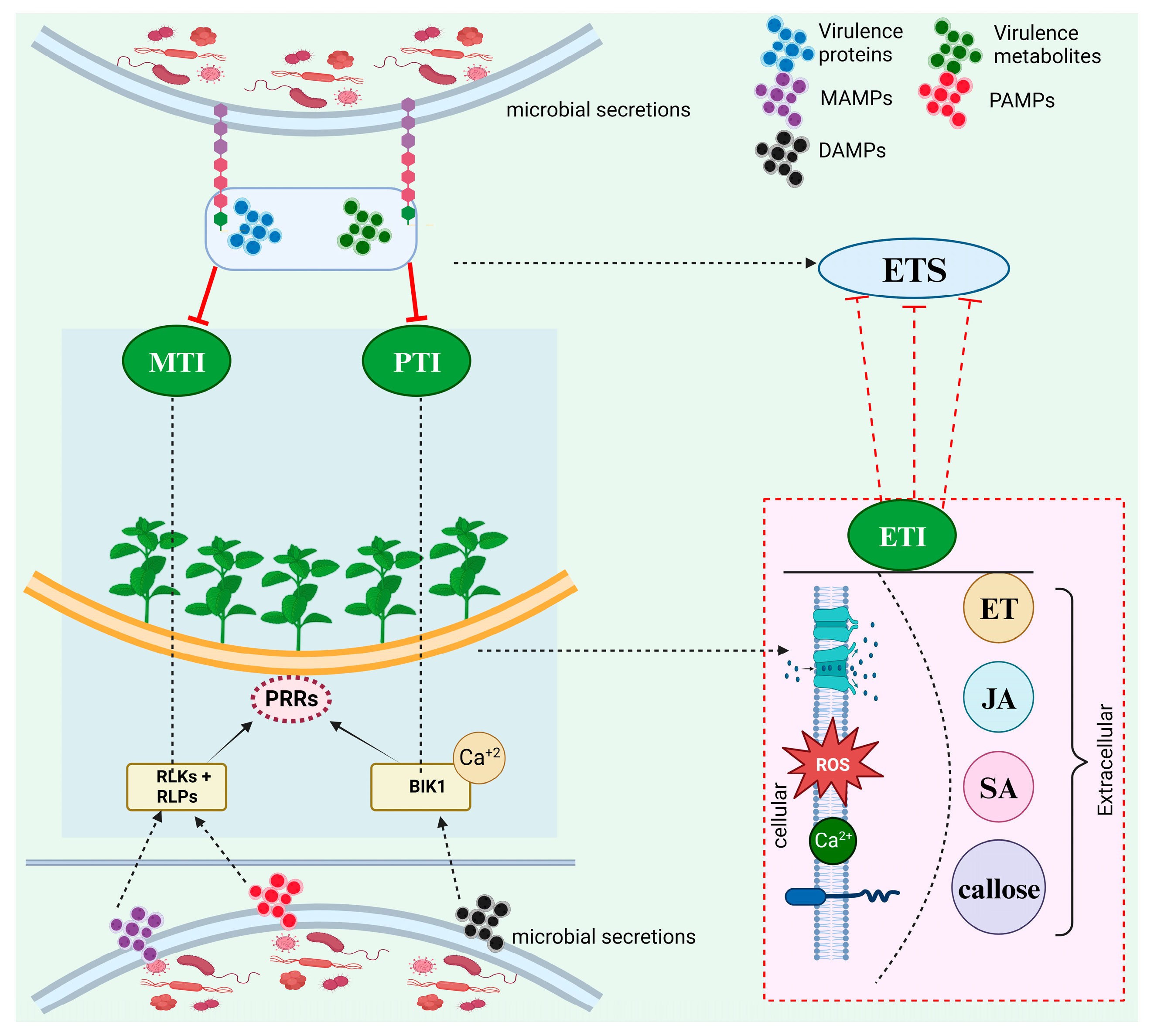
2. Plant Immune Response; An Overview
3. Biochemical Sensing Between Biocontrol Fungi and Host Plant
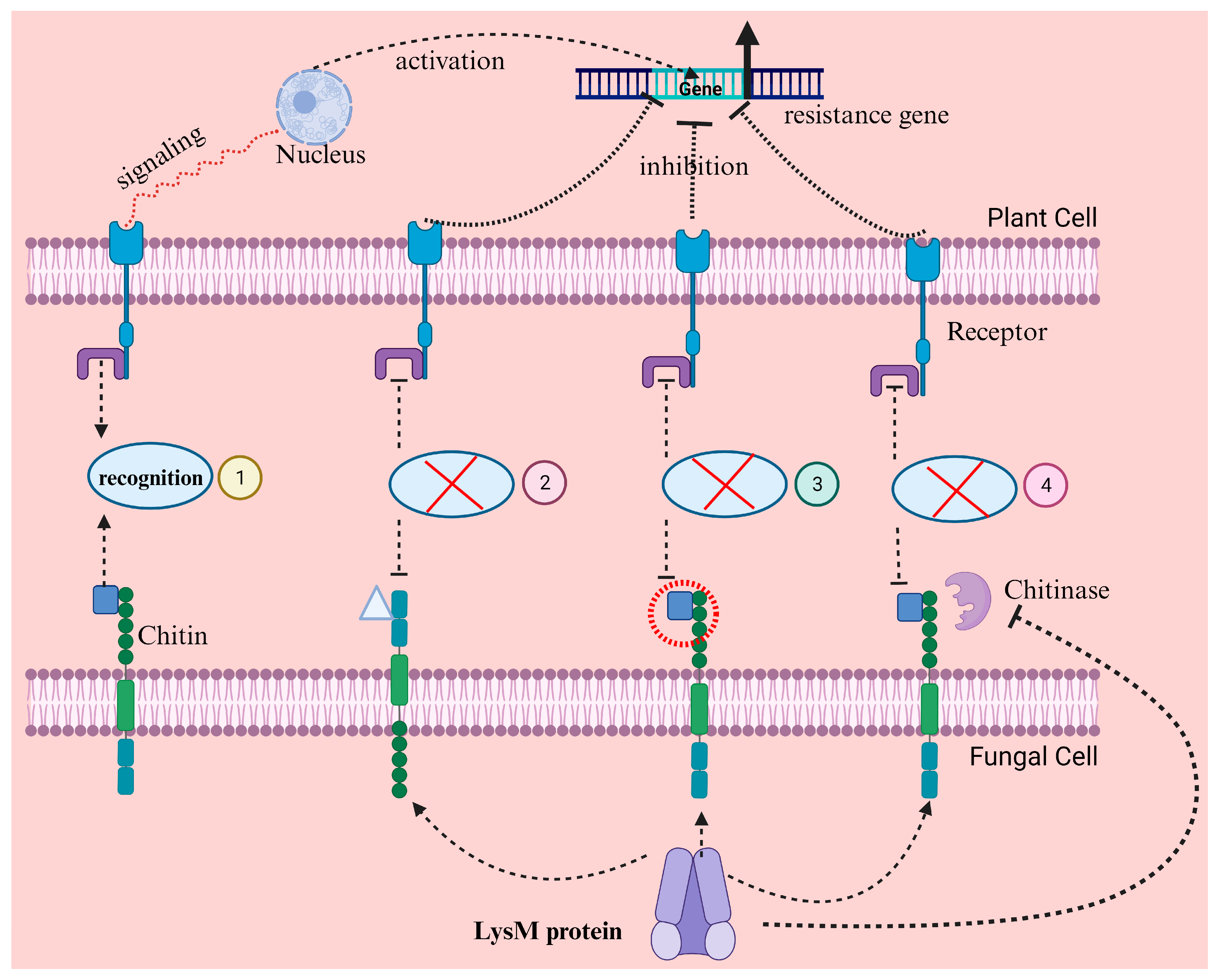
4. Molecular Mimicking to Interfere with Plant Immune Functions
5. Secretion of Apoplastic Proteins and Nucleotides to Promote Colonization
| Effector | Location | Function | Reference |
|---|---|---|---|
| CFEM proteins | Fungal extracellular membrane | Iron integration, appressorium expansion, redox homeostasis, sensing plant surface | [53] |
| LysM-like effectors | Cell membrane | Chitin recognition during fungal development, masking chitin oligomer Avoid ligand-PRR bonding Self-defense against chitinases | [54] |
| CP | Cell wall surface | Bind chitin and masking fungal cell wall recognition | [53] |
| Expansion-like proteins | Outer membrane | Assisting ingress into the host roots Soften the host cell wall with hydrolytic enzymes | [53] |
| Hydrophobins | Cell wall proteins Surface active proteins | Fungal adherence Altering root architecture | [55] |
| Lectin | Middle lamella | Alter fungal cell wall compositions and properties | [56] |
| WSC-proteins | Cell wall proteins | Increase cellular resistance to cell wall perturbation, oxidation, high osmolarity, and metal ions. | [57] |
6. Transcriptional Reprogramming of the Plant Hormone Defense Signal Genes
7. Secretion of the Common Symbiosis Signaling Molecule
8. Strategies to Tackle the Rigid Cell Wall of the Host
9. Host Gene Silencing Through Inter-Kingdom miRNA
10. Host-Fungal Genetics Contribute to Colonization
11. Factors Affecting Root Colonization
12. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chauhan, P.; Sharma, N.; Tapwal, A.; Kumar, A.; Verma, G.S.; Meena, M.; Seth, C.S.; Swapnil, P. Soil Microbiome: Diversity, Benefits and Interactions with Plants. Sustainability 2023, 15, 14643. [Google Scholar] [CrossRef]
- Ayaz, M.; Li, C.H.; Ali, Q.; Zhao, W.; Chi, Y.K.; Shafiq, M.; Ali, F.; Yu, X.Y.; Yu, Q.; Zhao, J.T.; et al. Bacterial and Fungal Biocontrol Agents for Plant Disease Protection: Journey from Lab to Field, Current Status, Challenges, and Global Perspectives. Molecules 2023, 28, 6735. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, G.; He, T.; Saleem, M. Signaling and Detoxification Strategies in Plant-Microbes Symbiosis under Heavy Metal Stress: A Mechanistic Understanding. Microorganisms 2022, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Pantigoso, H.A.; Newberger, D.; Vivanco, J.M. The rhizosphere microbiome: Plant–microbial interactions for resource acquisition. J. App. Microbiol. 2022, 133, 2864–2876. [Google Scholar] [CrossRef]
- Mohanram, S.; Kumar, P. Rhizosphere microbiome: Revisiting the synergy of plant-microbe interactions. Ann. Microbiol. 2019, 69, 307–320. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Liang, W.; Liu, S. Rhizosphere microbiome: The emerging barrier in plant-pathogen interactions. Front. Microbiol. 2021, 12, 772420. [Google Scholar] [CrossRef]
- Chapelle, E.; Mendes, R.; Bakker, P.A.; Raaijmakers, J.M. Fungal invasion of the rhizosphere microbiome. ISME J. 2016, 10, 265–268. [Google Scholar] [CrossRef]
- Thapa, S.; Prasanna, R. Prospecting the characteristics and significance of the phyllosphere microbiome. Ann. Microbiol. 2018, 68, 229–245. [Google Scholar] [CrossRef]
- Gong, T.; Xin, X.F. Phyllosphere microbiota: Community dynamics and its interaction with plant hosts. J. Integr. Plant Biol. 2021, 63, 297–304. [Google Scholar] [CrossRef]
- Stone, B.W.; Weingarten, E.A.; Jackson, C.R. The role of the phyllosphere microbiome in plant health and function. Ann. Plant Rev. 2018, 1, 533–556. [Google Scholar]
- Saleem, B. Phyllosphere microbiome: Plant defense strategies. In Microbiomes and the Global Climate Change; Springer: Singapore, 2021; pp. 173–201. [Google Scholar]
- Van der Burgh, A.M.; Joosten, M.H. Plant immunity: Thinking outside and inside the box. Trends Plant Sci. 2019, 24, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Galili, G. Tuning the orchestra: miRNAs in plant immunity. Trends Plant Sci. 2019, 24, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Zhang, Y. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Snelders, N.C.; Kettles, G.J.; Rudd, J.J.; Thomma, B.P. Plant pathogen effector proteins as manipulators of host microbiomes? Mol. Plant Pathol. 2018, 19, 257–259. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Wang, J.; Hu, Y.H.; Sun, Q.S.; Geng, R.; Ding, L.N. Antimicrobial Peptides: Classification, Mechanism, and Application in Plant Disease Resistance. Probiotics Antimicrob. Proteins 2025, 17, 1432–1446. [Google Scholar] [CrossRef]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.I.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P.I.; Yasuda, S. Pattern recognition receptors and signaling in plant–microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Varalakshmi, B.; Suganya, V.; Shanmugapriya, A.; Karpagam, T.; Firdous, S.J.; Manikandan, R.; Sridevi, R.; Saradhasri, V.; Abinaya, M. Manipulation of cell wall components and enzymes on plant-microbe interactions. In Plant-Microbe Interaction-Recent Advances in Molecular and Biochemical Approaches; Academic Press: Cambridge, MA, USA, 2023; pp. 303–326. [Google Scholar]
- Doughari, J. An overview of plant immunity. J. Plant Pathol. Microbiol. 2015, 6, 10–4172. [Google Scholar]
- Thomma, B.P.; Nürnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef]
- Resjö, S.; Zahid, M.A.; Burra, D.D.; Lenman, M.; Levander, F.; Andreasson, E. Proteomics of PTI and two ETI immune reactions in potato leaves. Int. J. Mol. Sci. 2019, 20, 4726. [Google Scholar] [CrossRef] [PubMed]
- Rutter, B.D.; Innes, R.W. Extracellular vesicles as key mediators of plant–microbe interactions. Curr. Opin. Plant Biol. 2018, 44, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Jiang, T.; Liu, Y.X.; Bai, Y.C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.W.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364, eaau6389. [Google Scholar] [CrossRef] [PubMed]
- Enebe, M.C.; Babalola, O.O. The impact of microbes in the orchestration of plants’ resistance to biotic stress: A disease management approach. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef]
- Romani, F.; Banić, E.; Florent, S.N.; Kanazawa, T.; Goodger, J.Q.; Mentink, R.A. Oil body formation in Marchantia polymorpha is controlled by MpC1HDZ and serves as a defense against arthropod herbivores. Curr. Biol. 2020, 30, 2815–2828. [Google Scholar] [CrossRef]
- Fujita, M.; Kusajima, M.; Fukagawa, M.; Okumura, Y.; Nakajima, M.; Akiyama, K. Response of tomatoes primed by mycorrhizal colonization to virulent and avirulent bacterial pathogens. Sci. Rep. 2022, 12, 4686. [Google Scholar] [CrossRef]
- Yang, C.; Dolatabadian, A.; Fernando, W.D. The wonderful world of intrinsic and intricate immunity responses in plants against pathogens. Can. J. Plant Pathol. 2022, 44, 1–20. [Google Scholar] [CrossRef]
- Métraux, J.P.; Jackson, R.W.; Schnettler, E.; Goldbach, R.W. Plant pathogens as suppressors of host defense. Adv. Bot. Res. 2009, 51, 39–89. [Google Scholar]
- Shah, A.A.; Gupta, A. Secondary metabolite basis of elicitor-and effector-triggered immunity in pathogen elicitation amid infections. In Genetic Manipulation of Secondary Metabolites in Medicinal Plant; Springer: Singapore, 2023; pp. 225–251. [Google Scholar]
- Vergara, C.; Araujo, K.E.C.; Souza, S.R.D.; Schultz, N.; Saggin, O.J.; Sperandio, M.V.L.; Zilli, J.É. Plant-mycorrhizal fungi interaction and response to inoculation with different growth-promoting fungi. Pesqui. Agropecuária Bras. 2018, 54, e25140. [Google Scholar] [CrossRef]
- Lombardi, N.; Vitale, S.; Turrà, D.; Reverberi, M.; Fanelli, C.; Vinale, F.; Marra, R.; Ruocco, M.; Pascale, A.; d’Errico, G.; et al. Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol. Plant-Microbe Int. 2018, 31, 982–994. [Google Scholar] [CrossRef]
- Delaux, P.M.; Schornack, S. Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 2021, 371, eaba6605. [Google Scholar] [CrossRef] [PubMed]
- Sasse, J.; Simon, S.; Gübeli, C.; Liu, G.-W.; Cheng, X.; Friml, J. Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr. Biol. 2015, 25, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Banasiak, J.; Borghi, L.; Stec, N.; Martinoia, E.; Jasiński, M. The full-size ABCG transporter of Medicago truncatula is involved in strigolactone secretion, affecting arbuscular mycorrhiza. Front. Plant Sci. 2020, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Banasiak, J.; Jamruszka, T.; Murray, J.D.; Jasiński, M. A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume–rhizobium symbioses. Plant Physiol. 2021, 187, 2071–2091. [Google Scholar] [CrossRef]
- Anand, G.; Gupta, R.; Marash, I.; Leibman-Markus, M.; Bar, M. Cytokinin production and sensing in fungi. Microbiol. Res. 2022, 262, 127103. [Google Scholar] [CrossRef]
- Akhtar, S.S.; Mekureyaw, M.F.; Pandey, C.; Roitsch, T. Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front. Plant Sci. 2020, 10, 1777. [Google Scholar] [CrossRef]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef]
- Semchenko, M.; Barry, K.E.; de Vries, F.T.; Mommer, L.; Moora, M.; Maciá-Vicente, J.G. Deciphering the role of specialist and generalist plant–microbial interactions as drivers of plant–soil feedback. New Phytol. 2022, 234, 1929–1944. [Google Scholar] [CrossRef]
- Favre-Godal, Q.; Gourguillon, L.; Lordel-Madeleine, S.; Gindro, K.; Choisy, P. Orchids and their mycorrhizal fungi: An insufficiently explored relationship. Mycorrhiza 2020, 30, 5–22. [Google Scholar] [CrossRef]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Proietti, S.; Hickman, R.; Van Verk, M.C.; Zamioudis, C.; Pieterse, C.M.J. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018, 93, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Wawra, S.; Fesel, P.; Widmer, H.; Neumann, U.; Lahrmann, U.; Becker, S.; Hehemann, J.-H.; Langen, G.; Zuccaro, A. FGB1 and WSC3 are in planta-induced beta-glucan-binding fungal lectins with different functions. New Phytol. 2019, 222, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Emonet, A.; Denervaud Tendon, V.; Marhavy, P.; Wu, D.; Lahaye, T.; Geldner, N. Co-incidence of Damage and Microbial Patterns Controls Localized Immune Responses in Roots. Cell 2020, 180, 440–453.e18. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Rodriguez-Moreno, L.; Mansurkhodzaev, A.; Wang, P.; van den Berg, W.; Gasciolli, V. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2020, 225, 448–460. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef]
- Zeng, T.; Holmer, R.; Hontelez, J.; te Lintel-Hekkert, B.; Marufu, L.; de Zeeuw, T.; Wu, F.Y.; Schijlen, E.; Bisseling, T.; Limpens, E. Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J. 2018, 94, 411–425. [Google Scholar] [CrossRef]
- Nogueira-Lopez, G.; Greenwood, D.R.; Middleditch, M.; Winefield, C.; Eaton, C.; Steyaert, J.M.; Mendoza-Mendoza, A. The Apoplastic Secretome of Trichoderma virens During Interaction with Maize Roots Shows an Inhabition of Plant Defence and Scavenging Oxidative Stress Secreted Proteins. Front. Plant Sci. 2018, 9, 409. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a Plant Receptor for Extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef]
- Nizam, S.; Qiang, X.; Wawra, S.; Nostadt, R.; Getzke, F.; Schwanke, F.; Dreyer, I.; Langen, G.; Zuccaro, A. Serendipita indica E5′NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonization. EMBO Rep. 2019, 20, e47430. [Google Scholar] [CrossRef]
- Luti, S.; Martellini, F.; Bemporad, F.; Mazzoli, L.; Paoli, P.; Pazzagli, L. A single amino acid mutation affects elicitor and expansins-like activities of cerato-platanin, a noncatalytic fungal protein. PLoS ONE 2017, 12, e0178337. [Google Scholar] [CrossRef]
- Cheval, C.; Faulkner, C. Plasmodesmal regulation during plant–pathogen interactions. New Phytol. 2018, 217, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Mendoza, A.; Zaid, R.; Lawry, R.; Hermosa, R.; Monte, E.; Horwitz, B.A.; Mukherjee, P.K. Molecular dialogues between Trichoderma and roots: Role of the fungal secretome. Fungal Biol. Revi. 2018, 32, 62–85. [Google Scholar] [CrossRef]
- Rovenich, H.; Zuccaro, A.; Thomma, B.P. Convergent evolution of filamentous microbes towards evasion of glycantriggered immunity. New Phytol. 2016, 212, 896–901. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.M.; Chen, Y.; Zhu, J.; Ying, S.H.; Feng, M.G. Subcellular localization of five singular WSC domain-containing proteins and their roles in Beauveria bassiana responses to stress cues and metal ions. Environ. Microbiol. Rep. 2016, 8, 295–304. [Google Scholar] [CrossRef]
- Khan, R.A.A.; Najeeb, S.; Hussain, S.; Xie, B.; Li, Y. Bioactive secondary metabolites from Trichoderma spp. against phytopathogenic fungi. Microorganisms 2020, 8, 817. [Google Scholar] [CrossRef]
- Khan, R.A.A.; Najeeb, S.; Mao, Z.; Ling, J.; Yang, Y.; Li, Y.; Xie, B. Bioactive secondary metabolites from Trichoderma spp. against phytopathogenic bacteria and root-knot nematode. Microorganisms 2020, 8, 401. [Google Scholar] [CrossRef]
- Khan, R.A.A.; Najeeb, S.; Chen, J.; Wang, R.; Zhang, J.; Hou, J.; Liu, T. Insights into the molecular mechanism of Trichoderma stimulating plant growth and immunity against phytopathogens. Physiol. Plant. 2023, 175, e14133. [Google Scholar] [CrossRef]
- Fiorin, G.L.; Sanchéz-Vallet, A.; Thomma, B.P.H.J.; Mesarich, C.H. The Trichoderma atroviride LysM effector Tal6 suppresses plant immunity to promote fungal root colonization. New Phytol. 2018, 220, 585–598. [Google Scholar] [CrossRef]
- Hermosa, R.; Viterbo, A.; Chet, I.; Monte, E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 2012, 158, 17–25. [Google Scholar] [CrossRef]
- Karibasappa, C.S.; Singh, Y.; Aravind, T.; Singh, K.P. Concept of effectors and receptors in improving plant immunity. In Emerging Trends in Plant Pathology; Springer: Singapore, 2021; pp. 475–497. [Google Scholar] [CrossRef]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay Between Innate Immunity and the Plant Microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E. Plant signaling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pages, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Becard, G.; Bonfante, P. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.M.; Uroz, S.; Barker, D.G. Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science 2017, 356, eaad4501. [Google Scholar] [CrossRef]
- Li, W.C.; Huang, C.H.; Chen, C.L.; Chuang, Y.C.; Tung, S.Y.; Wang, T.F. Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol. Biofuels 2017, 10, 170. [Google Scholar] [CrossRef]
- Moran-Diez, E.; Hermosa, R.; Ambrosino, P.; Cardoza, R.E.; Gutierrez, S.; Lorito, M.; Monte, E. The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum-plant beneficial interaction. Mol. Plant Microbe Interact. 2009, 22, 1021–1031. [Google Scholar] [CrossRef]
- Moran-Diez, M.E.; Trushina, N.; Lamdan, N.L.; Rosenfelder, L.; Mukherjee, P.K.; Kenerley, C.M.; Horwitz, B.A. Host-specific transcriptomic pattern of Trichoderma virens during interaction with maize or tomato roots. BMC Genom. 2015, 16, 8. [Google Scholar] [CrossRef]
- Masachis, S.; Segorbe, D.; Turrà, D.; Leon-Ruiz, M.; Fürst, U.; El Ghalid, M.; Leonard, G.; López-Berges, M.S.; Richards, T.A.; Felix, G.; et al. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016, 1, 16043. [Google Scholar] [CrossRef]
- Pelagio-Flores, R.; Esparza-Reynoso, S.; Garnica-Vergara, A.; López-Bucio, J.; Herrera-Estrella, A. Trichoderma-induced acidification is an early trigger for changes in arabidopsis root growth and determines fungal phytostimulation. Front. Plant Sci. 2017, 8, 822. [Google Scholar] [CrossRef]
- Trushina, N.; Levin, M.; Mukherjee, P.K.; Horwitz, B.A. PacC and pH–dependent transcriptome of the mycotrophic fungus Trichoderma virens. BMC Genom. 2013, 14, 138. [Google Scholar] [CrossRef]
- Ku, Y.S.; Wong, J.W.H.; Mui, Z.; Liu, X.; Hui, J.H.L.; Chan, T.F.; Lam, H.M. Small RNAs in plant responses to abiotic stresses: Regulatory roles and study methods. Int. J. Mol. Sci. 2015, 16, 24532–24554. [Google Scholar] [CrossRef]
- Couzigou, J.M.; Lauressergues, D.; André, O.; Gutjahr, C.; Guillotin, B.; Bécard, G.; Combier, J.P. Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe 2017, 21, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Develop. 2004, 18, 1187–1197. [Google Scholar] [CrossRef]
- Silvestri, A.; Fiorilli, V.; Miozzi, L.; Accotto, G.P.; Turina, M.; Lanfranco, L. In silico analysis of fungal small RNA accumulation reveals putative plant mRNA targets in the symbiosis between an arbuscular mycorrhizal fungus and its host plant. BMC Genom. 2019, 20, 169. [Google Scholar] [CrossRef]
- Raman, V.; Simon, S.A.; Demirci, F.; Nakano, M.; Meyers, B.C.; Donofrio, N.M. Small RNA functions are required for growth and development of Magnaporthe oryzae. Mol. Plant-Microbe Int. 2017, 30, 517–530. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Wong-Bajracharya, J.; Singan, V.R.; Monti, R. The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2103527119. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Steindorff, A.S.; Chenthamara, K.; Manganiello, G.; Henrissat, B.; Zhang, J.; Cai, F.; Kopchinskiy, A.G.; Grigoriev, I.V.; Druzhinina, I.S. Evolution and comparative genomics of the most common Trichoderma species. BMC Gen. 2019, 20, 485. [Google Scholar] [CrossRef]
- Brotman, Y.; Landau, U.; Cuadros-Inostroza, A.; Takayuki, T.; Fernie, A.R.; Chet, I.; Viterbo, A.; Willmitzer, L. Trichoderma-plant root colonization: Escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 2013, 9, e1003221. [Google Scholar] [CrossRef]
- Sarrocco, S.; Matarese, F.; Baroncelli, R.; Vannacci, G.; Seidl-Seiboth, V.; Kubicek, C.P.; Vergara, M. The constitutive endopolygalacturonase TvPG2 regulates the induction of plant systemic resistance by Trichoderma virens. Phytopathology 2017, 107, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Atanasova, L.; Jensen, D.F.; Zeilinger, S. Necrotrophic mycoparasites and their genomes. Microbiol. Spectrosc. 2017, 5, 1005–1026. [Google Scholar] [CrossRef]
- Schmoll, M.; Dattenböck, C.; Carreras-Villaseñor, N.; Mendoza-Mendoza, A.; Tisch, D.; Alemán, M.I.; Baker, S.E.; Brown, C. The genomes of three uneven siblings: Footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 2016, 80, 205–327. [Google Scholar] [CrossRef] [PubMed]
- Crutcher, F.K.; Moran-Diez, M.E.; Ding, S.; Liu, J.; Horwitz, B.A.; Mukherjee, P.K.; Kenerley, C.M. A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 2015, 119, 476–486. [Google Scholar] [CrossRef]
- Martínez-Medina, A.; Appels, F.V.; van Wees, S.C. Impact of salicylic acid-and jasmonic acidregulated defences on root colonization by Trichoderma harzianum T-78. Plant Signal. Behav. 2017, 12, e1345404. [Google Scholar] [CrossRef]
- Nieto-Jacobo, M.F.; Steyaert, J.M.; Salazar-Badillo, F.B.; Nguyen, D.V.; Rostás, M.; Braithwaite, M.; De Souza, J.T.; Jimenez-Bremont, J.F.; Ohkura, M.; Stewart, A.; et al. Environmental growth conditions of Trichoderma spp. affects indole acetic acid derivatives, volatile organic compounds, and plant growth promotion. Front. Plant Sci. 2017, 8, 102. [Google Scholar] [CrossRef]
- González-Pérez, E.; Ortega-Amaro, M.A.; Salazar-Badillo, F.B.; Bautista, E.; Douterlungne, D.; Jiménez-Bremont, J.F. The Arabidopsis-Trichoderma interaction reveals that the fungal growth medium is an important factor in plant growth induction. Sci. Rep. 2018, 8, 16427. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Zhao, M.; Li, G.; Wang, Y.; Shen, Q.; Yang, J.; Asseri, T.A.Y.; Wang, Y.; Guo, M.; Ahmed, W. Molecular Tactics of Biocontrol Fungi to Hack Plant Immunity for Successful Host Colonization—A Focus on Trichoderma Fungi. Microorganisms 2025, 13, 1251. https://doi.org/10.3390/microorganisms13061251
Yang Y, Zhao M, Li G, Wang Y, Shen Q, Yang J, Asseri TAY, Wang Y, Guo M, Ahmed W. Molecular Tactics of Biocontrol Fungi to Hack Plant Immunity for Successful Host Colonization—A Focus on Trichoderma Fungi. Microorganisms. 2025; 13(6):1251. https://doi.org/10.3390/microorganisms13061251
Chicago/Turabian StyleYang, Yingfen, Meiwei Zhao, Guotao Li, Ying Wang, Qingqing Shen, Jun Yang, Tahani A. Y. Asseri, Yanjun Wang, Min Guo, and Waqar Ahmed. 2025. "Molecular Tactics of Biocontrol Fungi to Hack Plant Immunity for Successful Host Colonization—A Focus on Trichoderma Fungi" Microorganisms 13, no. 6: 1251. https://doi.org/10.3390/microorganisms13061251
APA StyleYang, Y., Zhao, M., Li, G., Wang, Y., Shen, Q., Yang, J., Asseri, T. A. Y., Wang, Y., Guo, M., & Ahmed, W. (2025). Molecular Tactics of Biocontrol Fungi to Hack Plant Immunity for Successful Host Colonization—A Focus on Trichoderma Fungi. Microorganisms, 13(6), 1251. https://doi.org/10.3390/microorganisms13061251






