Evaluation of Probiotic Bacillus velezensis for the Control of Pathogens That Cause Post-Weaning Diarrhea in Piglets—Results from In Vitro Testing and an In Vivo Model Using Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Cell Line, Bacterial Strains, Culture Conditions and Preparation of Cell-Free Supernatant
2.3. Pathogen Inhibition Assay
- ODa = OD600 of pathogen plus CFS, after 16 h of incubation;
- OD0 = OD600 of media alone, after 16 h of incubation (no inoculum and no CFS);
- ODb = OD600 of the pathogen and culture media alone, after 16 h of incubation (no CFS).
2.4. IPEC-J2 mRNA Expression Without or with Probiotic CFS Pretreatment
2.5. Wound-Healing Assay
2.6. Adhesion, Exclusion and Competitive Exclusion Assays
2.7. Nematode-Killing Assay
2.8. Statistical Analysis
3. Results
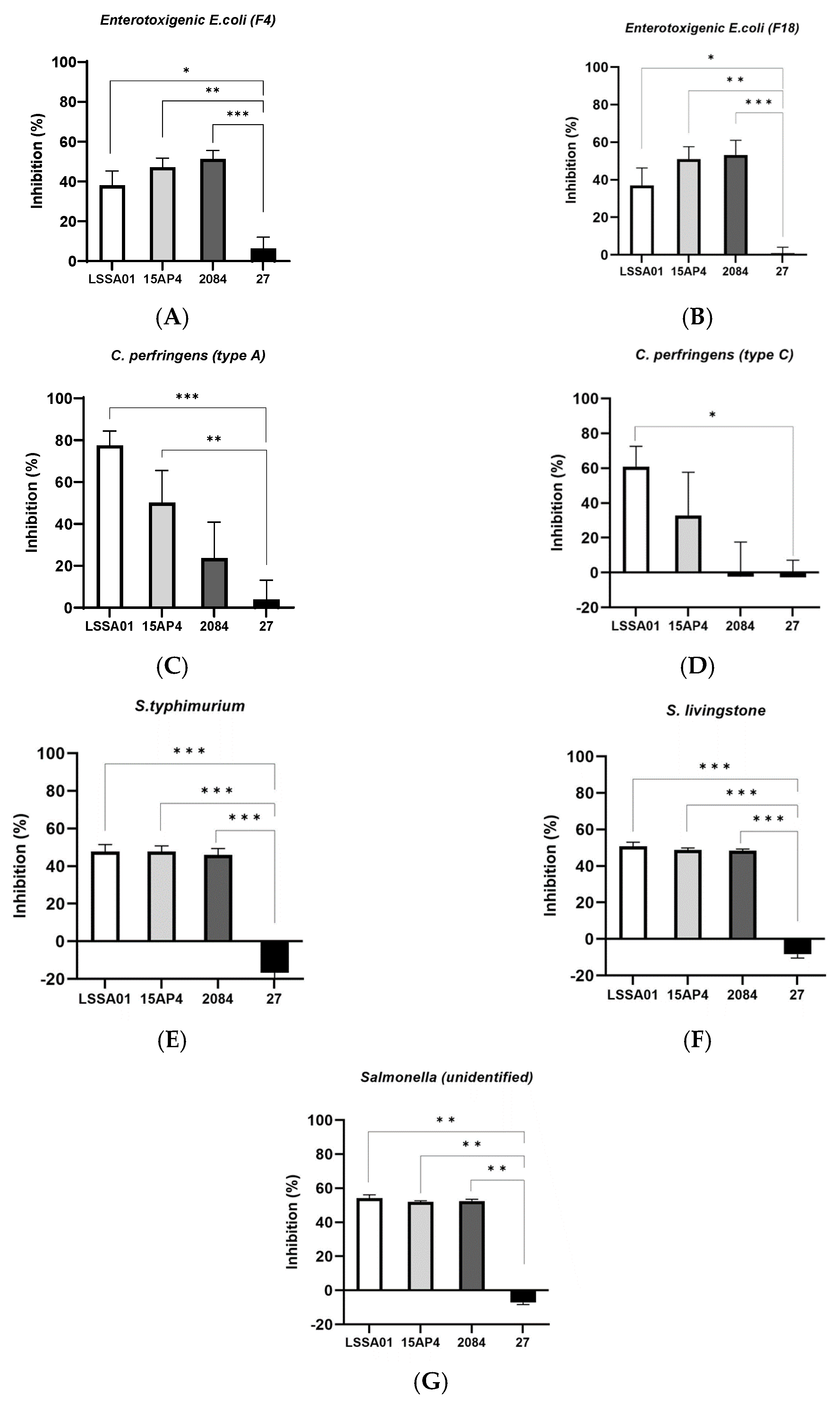
3.1. Cell-Free Supernatant from B. velezensis LSSA01, 15AP4 and 2084 Inhibited the Growth of ETEC, C. perfringens and Salmonella Isolates
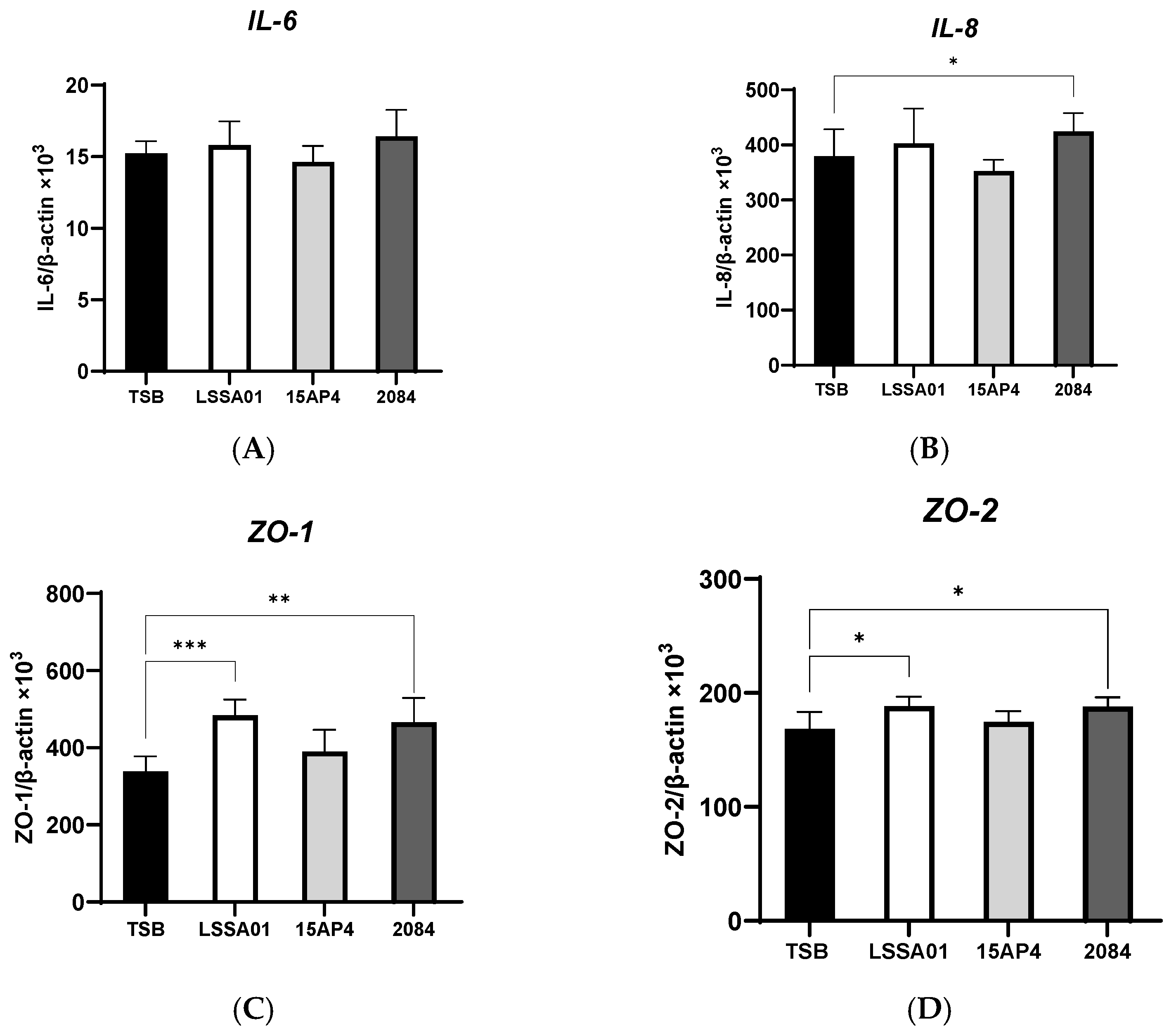
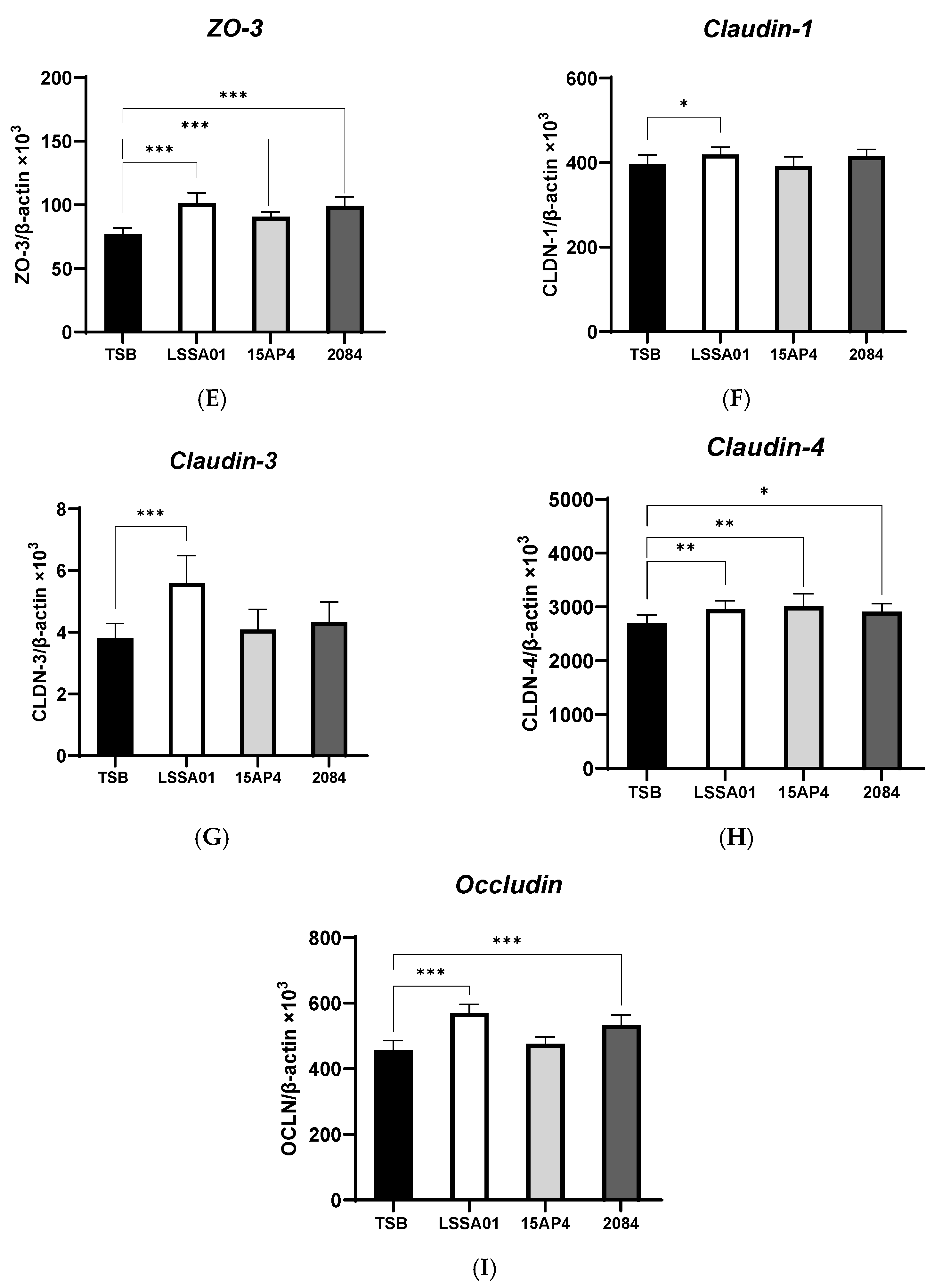
3.2. Upregulation of IPEC-J2 Expression of Cytokines and Tight Junction Proteins by B. velezensis Cell-Free Supernatant
3.3. Cell-Free Supernatant from B. velezensis Strain LSSA01 Facilitated Wound Healing
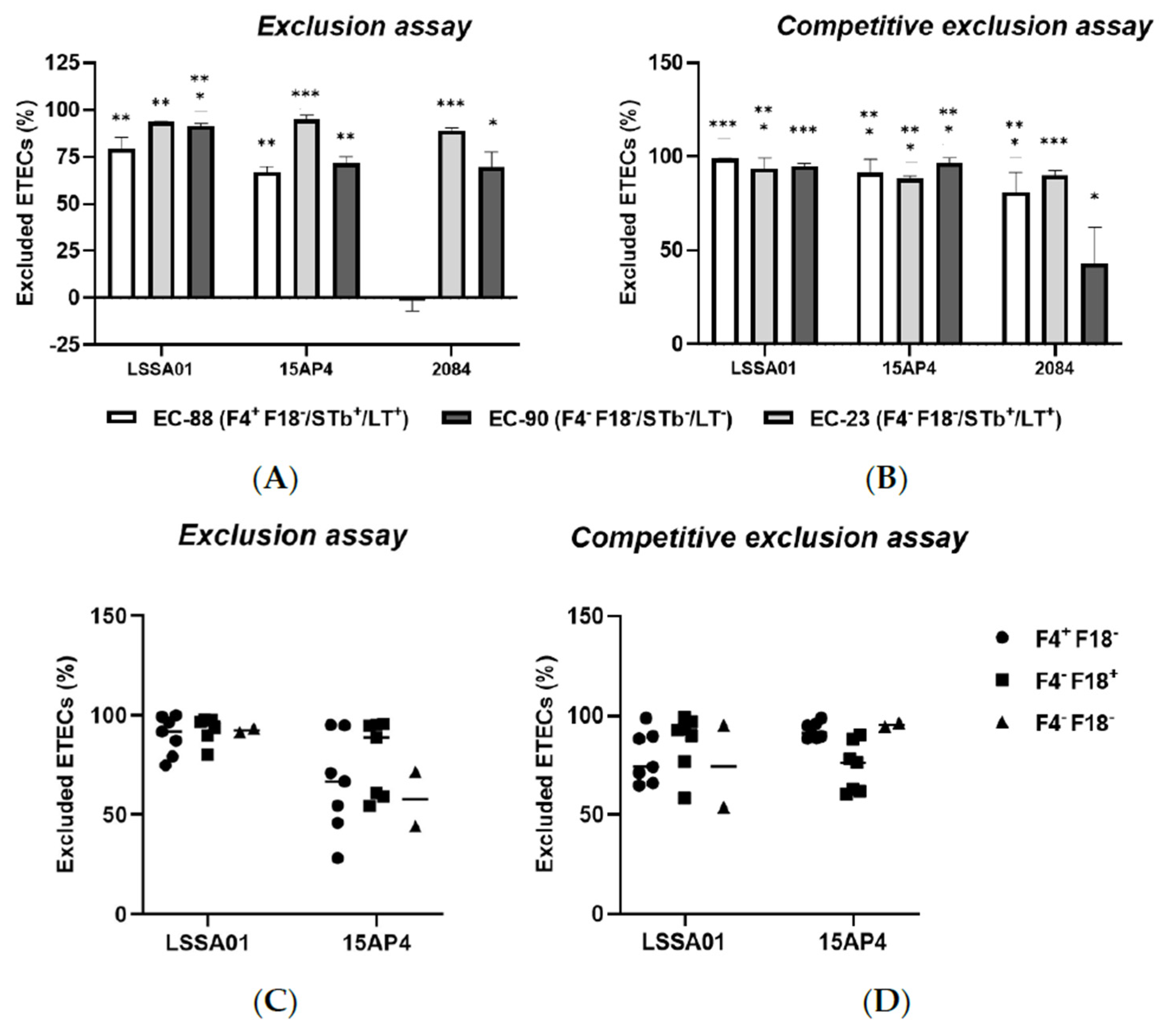
3.4. B. velezensis Colony-Forming Units Excluded ETEC Adhesion to IPEC-J2 Cells
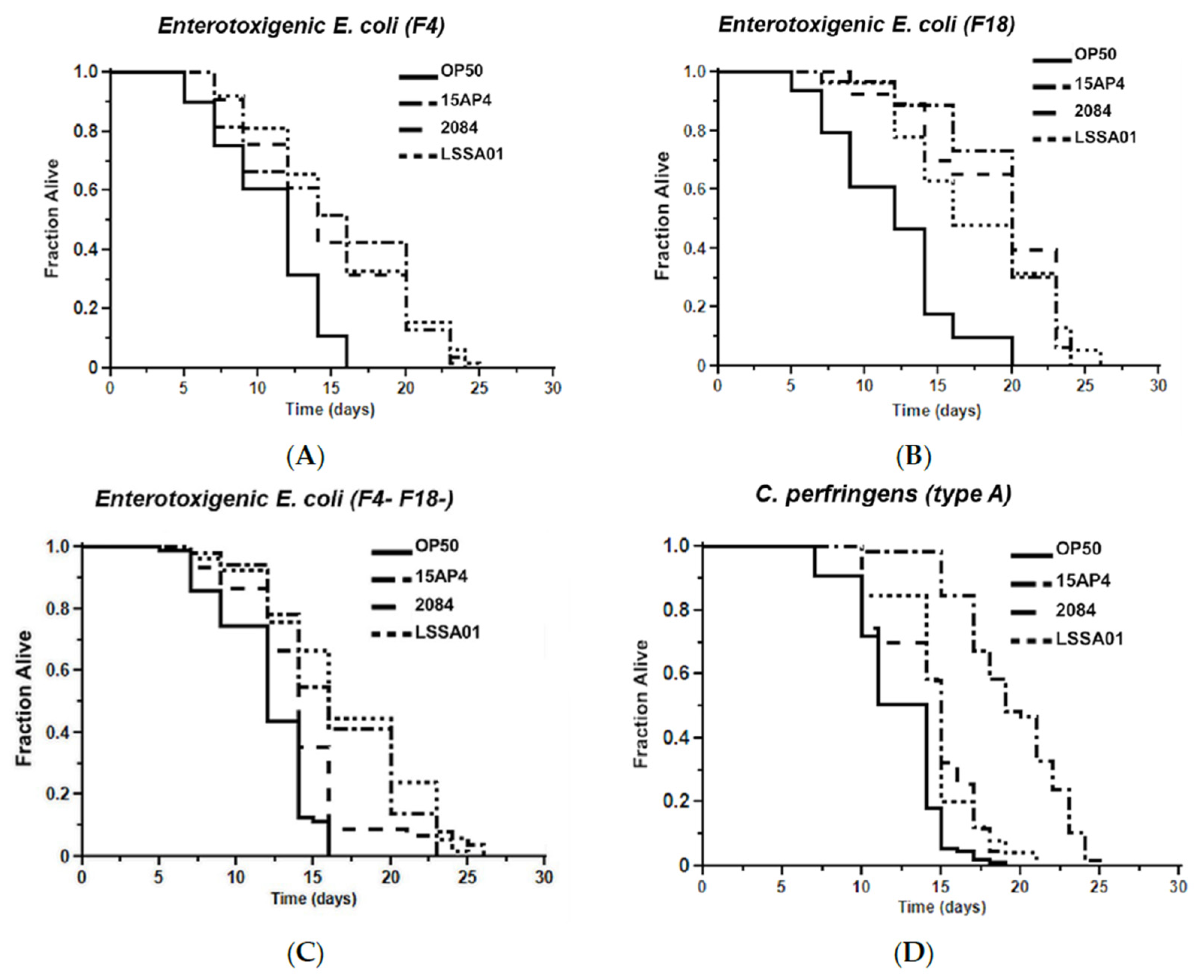
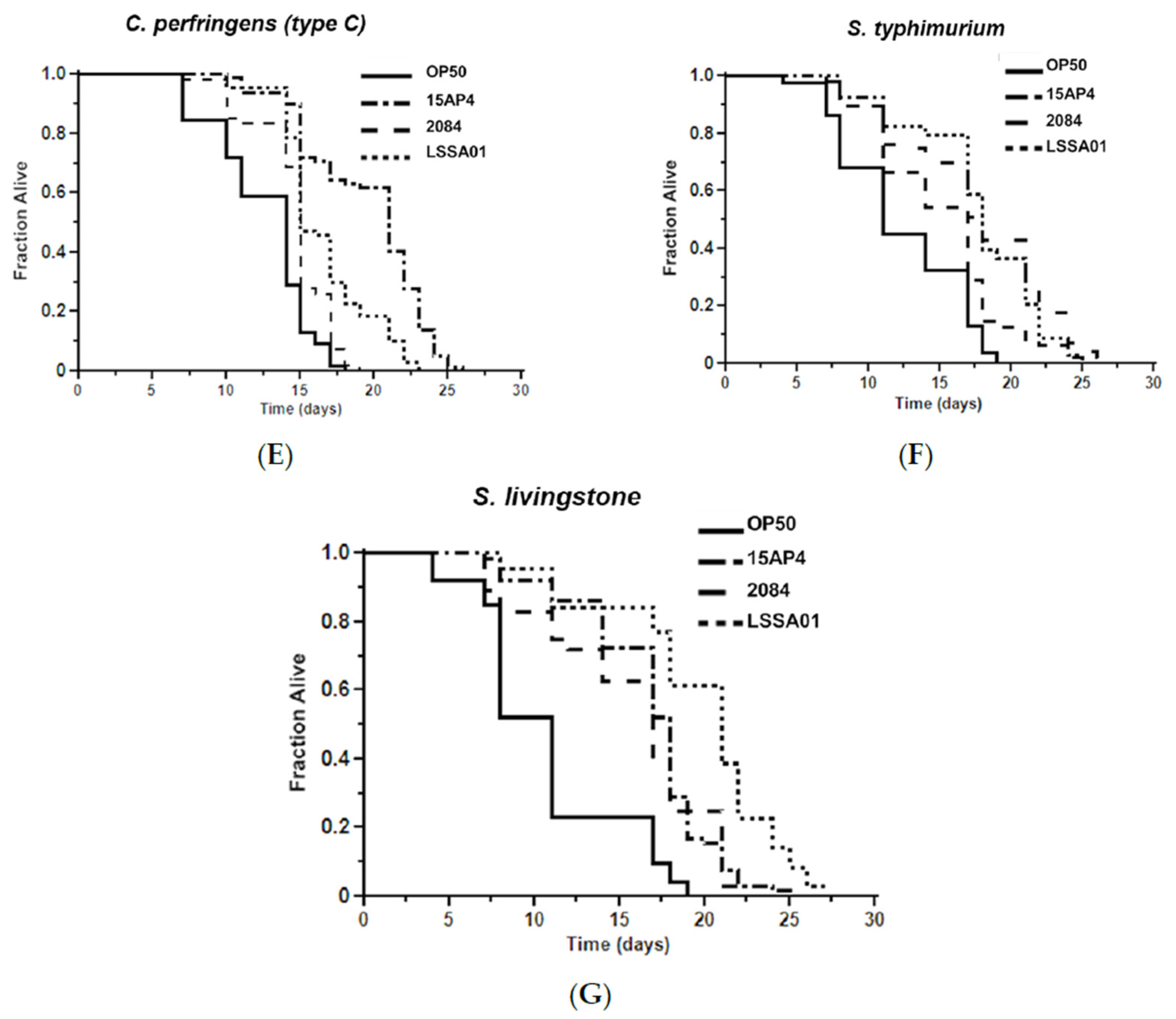
3.5. Effect of B. velezensis Strains on C. elegans Survival When Exposed to Pathogenic Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BHI | Brain heart infusion broth |
| CFU | Colony forming unit |
| ETEC | Enterotoxigenic Escherichia coli |
| FBS | Fetal bovine serum |
| LB | Luria Bertani broth |
| LT | Heat-labile toxin |
| NGM | Nematode growth medium |
| OD | Optical density |
| P/S | Penicillin/streptomycin |
| PWD | Post-weaning diarrhea |
| RT-qPCR | Reverse-transcription quantitative polymerase chain reaction |
| SCFA | Short-chain fatty acid |
| TSA | Tryptic soy broth |
References
- Tang, Q.; Lan, T.; Zhou, C.; Gao, J.; Wu, L.; Wei, H.; Li, W.; Tang, Z.; Tang, W.; Diao, H.; et al. Nutrition strategies to control post-weaning diarrhea of piglets: From the perspective of feeds. Anim. Nutr. 2024, 17, 2097–2311. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Hou, L.; Sun, L.; Gao, J.; Gao, K.; Yang, X.; Jiang, Z.; Wang, L. Intestinal morphology and immune profiles are altered in piglets by early-weaning. Int. Immunopharmacol. 2022, 105, 108520. [Google Scholar] [CrossRef] [PubMed]
- Wijtten, J.A.; van der Meulen, J.; Verstegen, M.W.A. Intestinal barrier function and absorption in pigs after weaning: A review. Br. J. Nutr. 2011, 105, 967–981. [Google Scholar] [CrossRef]
- Eriksen, E.Ø.; Kudirkiene, E.; Barington, K.; Goecke, N.B.; Blirup-Plum, A.; Nielsen, J.P.; Olsen, J.E.; Jensen, H.E.; Pankoke, K.; Larsen, L.E.; et al. An observational field study of porcine post-weaning diarrhea: Clinical and microbiological findings, and fecal pH-measurements as a potential diagnostic tool. Porc. Health Manag. 2023, 9, 33. [Google Scholar] [CrossRef]
- Lupi, A.; Gibellini, M.; Gin, T.; Vangroenweghe, F.; Vandenbroucke, V.; Bauerfeind, R.; Bonilauri, P.; Labarque, G.; Hidalgo, A. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea in Europe. Porc. Health Manag. 2016, 2, 39. [Google Scholar] [CrossRef]
- Canibe, N.; Højberg, O.; Kongsted, H.; Vodolazska, D.; Lauridsen, C.; Nielsen, T.S.; Schönherz, A.A. Review on preventive measures to reduce post-weaning diarrhoea in piglets. Animals 2022, 12, 2585. [Google Scholar] [CrossRef]
- FAO/WHO. Probiotics in Animal Nutrition. Production, Impact and Regulation. Food and Agriculture Organization of the United Nations. Rome. 2016. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/2fda2226-b000-4cee-8186-8cacab86316b/content#:~:text=The%20FAO%20and%20WHO%20definition,CLASSIFICATION (accessed on 7 April 2016).
- Gresse, R.; Chaucheyras-Durand, F.; Fleury, M.A.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut Microbiota Dysbiosis in Postweaning Piglets: Understanding the Keys to Health. Trends Microbiol. 2017, 25, 851–873. [Google Scholar] [CrossRef]
- Buntyn, J.O.; Schmidt, T.B.; Nisbet, D.J.; Callaway, T.R. The role of direct-fed microbials in conventional livestock production. Annu. Rev. Anim. Biosci. 2016, 4, 335–355. [Google Scholar] [CrossRef]
- Daudelin, J.F.; Lessard, M.; Beaudoin, F.; Nadeau, E.; Bissonnette, N.; Boutin, Y.; Brousseau, J.-P.; Lauzon, K.; Fairbrother, J.M. Administration of probiotics influences F4 (K88)-positive enterotoxigenic Escherichia coli attachment and intestinal cytokine expression in weaned pigs. Vet. Res. 2011, 42, 69. [Google Scholar] [CrossRef]
- Guerra-Ordaz, A.A.; González-Ortiz, G.; La Ragione, R.M.; Woodward, M.J.; Collines, J.W.; Pérez, J.F.; Martín-Orúe, S.M. actulose and Lactobacillus plantarum, a potential complementary synbiotic to control postweaning colibacillosis in piglets. Appl. Environ. Micro. 2014, 80, 4879–4886. [Google Scholar] [CrossRef]
- Baker, A.A.; Davis, E.; Spencer, J.D.; Moser, R.; Rehberger, T. The effect of a Bacillus-based direct-fed microbial supplemented to sows on the gastrointestinal microbiota of their neonatal piglets. J. Anim. Sci. 2013, 7, 3390–3399. [Google Scholar] [CrossRef] [PubMed]
- Mazur-Kuśnirek, M.; Kipiňski, K.; Jørgensen, J.N.; Hansen, L.H.B.; Antoszkiewicz, Z.; Zabeilski, R.; Konieczka, P. The effect of a Bacillus-based probiotic on sow and piglet performance in two production cycles. Animals 2023, 20, 3163. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yang, M.; Su, W.; Mei, L.; Li, Y.; Guo, Y.; Li, Y.; Liang, W.; Yang, B.; Huang, Z.; et al. Probiotics in piglet: From gut health to pathogen defense mechanisms. Front. Immunol. 2024, 15, 1468873. [Google Scholar] [CrossRef]
- Quach, N.T.; Vu, T.H.N.; Nguyen, N.A.; Nguyen, N.A.; Nguyen, V.T.; Bui, T.L.; Ky, S.C.; Le, T.L.; Hoang, H.; Ngo, C.C.; et al. Phenotypic features and analysis of genes supporting probiotic action unravel underlying perspectives of Bacillus velezensis VTX9 as a potential feed additive for swine. Ann. Microbiol. 2021, 71, 36. [Google Scholar] [CrossRef]
- Corsi, A.K.; Wightman, B.; Chalfie, M.A. Transparent Window into Biology: A Primer on Caenorhabditis Elegans. Genetics 2015, 200, 387–407. [Google Scholar] [CrossRef]
- Olsen, A.; Gill, M.S. Ageing: Lessons from C. elegans; Olsen, A., Gill, M.S., Eds.; Springer International: Cham, Switzerland, 2017. [Google Scholar]
- Cohen, L.B.; Troemel, E.R. Microbial pathogenesis and host defense in the nematode C. elegans. Curr. Opin. Microbiol. 2014, 23, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Ewbank, J.J.; Pujol, N. Local and long-range activation of innate immunity by infection and damage in C. elegans. Curr. Opin Immunol. 2016, 38, 1–7. [Google Scholar] [CrossRef]
- Christensen, K.; Morch, M.; Morthorst, T.; Lykkemark, S.; Olsen, A. Microbiota, probiotic bacteria and ageing. In Ageing: Lessons from C. elegans; Olsen, A., Gill, M.S., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 411–429. [Google Scholar]
- Kim, Y.; Mylonakis, E. Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances gram-positive immune responses. Infect. Immun. 2012, 80, 2500–2508. [Google Scholar] [CrossRef]
- Sarsour, A.H.; Koltes, D.A.; Kim, E.J.; Persia, M.E. Effects of a direct fed microbial (DFM) on broiler chickens exposed to acute and chronic cyclic heat stress in two consecutive experiments. Poult. Sci. 2022, 101, 101705. [Google Scholar] [CrossRef]
- EFSA. Safety and efficacy of a feed additive consisting of Bacillus velezensis PTA-6507, B. velezensis NRRL B-50013 and B. velezensis NRRL B-50104 (Enviva® PRO 202 GT) for turkeys for fattening (Danisco Animal Nutrition). EFSA J. 2021, 19, e06535. [Google Scholar]
- EFSA. Safety and efficacy of Enviva® PRO 202 GT (Bacillus amyloliquefaciens PTA-6507, Bacillus amyloliquefaciens NRRL B-50013 and Bacillus amyloliquefaciens NRRL B-50104) for chickens for fattening, chickens reared for laying and minor poultry species for fattening and to point of lay. EFSA J. 2016, 14, 304505. [Google Scholar]
- Fernández, S.M.; Cretenet, M.; Bernardeau, M. In vitro inhibition of avian pathogenic Enterococcus cecorum isolates by probiotic Bacillus strains. Poult. Sci. 2019, 98, 2338–2346. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Christensen, L.G.; Rasmussen, P.B.; Kragh, K.M. In vitro immunomodulatory effects of thymol and cinnamaldehyde in a pig intestinal epithelial cell line (IPEC-J2). J. Appl. Anim. Nutr. 2020, 8, 127–134. [Google Scholar] [CrossRef]
- Kadedar, D.; Udrea, A.C.; Bak, S.Y.; Christensen, N.; Gibbs, K.; Shen, C.; Bernardeau, M. Cell-Free Culture Supernatant of Lactobacillus acidophilus AG01 and Bifidobacterium animalis subsp. lactis AG02 Reduces the Pathogenicity of NetB-Positive Clostridium perfringens in a Chicken Intestinal Epithelial Cell Line. Microorganisms 2024, 12, 839. [Google Scholar]
- Konieczka, P.; Ferenc, K.; Jørgensen, J.N.; Hansen, L.H.B.; Zabielski, R.; Olszewski, J.; Gajewski, Z.; Mazur-Kuśnirek, M.; Szkopek, D.; Szyryńska, N.; et al. Feeding Bacillus-based probiotics to gestating and lactating sows is an efficient method for improving immunity, gut functional status, and biofilm formation by probiotic bacteria in piglets at weaning. Anim. Nutr. 2023, 13, 361–372. [Google Scholar] [CrossRef]
- Ripert, G.; Racedo, S.M.; Elie, A.M.; Jacquot, C.; Bressollier, P.; Urdaci, M.C. Secreted compounds of the probiotic Bacillus clausii strain O/C inhibit the cytotoxic effects induced by Clostridium difficile and Bacillus cereus toxins. Antimicrob. Agents Chemother. 2016, 60, 3445–3454. [Google Scholar] [CrossRef]
- Shobharani, P.; Padmaja, R.J.; Halami, P.M. Diversity in the antibacterial potential of probiotic cultures Bacillus licheniformis MCC2514 and Bacillus licheniformis MCC2512. Res. Microbiol. 2015, 166, 546–554. [Google Scholar] [CrossRef]
- Kiernan, D.P.; O’Doherty, J.V.; Sweeney, T. The Effect of Maternal Probiotic or Synbiotic Supplementation on Sow and Offspring Gastrointestinal Microbiota, Health, and Performance. Animals 2023, 13, 2996. [Google Scholar] [CrossRef]
- Drumond, M.M.; Tapia-Costa, A.P.; Neumann, E.; Nunes, A.C.; Barbosa, J.W.; Kassuha, D.E.; Mancha-Agresti, P. Cell-free supernatant of probiotic bacteria exerted antibiofilm and antibacterial activities against Pseudomonas aeruginosa: A novel biotic therapy. Front. Pharmacol. 2023, 14, 1152588. [Google Scholar] [CrossRef]
- Kaewchomphunuch, T.; Charoenpichitnunt, T.; Thongbaiyai, V.; Ngamwongsatit, N.; Kaeoket, K. Cell-free culture supernatants of Lactobacillus spp. and Pediococcus spp. inhibit growth of pathogenic Escherichia coli isolated from pigs in Thailand. BMC Vet. Res. 2020, 18, 60. [Google Scholar] [CrossRef]
- Xie, K.; Xie, H.; Su, G.; Chen, D.; Yu, B.; Mao, X.; Huang, Z.; Yu, J.; Luo, J.; Zheng, P.; et al. β-defensin 129 attenuates bacterial endotoxin-induced inflammation and intestinal epithelial cell apoptosis. Front. Immunol. 2019, 10, 2333. [Google Scholar] [CrossRef] [PubMed]
- Bassler, M.B.; Miller, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar]
- De Groot, N.; Fariñas, F.; Cabrera-Gómez, C.G.; Pallares, F.J.; Ramis, G. Weaning causes a prolonged but transient change in immune gene expression in the intestine of piglets. J. Anim. Sci. 2021, 99, skab065. [Google Scholar] [CrossRef]
- Eckmann, L.; Kagnoff, M.F.; Fierer, J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect. Immun. 1993, 61, 4569–4574. [Google Scholar] [CrossRef]
- Yu, H.; Huang, X.; Ma, U.; Gao, M.; Wang, O.; Gao, T.; Shen, Y.; Liu, X. Interleukin-8 regulates endothelial permeability by down-regulation of tight junction but not dependent on integrins induced focal adhesions. Int. J. Biol. Sci. 2013, 9, 966–979. [Google Scholar] [CrossRef]
- Wang, J.; Ji, H. Tight junction proteins in the weaned piglet intestine: Roles and regulation. Curr. Protein Pept. Sci. 2019, 20, 7. [Google Scholar] [CrossRef]
- Andrews, C.; McLean, M.H.; Durum, S.K. Cytokine Tuning of Intestinal Epithelial Function. Front. Immunol. 2018, 9, 1270. [Google Scholar] [CrossRef]
- Findley, M.K.; Koval, M. Regulation and roles for claudin-family tight junction proteins. UBMB Life 2009, 61, 431–437. [Google Scholar] [CrossRef]
- Umeda, K.; Ikenouchi, J.; Katahira-Tayama, S.; Furuse, K.; Sasaki, H.; Nakayama, M.; Matsui, T.; Tsukita, S.; Furuse, M.; Tsukita, S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 2006, 126, 741–754. [Google Scholar] [CrossRef]
- Garcia-Hernandez, V.; Quiros, M.; Nusrat, A. Intestinal epithelial claudins: Expression and regulation in homeostasis and inflammation. Ann. N. Y. Acad. Sci. 2017, 1397, 66–79. [Google Scholar] [CrossRef]
- Moeser, A.J.; Pohl, C.S.; Rajput, M. Weaning stress and gastrointestinal barrier development: Implications for lifelong gut health in pigs. Anim. Nutr. 2017, 3, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hollander, D.; Kaunitz, J.D. The “Leaky Gut”: Tight Junctions but Loose Associations? Dig. Dis. Sci. 2020, 65, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Krishna Rao, R.; Samak, G. Protection and Restitution of Gut Barrier by Probiotics: Nutritional and Clinical Implications. Curr. Nutr. Food Sci. 2013, 9, 99–107. [Google Scholar] [CrossRef]
- Barros, C.P.; Guimarães, J.T.; Esmerino, E.A.; Duarte, M.C.K.; Silva, M.C.; Silva, R.; Ferreira, B.M.; Sant’Ana, A.S.; Freitas, M.Q.; Cruz, A.G. Paraprobiotics and postbiotics: Concepts and potential applications in dairy products. Curr. Opin. Food Sci. 2020, 32, 1–8. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Leoni, G.; Neumann, P.A.; Sumagin, R.; Denning, T.L.; Nusrat, A. Wound repair: Role of immune-epithelial interactions. Mucosal Immunol. 2015, 8, 959–968. [Google Scholar] [CrossRef]
- Sakellaris, T.; Madhavan, V.; Harry, S. Colonization factors of enterotoxigenic Escherichia coli. Adv. Appl. Microbiol. 2015, 90, 155–197. [Google Scholar]
- Xia, P.; Quan, G.; Yang, Y.; Zhao, J.; Wang, Y.; Zhou, M.; Wang, Y.; Zhou, M.; Hardwidge, P.R.; Zhu, J.; et al. Binding determinants in the interplay between porcine aminopeptidase N and enterotoxigenic Escherichia coli F4 fimbriae. Vet. Res. 2018, 49, 23. [Google Scholar] [CrossRef]
- Xia, R.W.; Qin, W.Y.; Gan, L.N.; Wang, J.; Wu, S.L.; Bao, W.B. Differential expression of FUT1 and FUT2 in large white, Meishan, and Sutai porcine breeds. Genet. Mol. Res. 2016, 15, 15017613. [Google Scholar] [CrossRef]
- Elsinghorst, E.A.; Kopecko, D.J. Molecular cloning of epithelial cell invasion determinants from enterotoxigenic Escherichia coli. Infect. Immun. 1992, 60, 2409–2417. [Google Scholar] [CrossRef]
- Elsinghorst, E.A.; Weitz, J.A. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 1994, 62, 3463–3471. [Google Scholar] [CrossRef] [PubMed]
- Fleckenstein, J.M.; Kopecko, D.J.; Warren, R.L.; Elsinghorst, E.A. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 1996, 64, 2256–2265. [Google Scholar] [CrossRef] [PubMed]
- Lindenthal, C.; Elsinghorst, E.A. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 1999, 67, 4084–4091. [Google Scholar] [CrossRef]
- Ikeda, T.; Yasui, C.; Hoshino, K.; Arikawa, K.; Nishikawa, Y. Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 2007, 73, 6404–6409. [Google Scholar] [CrossRef]
- Park, M.R.; Ryu, S.; Maburutse, B.E.; Oh, N.S.; Kim, S.H.; Oh, S.; Jeong, S.-Y.; Jeong, D.-Y.; Oh, S.; Kim, Y. Probiotic Lactobacillus fermentum strain JDFM216 stimulates the longevity and immune response of Caenorhabditis elegans through a nuclear hormone receptor. Sci. Rep. 2018, 8, 7441. [Google Scholar] [CrossRef]
- Nakagawa, H.; Shiozaki, T.; Kobatake, E.; Hosoya, T.; Moriya, T.; Sakai, F.; Taru, H.; Miyazaki, T. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell 2016, 15, 227–236. [Google Scholar] [CrossRef]
- Gumienny, T.L.; Savage-Dunn, C. TGF-β signaling in C. elegans. In WormBook: The Online Review of C. elegans [Internet]; WormBook: Pasadena, CA, USA, 2013. [Google Scholar]
- Kamaladevi, K.; Balamurugan, A. Lactobacillus casei triggers a TLR mediated RACK-1 dependent p38 MAPK pathway in Caenorhabditis elegans to resist Klebsiella pneumoniae infection. Food Funct. 2016, 7, 3211–3223. [Google Scholar] [CrossRef]
- Donato, V.; Rodríguez, A.F.; Cogliati, S.; Bauman, C.; Costa, J.G.; Leñini, C.; Grau, R. Bacillus subtilis biofilm extends Caenorhabditis elegans longevity through downregulation of the insulin-like signaling pathway. Nat. Commun. 2017, 8, 14332. [Google Scholar] [CrossRef]

| Ecotoxigenic Escherichia coli (ETEC) Isolate F4 and F18 Fimbriae Genotype | Binding Affinity (%) | |
|---|---|---|
| B. velezensis strain | ||
| LSSA01 | Not applicable | 0.298 |
| 15AP4 | Not applicable | 0.216 |
| 2084 | Not applicable | 2.607 |
| ETEC isolate | ||
| EC-23 | F4−F18+ | 0.003 |
| EC-50 | F4−F18+ | 0.009 |
| EC-58 | F4−F18+ | 0.053 |
| EC-61 | F4−F18+ | 0.052 |
| EC-62 | F4−F18+ | 0.009 |
| EC-63 | F4−F18+ | 0.000 |
| EC-65 | F4−F18− | 0.006 |
| EC-88 | F4+F18− | 0.172 |
| EC-89 | F4+F18− | 0.000 |
| EC-90 | F4+F18− | 0.001 |
| EC-91 | F4−F18− | 0.002 |
| EC-92 | F4+F18− | 0.145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasmussen, P.B.; Walker, J.; Stubbs, S.R.; Udrea, A.C.; Shen, C. Evaluation of Probiotic Bacillus velezensis for the Control of Pathogens That Cause Post-Weaning Diarrhea in Piglets—Results from In Vitro Testing and an In Vivo Model Using Caenorhabditis elegans. Microorganisms 2025, 13, 1247. https://doi.org/10.3390/microorganisms13061247
Rasmussen PB, Walker J, Stubbs SR, Udrea AC, Shen C. Evaluation of Probiotic Bacillus velezensis for the Control of Pathogens That Cause Post-Weaning Diarrhea in Piglets—Results from In Vitro Testing and an In Vivo Model Using Caenorhabditis elegans. Microorganisms. 2025; 13(6):1247. https://doi.org/10.3390/microorganisms13061247
Chicago/Turabian StyleRasmussen, Pia Bilde, Josh Walker, Stacey Robida Stubbs, Andreea Cornelia Udrea, and Chong Shen. 2025. "Evaluation of Probiotic Bacillus velezensis for the Control of Pathogens That Cause Post-Weaning Diarrhea in Piglets—Results from In Vitro Testing and an In Vivo Model Using Caenorhabditis elegans" Microorganisms 13, no. 6: 1247. https://doi.org/10.3390/microorganisms13061247
APA StyleRasmussen, P. B., Walker, J., Stubbs, S. R., Udrea, A. C., & Shen, C. (2025). Evaluation of Probiotic Bacillus velezensis for the Control of Pathogens That Cause Post-Weaning Diarrhea in Piglets—Results from In Vitro Testing and an In Vivo Model Using Caenorhabditis elegans. Microorganisms, 13(6), 1247. https://doi.org/10.3390/microorganisms13061247






