Dolichocephalovirinae Phages Exist as Episomal Pseudolysogens Across Diverse Soil Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Culture Preparation
2.2. Isolation of Bacteriophages from Bacterial Strains
2.3. Independent Isolation of Bacteriophages from Soil and Plant Root Samples
2.4. Phage Genome Size
2.5. Lysogeny Detection
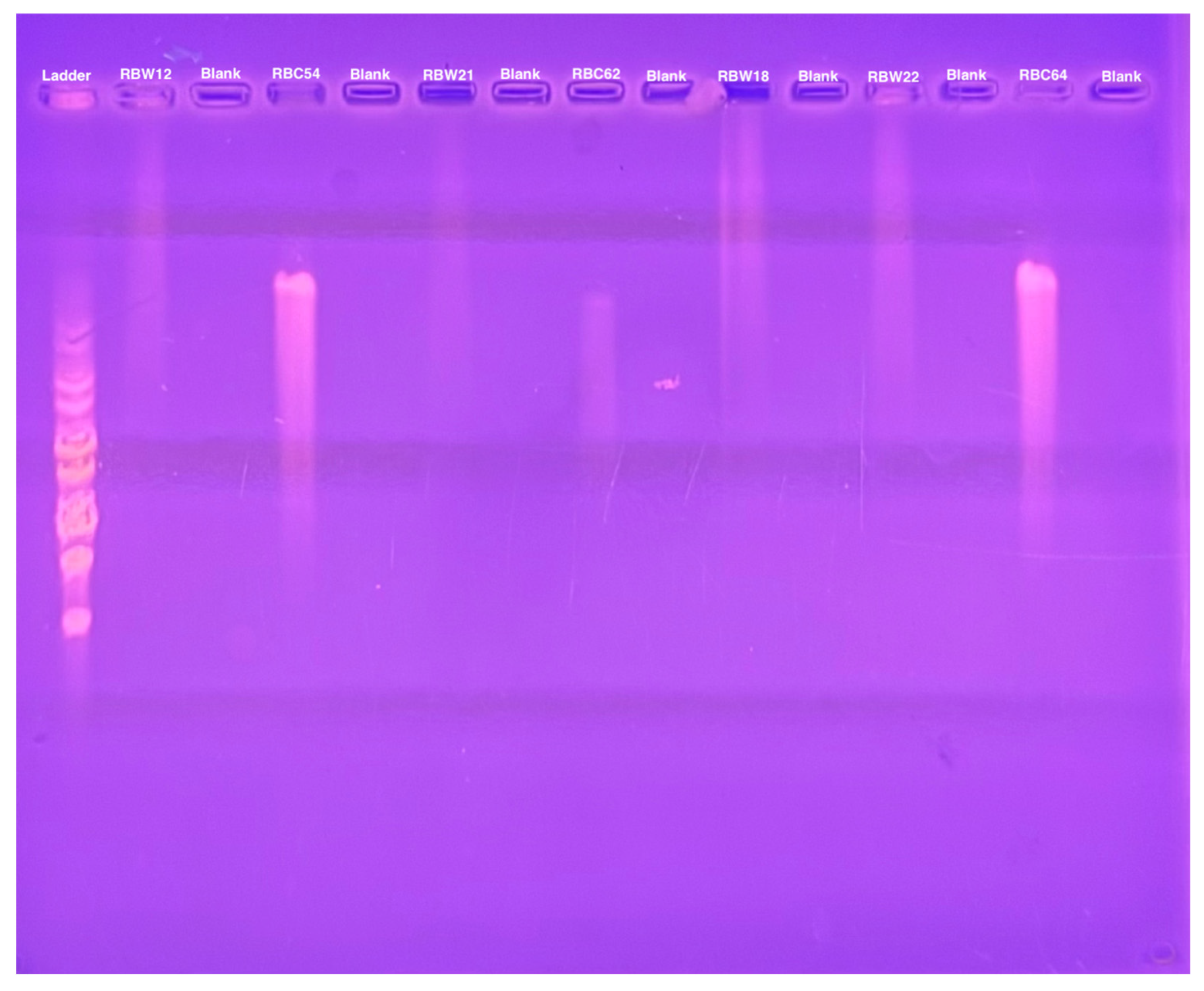
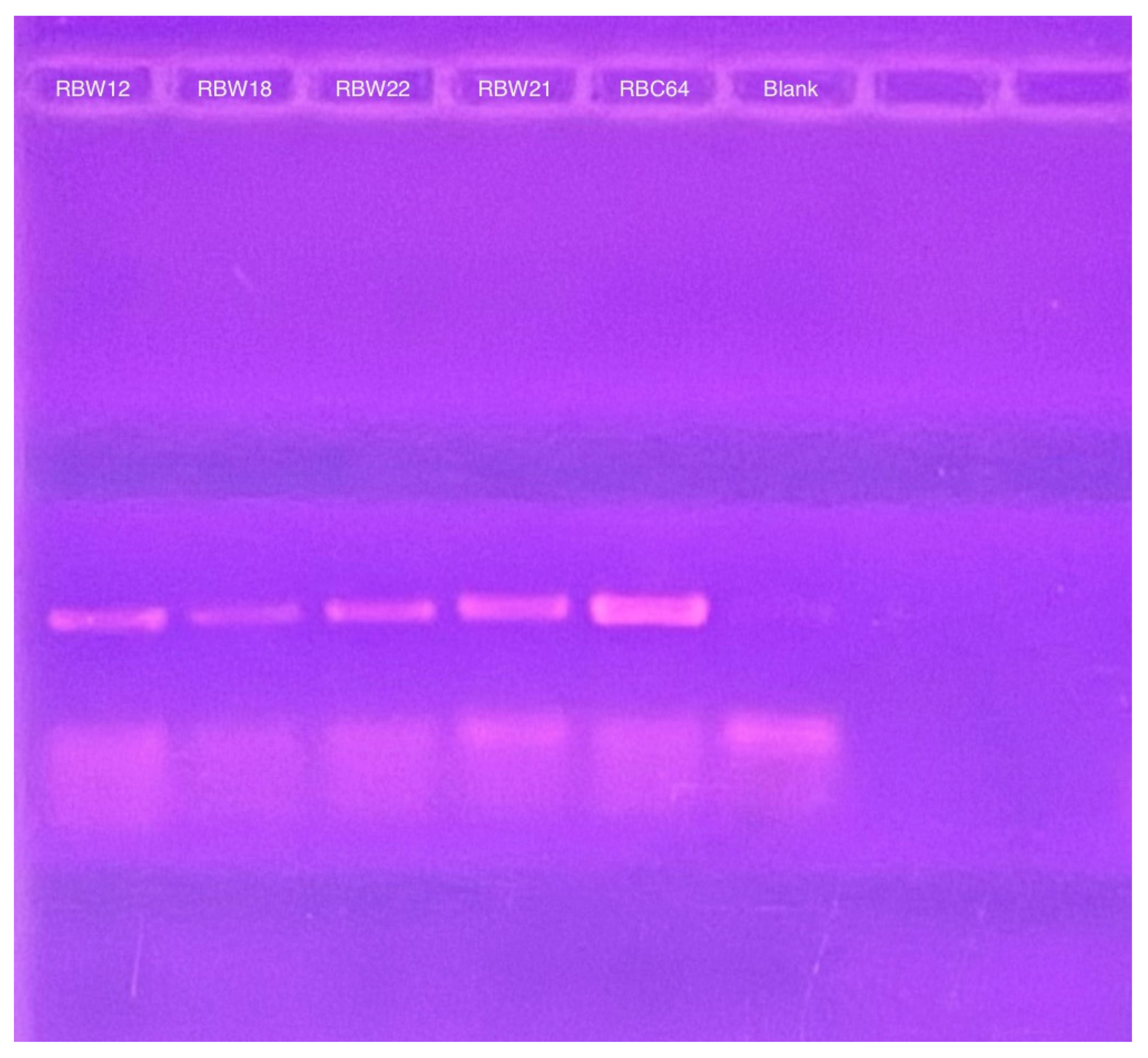
2.6. Episomal Bacteriophage Genome Detection
2.7. PCR Analysis of Additional Phage Genomes
2.8. Phage Genome Sequencing and Annotation
2.9. Transmission Electron Microscopy
3. Results
3.1. Isolation and Characterization of Bacteriophages
3.2. Bacteriophage Genome Sizes
3.3. Phage Genomes Are Present in the Cytoplasm of the Host Strains
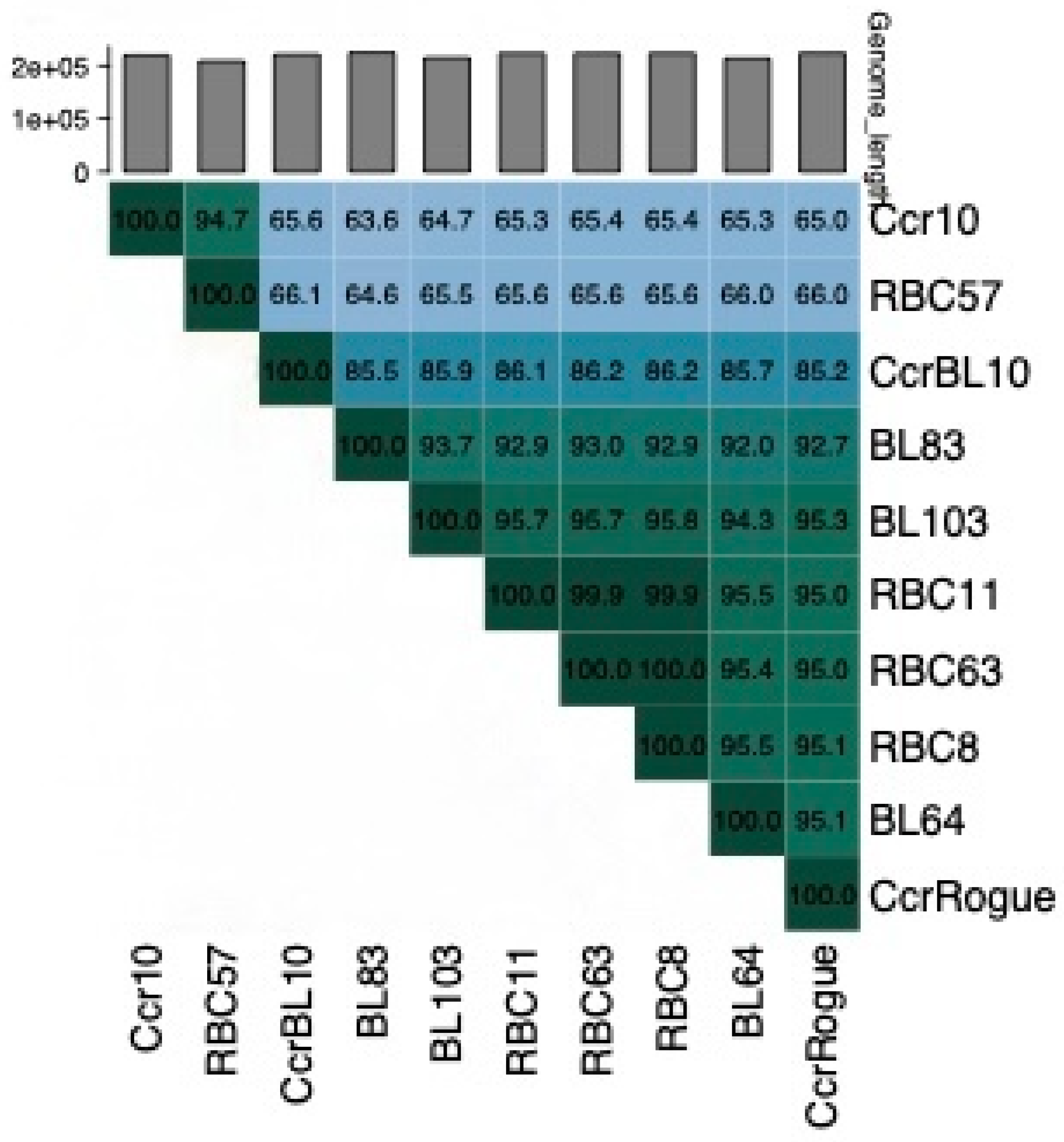
3.4. Genome Nucleotide Sequences
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Oerke, E.-C.; Dehne, H.-W. Safeguarding production—Losses in major crops and the role of crop protection. Crop. Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations, FAOSTAT Statistics Database 2020. Available online: www.fao.org/faostat/en/ (accessed on 10 June 2021).
- Lehti, T.A.; Pajunen, M.I.; Skog, M.S.; Finne, J. Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat. Commun. 2017, 8, 1915. [Google Scholar] [CrossRef] [PubMed]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Ely, B.; Lenski, J.; Mohammadi, T. Structural and Genomic Diversity of Bacteriophages. Methods Mol. Biol. 2024, 2738, 3–16. [Google Scholar] [PubMed]
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef]
- Ghosh, D.; Roy, K.; Williamson, K.E.; White, D.C.; Wommack, K.E.; Sublette, K.L.; Radosevich, M. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl. Environ. Microbiol. 2008, 74, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Danovaro, R.; Dell’anno, A.; Trucco, A.; Serresi, M.; Vanucci, S. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 2001, 67, 1384–1387. [Google Scholar] [CrossRef] [PubMed]
- Danovaro, R.; Manini, E.; Dell’Anno, A. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 2002, 68, 1468–1472. [Google Scholar] [CrossRef]
- Williamson, K.E.; Schnitker, J.B.; Radosevich, M.; Smith, D.W.; Wommack, K.E. Cultivation-based assessment of lysogeny among soil bacteria. Microb. Ecol. 2008, 56, 437–447. [Google Scholar] [CrossRef]
- Mäntynen, S.; Laanto, E.; Oksanen, H.M.; Poranen, M.M.; Díaz-Muñoz, S.L. Black box of phage-bacterium interactions: Exploring alternative phage infection strategies. Open Biol. 2021, 11, 210188. [Google Scholar] [CrossRef]
- Sun, M.; Yuan, S.; Xia, R.; Ye, M.; Balcazar, J.L. Underexplored viral auxiliary metabolic genes in soil: Diversity and eco-evolutionary significance. Environ. Microbiol. 2023, 25, 800–810. [Google Scholar] [CrossRef]
- Van Goethem, M.W.; Swenson, T.L.; Trubl, G.; Roux, S.; Northen, T.R. Characteristics of wetting-induced bacteriophage blooms in biological soil crust. mBio 2019, 10, 10-1128. [Google Scholar] [CrossRef]
- Emerson, J.B.; Roux, S.; Brum, J.R.; Bolduc, B.; Woodcroft, B.J.; Jang, H.B.; Singleton, C.M.; Solden, L.M.; Naas, A.E.; Boyd, J.A.; et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 2018, 3, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, Z.; Liu, H.; Li, Y.; Ge, T.; Tang, X.; He, Y.; Ma, B.; Xu, J.; Anantharaman, K.; et al. Soil nutrient conditions alter viral lifestyle strategy and potential function in phosphorous and nitrogen metabolisms. Soil Biol. Biochem. 2024, 189, 109279. [Google Scholar] [CrossRef]
- Zheng, X.; Jahn, M.T.; Sun, M.; Friman, V.P.; Balcazar, J.L.; Wang, J.; Shi, Y.; Gong, X.; Hu, F.; Zhu, Y. Organochlorine contamination enriches virus-encoded metabolism and pesticide degradation associated auxiliary genes in soil microbiomes. ISME J. 2022, 16, 1397–1408. [Google Scholar] [CrossRef]
- Huang, D.; Yu, P.; Ye, M.; Schwarz, C.; Jiang, X.; Alvarez, P.J.J. Enhanced mutualisticsymbiosis between soil phages and bacteria with elevated chromium-induced environmental stress. Microbiome 2021, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Ripp, S.; Miller, R.V. The role of pseudolysogeny in bacteriophage-host interactions in a natural freshwater environment. Microbiology 1997, 143, 2065–2070. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Haarmann, N.; Schwidder, M.; Muniesa, M.; Schmidt, H. Bacteriophages of shiga toxin-producing and their contribution to pathogenicity. Pathogens 2021, 29, 404. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F.; Depardieu, F.; Miele, S.; Dowding, J.; Laval, A.L.; Lieberman, E.; Garry, D.; Rocha, E.P.C.; Bernheim, A.; Bikard, D. Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe 2022, 11, 740–753. [Google Scholar] [CrossRef]
- Dougherty, P.E.; Nielsen, T.K.; Riber, L.; Lading, H.L.; Forero-Junco, L.M.; Kot, W.; Raaijmakers, J.M.; Hansen, L.H. Widespread and largely unknown prophage activity, diversity, and function in two genera of wheat phyllosphere bacteria. ISME J. 2023, 17, 2415–2425. [Google Scholar] [CrossRef]
- Rakonjac, J.; Bennett, N.J.; Spagnuolo, J.; Gagic, D.; Russel, M. Filamentous Bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011, 13, 51–75. [Google Scholar]
- Abedon, S.T. Disambiguating Bacteriophage Pseudolysogeny: An historical analysis of lysogeny, pseudolysogeny, and the phage carrier state. In Contemporary Trends in Bacteriophage Research; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2009; pp. 285–307. [Google Scholar]
- Siringan, P.; Connerton, P.L.; Cummings, N.J.; Connerton, I.F. Alternative bacteriophage life cycles: The carrier state of Campylobacter jejuni. Open Biol. 2014, 26, 130200. [Google Scholar] [CrossRef]
- Ripp, S.; Miller, R.V. Dynamics of the pseudolysogenic response in slowly growing cells of Pseudomonas aeruginosa. Microbiology 1998, 144, 2225–2232. [Google Scholar] [CrossRef]
- Cenens, W.; Makumi, A.; Mebrhatu, M.T.; Lavigne, R.; Aertsen, A. Phage-host interactions during pseudolysogeny: Lessons from the Pid/dgo interaction. Bacteriophage 2013, 3, e25029. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, T.; Ely, B. The Isolation and Characterization of Novel Caulobacter and Non-Caulobacter Lysogenic Bacteria from Soil and the Discovery of Broad-Host-Range Phages Infecting Multiple Genera. Microorganisms 2024, 12, 1894. [Google Scholar] [CrossRef] [PubMed]
- Lood, R.; Collin, M. Characterization and genome sequencing of two Propionibacterium acnes phages displaying pseudolysogeny. BMC Genom. 2011, 12, 198. [Google Scholar] [CrossRef]
- Wottrich, S.; Mendonca, S.; Safarpour, C.; Nguyen, C.; Marinelli, L.J.; Hancock, S.P.; Modlin, R.L.; Parker, J.M. Putative pseudolysogeny-dependent phage gene implicated in the superinfection resistance of Cutibacterium acnes. Microbiome Res. Rep. 2024, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Dingwall, A.; Shapiro, L.; Ely, B. Analysis of bacterial genome organization and replication using pulsed-field gel electrophoresis. Methods 1990, 1, 160–168. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the rapid annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2013, 42, D206–D214. [Google Scholar] [CrossRef]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 10, 944–945. [Google Scholar] [CrossRef]
- Darling, A.C.; Bau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Sisto, I.; Ely, B. Caulobacter strains code for novel restriction endonucleases that protect them from bacteriophage infections. Viruses 2015, 17, 311. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Wilson, K.; Ely, B. Analyses of four new Caulobacter PhiCbKviruses indicate independent lineages. J. Gen. Virol. 2019, 100, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Ely, B.; Thomas, Q.; Mohammadi, T. Genetic diversity among independent isolates of the Dolichocephalovirinae subfamily. Bacteria 2025, 4, 8. [Google Scholar] [CrossRef]
- Santamaría, R.I.; Bustos, P.; Sepúlveda-Robles, O.; Lozano, L.; Rodríguez, C.; Fernández, J.L.; Juárez, S.; Kameyama, L.; Guarneros, G.; Dávila, G.; et al. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl. Environ. Microbiol. 2014, 80, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef]
- Ofir, G.; Sorek, R. Contemporary phage biology: From classic models to new insights. Cell 2018, 172, 1260–1270. [Google Scholar] [CrossRef]
- Paul, J.H. Prophages in marine bacteria: Dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008, 2, 579–589. [Google Scholar] [CrossRef]
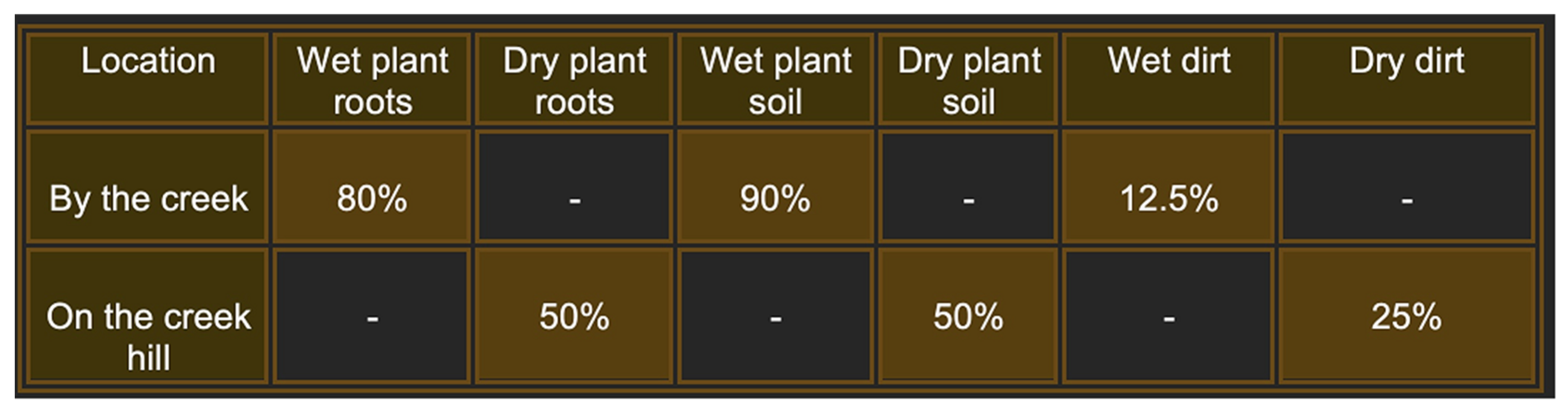
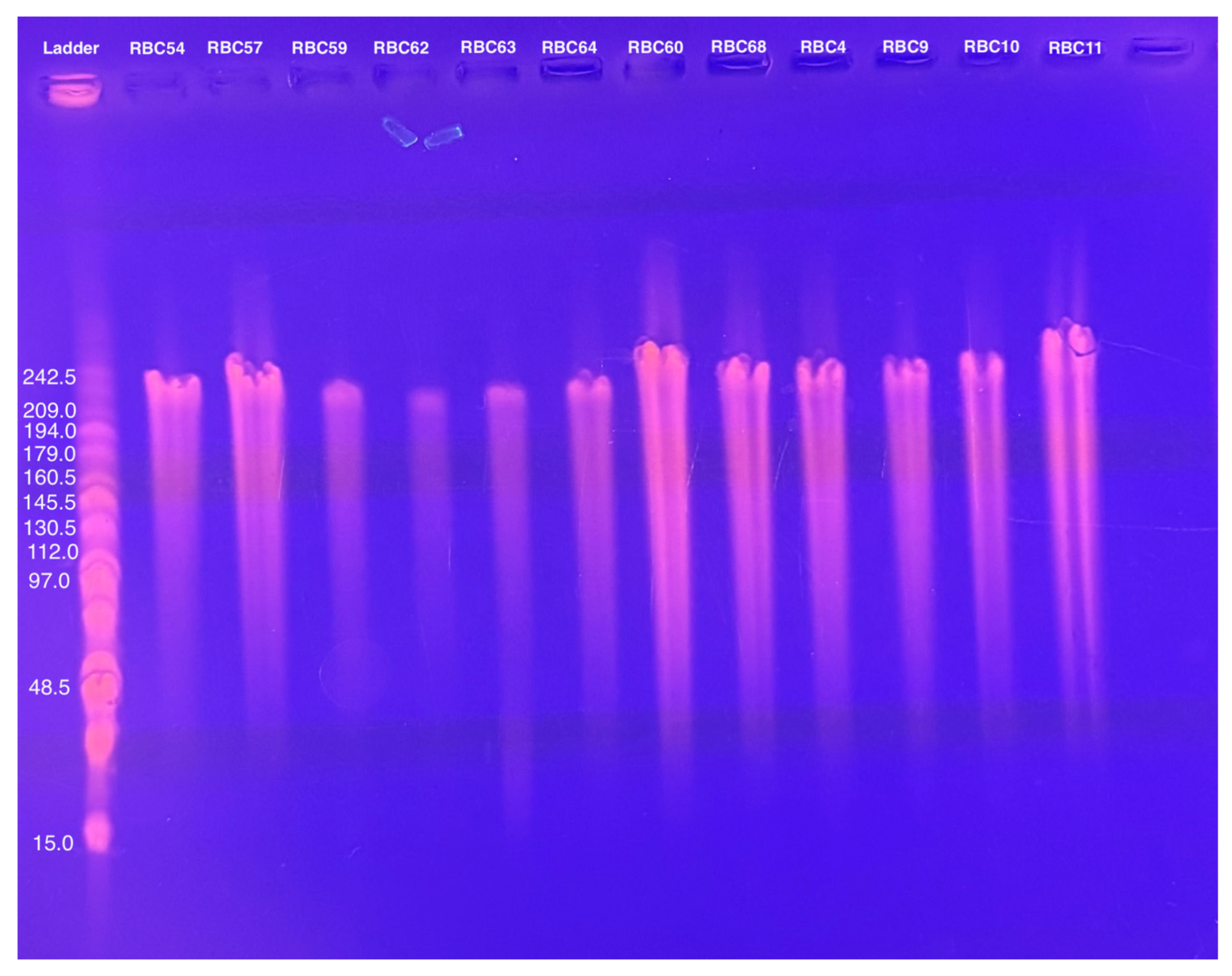
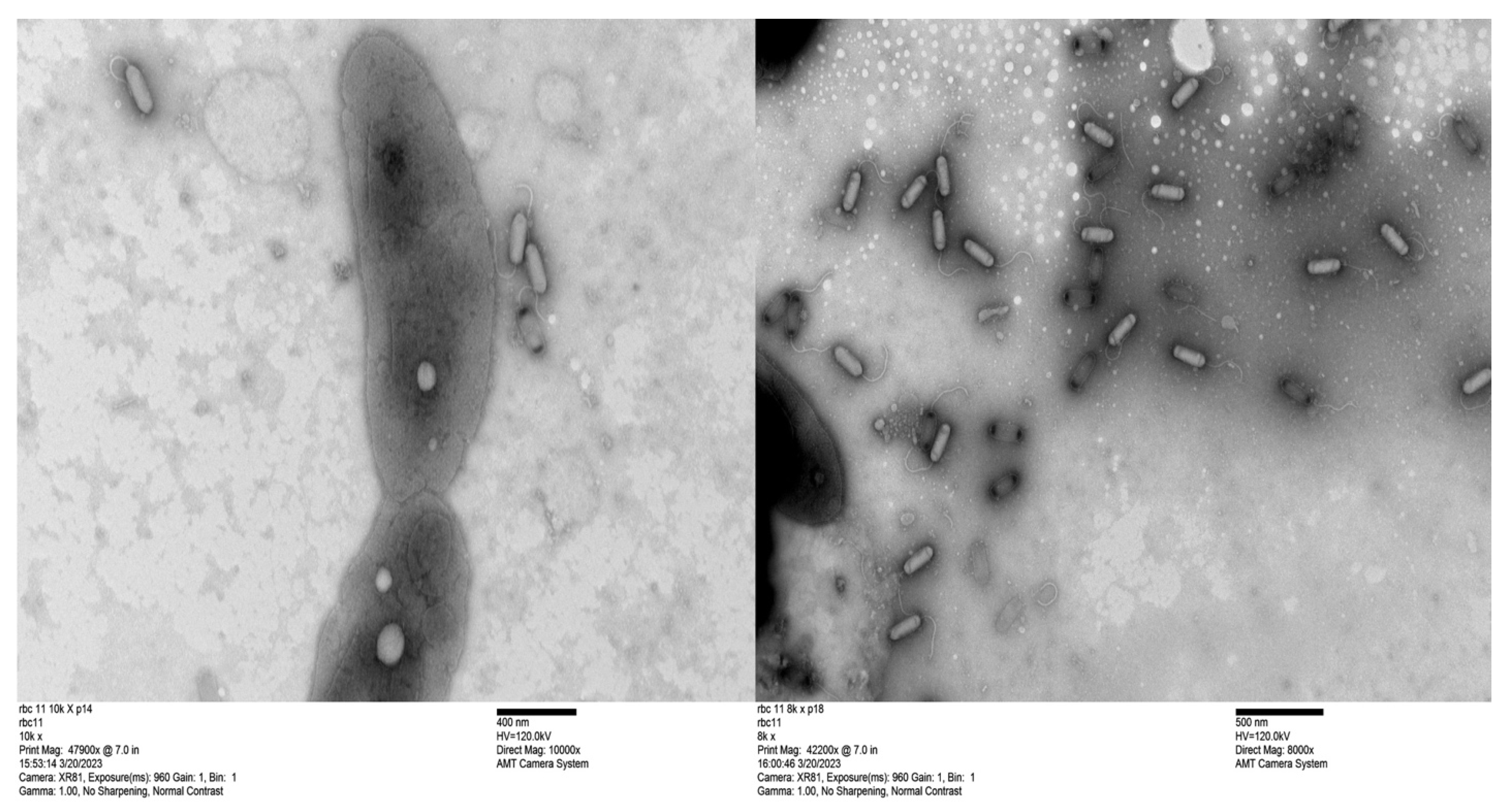
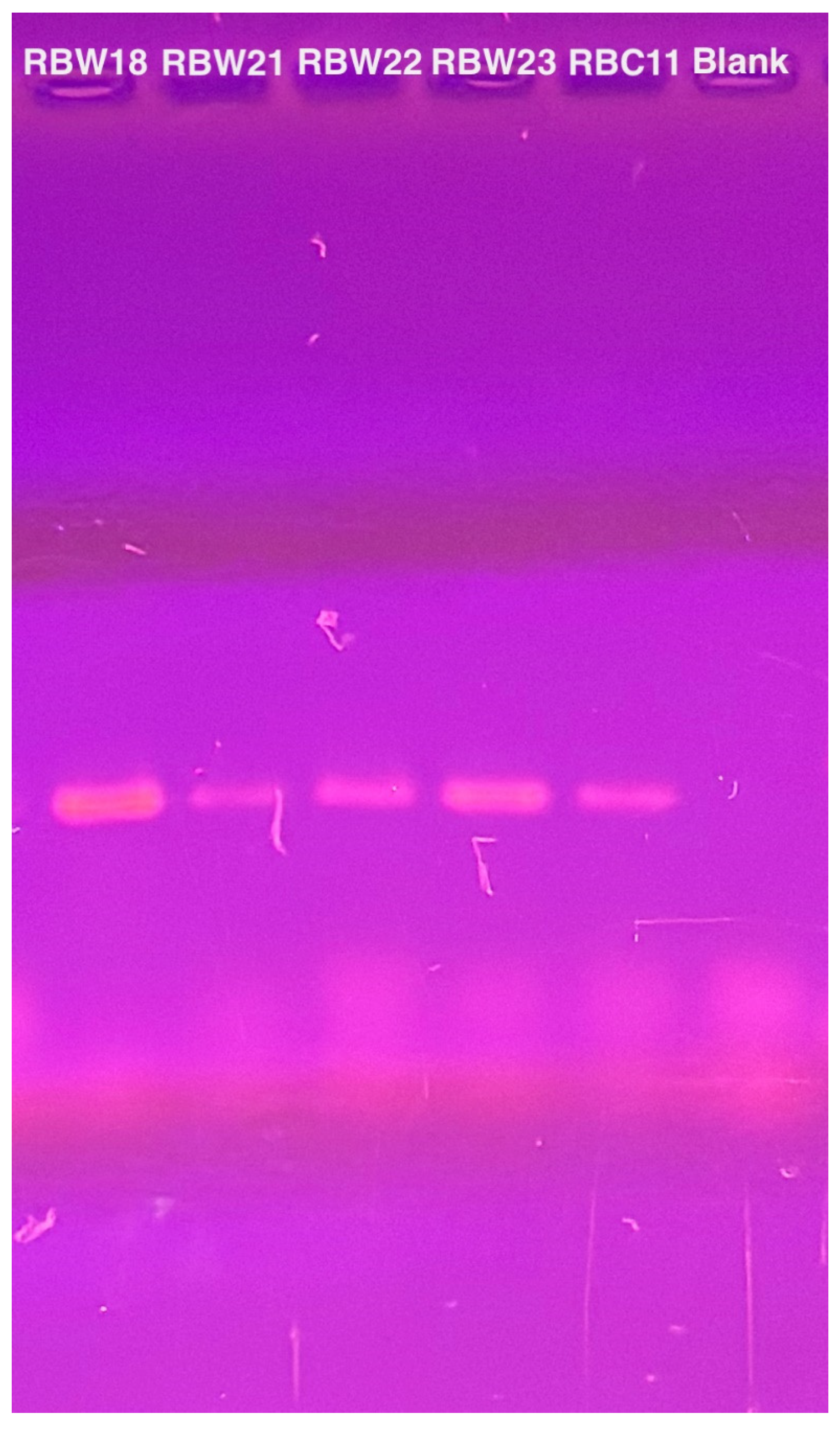
| Bacteriophage Name | RBC50 | RBC67 | RBC68 | RBC69 | RBC71 | RBC73 |
|---|---|---|---|---|---|---|
| Original Source | RBW1 Sphingomonas frigidaeris | RBW6 pseudomonas | RBW26 Caulobacter sp. | RBW29 Caulobacter vibrioides | RBW18 Brevundimonas | RBW25 Caulobacter rhizosphaerae |
| New host strain | RBW1 | RBW21 | SC1004 | CB13 | CBR1 | CBR1 |
| Plaque Morphology | Clear | Clear | Clear | Clear | Clear | Clear |
| Bacteriophage Name | RBC4 | RBC7 | RBC8 | RBC9 | RBC10 | RBC11 | RBC12 | RBC15 | RBC17 | RBC18 | RBC22 | RBC29 | RBC31 | RBC32 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date collected | 10/21 | 12/21 | 12/21 | 1/22 | 1/22 | 1/22 | 1/22 | 1/22 | 3/22 | 3/22 | 3/22 | 3/22 | 3/22 | 4/22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi, T.; Ely, B. Dolichocephalovirinae Phages Exist as Episomal Pseudolysogens Across Diverse Soil Bacteria. Microorganisms 2025, 13, 1239. https://doi.org/10.3390/microorganisms13061239
Mohammadi T, Ely B. Dolichocephalovirinae Phages Exist as Episomal Pseudolysogens Across Diverse Soil Bacteria. Microorganisms. 2025; 13(6):1239. https://doi.org/10.3390/microorganisms13061239
Chicago/Turabian StyleMohammadi, Tannaz, and Bert Ely. 2025. "Dolichocephalovirinae Phages Exist as Episomal Pseudolysogens Across Diverse Soil Bacteria" Microorganisms 13, no. 6: 1239. https://doi.org/10.3390/microorganisms13061239
APA StyleMohammadi, T., & Ely, B. (2025). Dolichocephalovirinae Phages Exist as Episomal Pseudolysogens Across Diverse Soil Bacteria. Microorganisms, 13(6), 1239. https://doi.org/10.3390/microorganisms13061239







