Population Genomics, Virulence Traits, and Antimicrobial Resistance of Streptococcus suis Isolated in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Streptococcus suis Isolate Collection
2.2. Phylogenetic Analysis
2.3. MLST Analysis and Molecular Serotype Prediction
2.4. Identification of Virulence Factors (VFs) and Antimicrobial Resistance Genes (ARGs)
2.5. Annotation and Visualization of Bacterial Integrative and Conjugative Elements (ICEs)
2.6. Antimicrobial Susceptibility Testing
2.7. Mouse Model of Infection
2.8. Statistical Analysis
3. Results
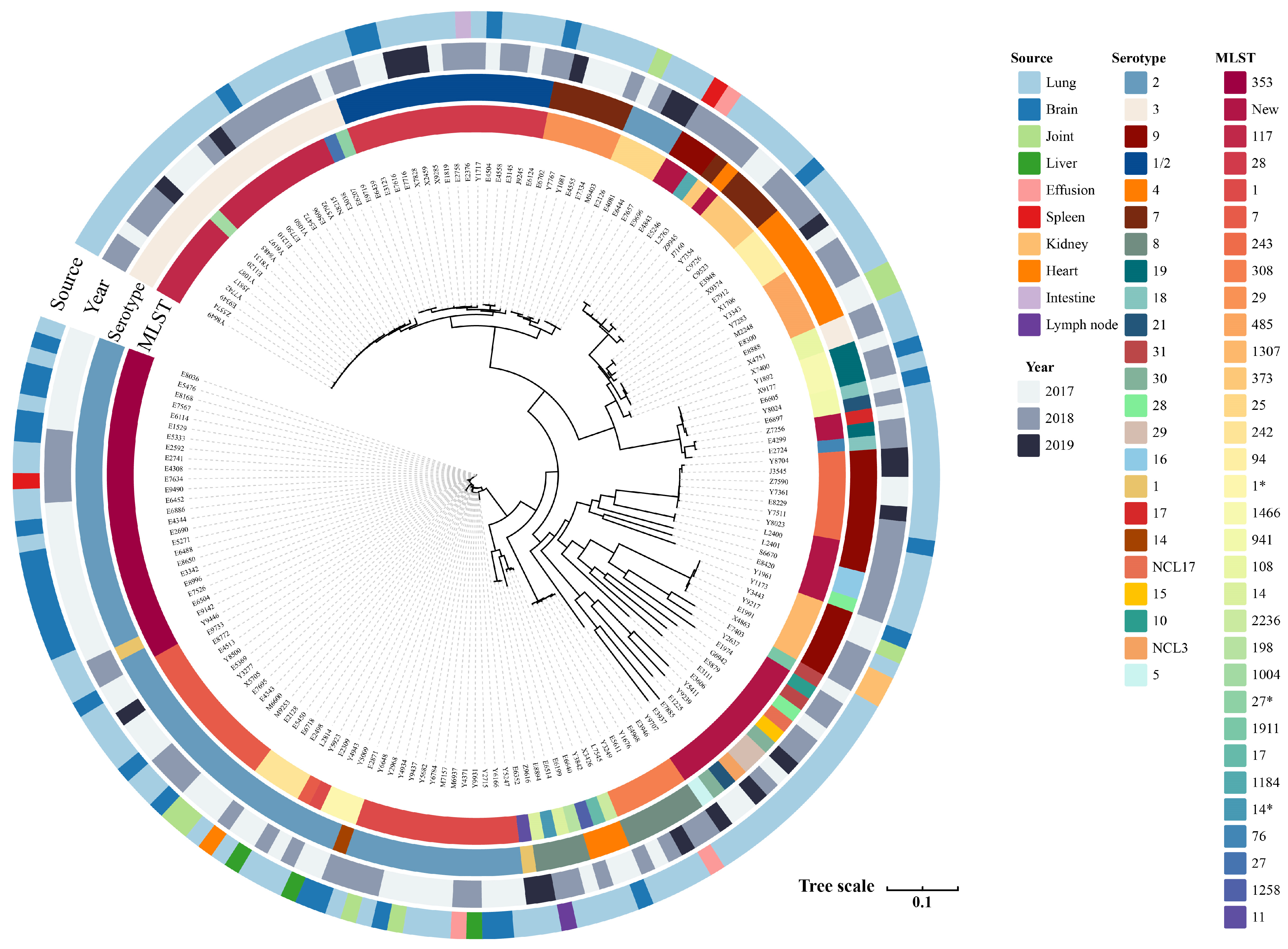
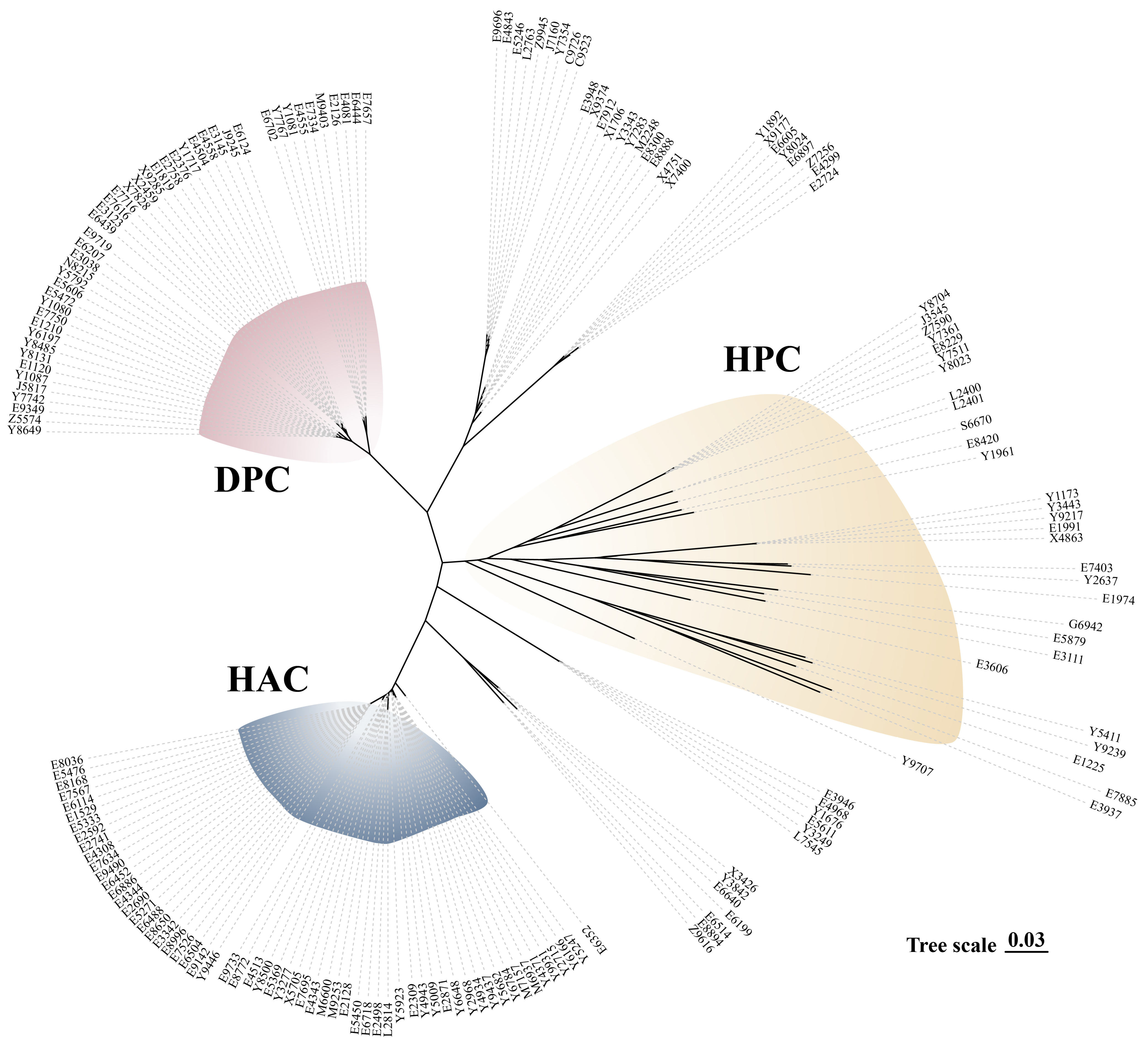
3.1. Serotype Distribution, MLST Identification, and Differential Clades
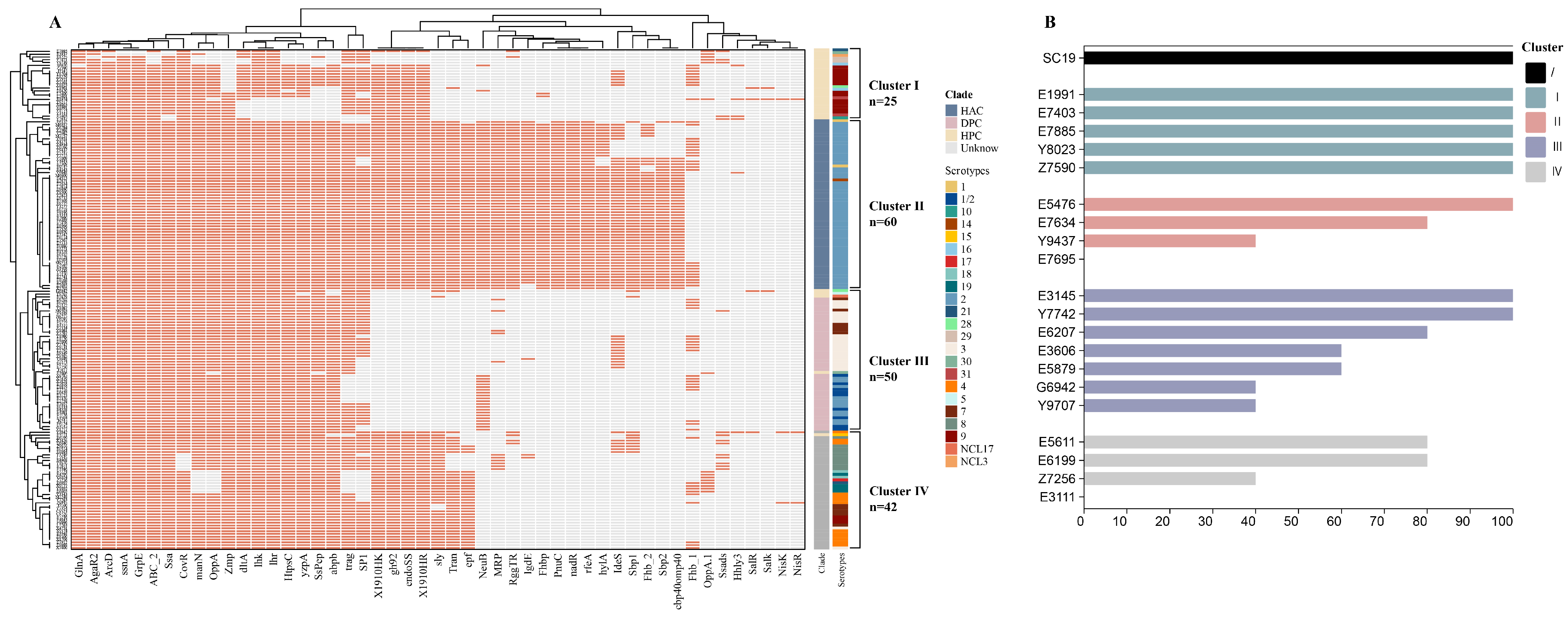
3.2. Analysis of Virulence-Related Genes and Virulence Phenotypes
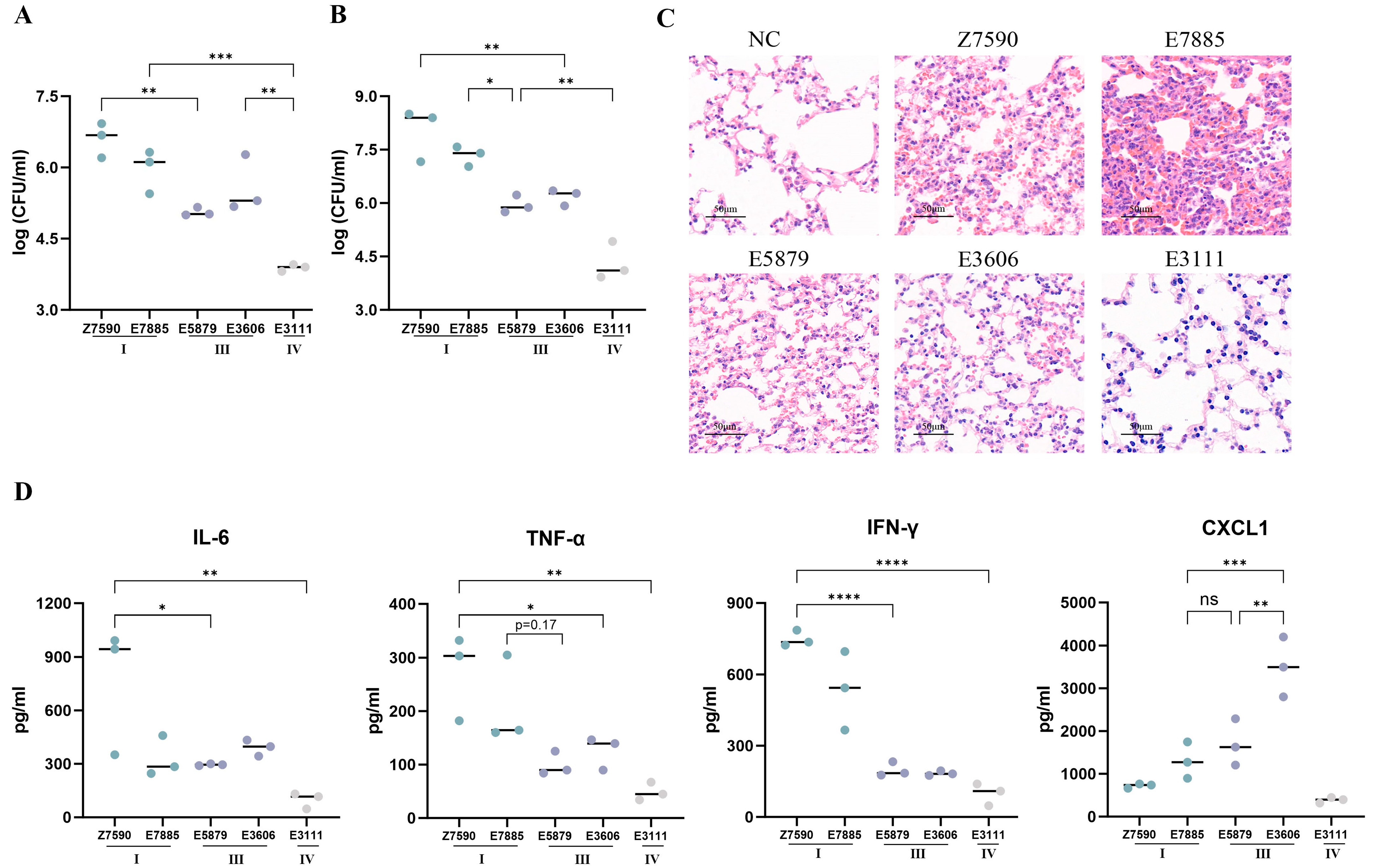
3.3. Mouse Model of Infection
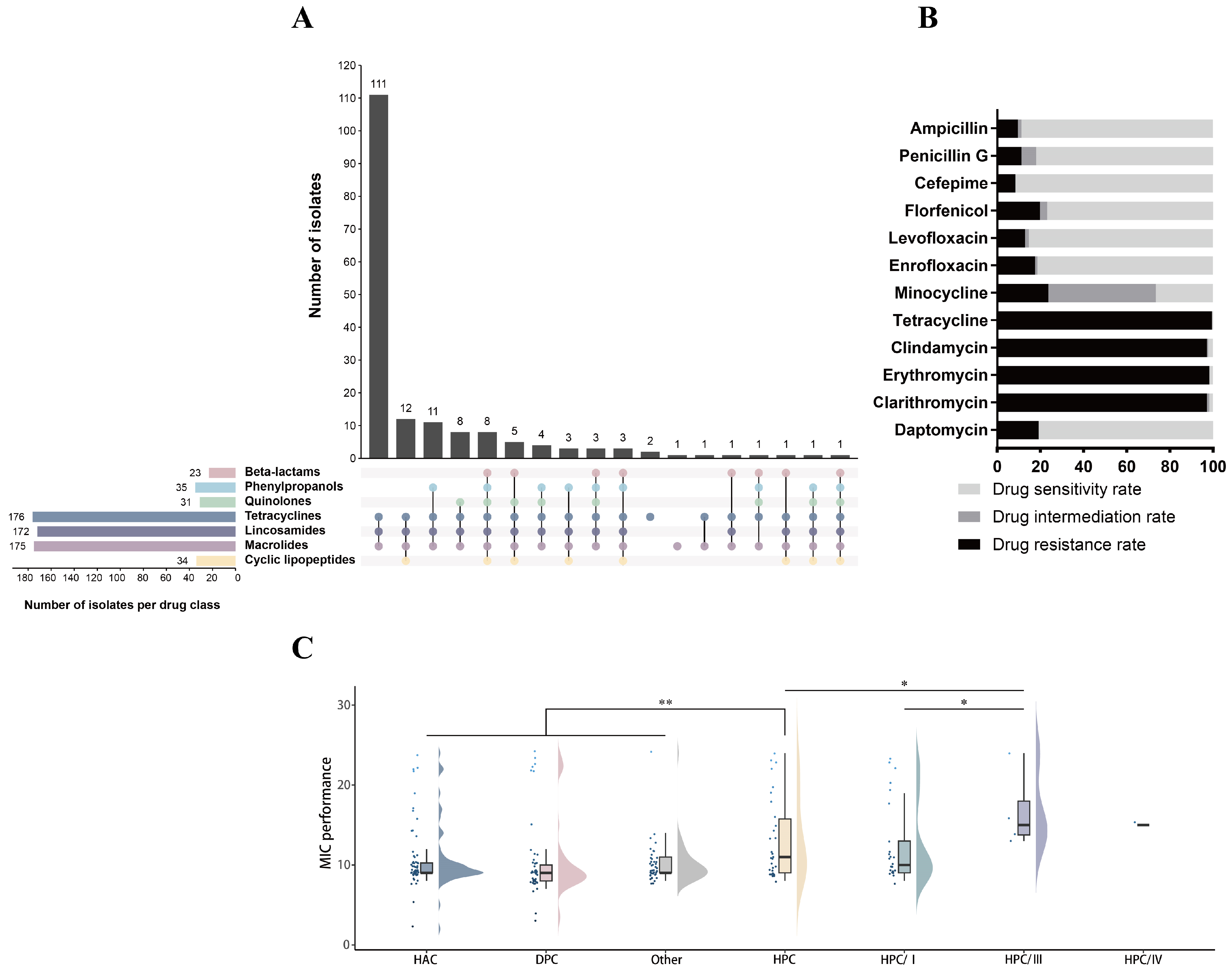
3.4. Antibiotic Resistance Phenotypes and Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bojarska, A.; Molska, E.; Janas, K.; Skoczyńska, A.; Stefaniuk, E.; Hryniewicz, W.; Sadowy, E. Streptococcus suis in invasive human infections in Poland: Clonality and determinants of virulence and antimicrobial resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Zajac, V.; Sroka, J.; Wasinski, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wójcik-Fatla, A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II–Pathogenesis. Ann. Agric. Environ. Med. 2018, 25, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, M.L.; De Greeff, A.; van Rooijen, W.J.; Stockhofe-Zurwieden, N.; Nielsen, J.; Wichgers Schreur, P.J.; Pannekoek, Y.; Heuvelink, A.; Van Der Ende, A.; Smith, H. Host-pathogen interaction at the intestinal mucosa correlates with zoonotic potential of Streptococcus suis. J. Infect. Dis. 2015, 212, 95–105. [Google Scholar] [CrossRef]
- Dong, X.; Chao, Y.; Zhou, Y.; Zhou, R.; Zhang, W.; Fischetti, V.A.; Wang, X.; Feng, Y.; Li, J. The global emergence of a novel Streptococcus suis clade associated with human infections. EMBO Mol. Med. 2021, 13, e13810. [Google Scholar] [CrossRef]
- Goyette-Desjardins, G.; Auger, J.-P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—An update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, 1–20. [Google Scholar] [CrossRef]
- Libante, V.; Nombre, Y.; Coluzzi, C.; Staub, J.; Guédon, G.; Gottschalk, M.; Teatero, S.; Fittipaldi, N.; Leblond-Bourget, N.; Payot, S. Chromosomal conjugative and mobilizable elements in Streptococcus suis: Major actors in the spreading of antimicrobial resistance and bacteriocin synthesis genes. Pathogens 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Arndt, E.R.; Farzan, A.; MacInnes, J.I.; Friendship, R.M. Antimicrobial resistance of Streptococcus suis isolates recovered from clinically ill nursery pigs and from healthy pigs at different stages of production. Can. Vet. J. 2019, 60, 519–522. [Google Scholar]
- Uruén, C.; García, C.; Fraile, L.; Tommassen, J.; Arenas, J. How Streptococcus suis escapes antibiotic treatments. Vet. Res. 2022, 53, 91. [Google Scholar] [CrossRef]
- Huang, J.; Ma, J.; Shang, K.; Hu, X.; Liang, Y.; Li, D.; Wu, Z.; Dai, L.; Chen, L.; Wang, L. Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: A probable mobile genetic elements reservoir for other streptococci. Front. Cell. Infect. Microbiol. 2016, 6, 118. [Google Scholar] [CrossRef]
- Huang, J.; Liang, Y.; Guo, D.; Shang, K.; Ge, L.; Kashif, J.; Wang, L. Comparative genomic analysis of the ICE Sa 2603 family ICEs and spread of erm (B)-and tet (O)-carrying transferable 89K-subtype ICEs in swine and bovine isolates in China. Front. Microbiol. 2016, 7, 55. [Google Scholar] [CrossRef]
- Aradanas, M.; Poljak, Z.; Fittipaldi, N.; Ricker, N.; Farzan, A. Serotypes, virulence-associated factors, and antimicrobial resistance of Streptococcus suis isolates recovered from sick and healthy pigs determined by whole-genome sequencing. Front. Vet. Sci. 2021, 8, 742345. [Google Scholar] [CrossRef]
- Zou, G.; Zhou, J.; Xiao, R.; Zhang, L.; Cheng, Y.; Jin, H.; Li, L.; Zhang, L.; Wu, B.; Qian, P. Effects of environmental and management-associated factors on prevalence and diversity of Streptococcus suis in clinically healthy pig herds in China and the United Kingdom. Appl. Environ. Microbiol. 2018, 84, e02590-17. [Google Scholar] [CrossRef] [PubMed]
- Uruen, C.; Gimeno, J.; Sanz, M.; Fraile, L.; Marín, C.M.; Arenas, J. Invasive Streptococcus suis isolated in Spain contain a highly promiscuous and dynamic resistome. Front. Cell. Infect. Microbiol. 2024, 13, 1329632. [Google Scholar] [CrossRef]
- King, S.J.; Leigh, J.A.; Heath, P.J.; Luque, I.; Tarradas, C.; Dowson, C.G.; Whatmore, A.M. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: Identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 2002, 40, 3671–3680. [Google Scholar] [CrossRef] [PubMed]
- Callejo, R.; Zheng, H.; Du, P.; Prieto, M.; Xu, J.; Zielinski, G.; Auger, J.-P.; Gottschalk, M. Streptococcus suis serotype 2 strains isolated in Argentina (South America) are different from those recovered in North America and present a higher risk for humans. JMM Case Rep. 2016, 3, e005066. [Google Scholar] [CrossRef]
- Fittipaldi, N.; Xu, J.; Lacouture, S.; Tharavichitkul, P.; Osaki, M.; Sekizaki, T.; Takamatsu, D.; Gottschalk, M. Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. Emerg. Infect. Dis. 2011, 17, 2239–2244. [Google Scholar] [CrossRef]
- Estrada, A.A.; Gottschalk, M.; Rossow, S.; Rendahl, A.; Gebhart, C.; Marthaler, D.G. Serotype and genotype (multilocus sequence type) of Streptococcus suis isolates from the United States serve as predictors of pathotype. J. Clin. Microbiol. 2019, 57, e00377-19. [Google Scholar] [CrossRef]
- Huang, J.; Shang, K.; Kashif, J.; Wang, L. Genetic diversity of Streptococcus suis isolated from three pig farms of China obtained by acquiring antibiotic resistance genes. J. Sci. Food Agric. 2015, 95, 1454–1460. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zhu, X.; Jing, H.; Du, H.; Segura, M.; Zheng, H.; Kan, B.; Wang, L.; Bai, X.; Zhou, Y. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg. Infect. Dis. 2006, 12, 1203–1208. [Google Scholar] [CrossRef]
- Okura, M.; Osaki, M.; Nomoto, R.; Arai, S.; Osawa, R.; Sekizaki, T.; Takamatsu, D. Current taxonomical situation of Streptococcus suis. Pathogens 2016, 5, 45. [Google Scholar] [CrossRef]
- Zheng, H.; Ji, S.; Liu, Z.; Lan, R.; Huang, Y.; Bai, X.; Gottschalk, M.; Xu, J. Eight novel capsular polysaccharide synthesis gene loci identified in nontypeable Streptococcus suis isolates. Appl. Environ. Microbiol. 2015, 81, 4111–4119. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Bai, X.; Lan, R.; Zheng, H.; Xu, J. Novel capsular polysaccharide loci and new diagnostic tools for high-throughput capsular gene typing in Streptococcus suis. Appl. Environ. Microbiol. 2016, 82, 7102–7112. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Qiu, X.; Roy, D.; Segura, M.; Du, P.; Xu, J.; Gottschalk, M. Genotyping and investigating capsular polysaccharide synthesis gene loci of non-serotypeable Streptococcus suis isolated from diseased pigs in Canada. Vet. Res. Commun. 2017, 48, 10. [Google Scholar] [CrossRef]
- Huang, J.; Liu, X.; Chen, H.; Chen, L.; Gao, X.; Pan, Z.; Wang, J.; Lu, C.; Yao, H.; Wang, L. Identification of six novel capsular polysaccharide loci (NCL) from Streptococcus suis multidrug resistant non-typeable strains and the pathogenic characteristic of strains carrying new NCL s. Transbound. Emerg. Dis. 2019, 66, 995–1003. [Google Scholar] [CrossRef]
- Segura, M. Streptococcus suis research: Progress and challenges. Pathogens 2020, 9, 707. [Google Scholar] [CrossRef]
- Petrocchi-Rilo, M.; Martínez-Martínez, S.; Aguarón-Turrientes, Á.; Roca-Martínez, E.; García-Iglesias, M.-J.; Pérez-Fernández, E.; González-Fernández, A.; Herencia-Lagunar, E.; Gutiérrez-Martín, C.-B. Anatomical site, typing, virulence gene profiling, antimicrobial susceptibility and resistance genes of Streptococcus suis isolates recovered from pigs in Spain. Antibiotics 2021, 10, 707. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Lacouture, S.; Bonifait, L.; Roy, D.; Fittipaldi, N.; Grenier, D. Characterization of Streptococcus suis isolates recovered between 2008 and 2011 from diseased pigs in Quebec, Canada. Vet. Microbiol. 2013, 162, 819–825. [Google Scholar] [CrossRef]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis virulence factors: Are they all really critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Athey, T.B.; Auger, J.-P.; Teatero, S.; Dumesnil, A.; Takamatsu, D.; Wasserscheid, J.; Dewar, K.; Gottschalk, M.; Fittipaldi, N. Complex population structure and virulence differences among serotype 2 Streptococcus suis strains belonging to sequence type 28. PLoS ONE 2015, 10, e0137760. [Google Scholar] [CrossRef]
- Auger, J.-P.; Fittipaldi, N.; Benoit-Biancamano, M.-O.; Segura, M.; Gottschalk, M. Virulence studies of different sequence types and geographical origins of Streptococcus suis serotype 2 in a mouse model of infection. Pathogens 2016, 5, 48. [Google Scholar] [CrossRef]
- Estrada, A.A.; Gottschalk, M.; Rendahl, A.; Rossow, S.; Marshall-Lund, L.; Marthaler, D.G.; Gebhart, C.J. Proposed virulence-associated genes of Streptococcus suis isolates from the United States serve as predictors of pathogenicity. Porc. Health Manag. 2021, 7, 22. [Google Scholar] [CrossRef]
- Li, Q.; Fu, Y.; Ma, C.; He, Y.; Yu, Y.; Du, D.; Yao, H.; Lu, C.; Zhang, W. The non-conserved region of MRP is involved in the virulence of Streptococcus suis serotype 2. Virulence 2017, 8, 1274–1289. [Google Scholar] [CrossRef]
- Xia, X.; Qin, W.; Zhu, H.; Wang, X.; Jiang, J.; Hu, J. How Streptococcus suis serotype 2 attempts to avoid attack by host immune defenses. Immunol. Infect. 2019, 52, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Lun, S.; Perez-Casal, J.; Connor, W.; Willson, P. Role of suilysin in pathogenesis of Streptococcus suis capsular serotype 2. Microb. Pathog. 2003, 34, 27–37. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Pian, Y.; Ren, Z.; Bi, L.; Yuan, Y.; Zheng, Y.; Jiang, Y.; Wang, F. Increased production of suilysin contributes to invasive infection of the Streptococcus suis strain 05ZYH33. Mol. Med. Rep. 2014, 10, 2819–2826. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.E.; Vecht, U.; Wisselink, H.J.; Stockhofe-Zurwieden, N.; Biermann, Y.; Smits, M.A. Mutants of Streptococcus suis types 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect. Immun. 1996, 64, 4409–4412. [Google Scholar] [CrossRef]
- O’Connor, T.J.; Boyd, D.; Dorer, M.S.; Isberg, R.R. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 2012, 338, 1440–1444. [Google Scholar] [CrossRef]
- Wang, J.; Liang, P.; Sun, H.; Wu, Z.; Gottschalk, M.; Qi, K.; Zheng, H. Comparative transcriptomic analysis reveal genes involved in the pathogenicity increase of Streptococcus suis epidemic strains. Virulence 2022, 13, 1455–1470. [Google Scholar] [CrossRef]
- Roodsant, T.J.; Van Der Putten, B.C.; Tamminga, S.M.; Schultsz, C.; Van Der Ark, K.C. Identification of Streptococcus suis putative zoonotic virulence factors: A systematic review and genomic meta-analysis. Virulence 2021, 12, 2787–2797. [Google Scholar] [CrossRef]
- Segura, M.; Calzas, C.; Grenier, D.; Gottschalk, M. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: Fighting against nonspecific defenses. FEBS Lett. 2016, 590, 3772–3799. [Google Scholar] [CrossRef]
- Arenas, J.; Zomer, A.; Harders-Westerveen, J.; Bootsma, H.J.; De Jonge, M.I.; Stockhofe-Zurwieden, N.; Smith, H.E.; De Greeff, A. Identification of conditionally essential genes for Streptococcus suis infection in pigs. Virulence 2020, 11, 446–464. [Google Scholar] [CrossRef] [PubMed]
- Athey, T.B.; Teatero, S.; Lacouture, S.; Takamatsu, D.; Gottschalk, M.; Fittipaldi, N. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol. 2016, 16, 162. [Google Scholar] [CrossRef]
- Lun, Z.-R.; Wang, Q.-P.; Chen, X.-G.; Li, A.-X.; Zhu, X.-Q. Streptococcus suis: An emerging zoonotic pathogen. Lancet Infect. Dis. 2007, 7, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Prüfer, T.L.; Rohde, J.; Verspohl, J.; Rohde, M.; De Greeff, A.; Willenborg, J.; Valentin-Weigand, P. Molecular typing of Streptococcus suis strains isolated from diseased and healthy pigs between 1996–2016. PLoS ONE 2019, 14, e0210801. [Google Scholar] [CrossRef]
- Scherrer, S.; Rosato, G.; Spoerry Serrano, N.; Stevens, M.J.; Rademacher, F.; Schrenzel, J.; Gottschalk, M.; Stephan, R.; Peterhans, S. Population structure, genetic diversity and pathotypes of Streptococcus suis isolated during the last 13 years from diseased pigs in Switzerland. Vet. Res. 2020, 51, 85. [Google Scholar] [CrossRef]
- Dong, W.; Zhu, Y.; Ma, Y.; Ma, J.; Zhang, Y.; Yuan, L.; Pan, Z.; Wu, Z.; Yao, H. Multilocus sequence typing and virulence genotyping of Streptococcus suis serotype 9 isolates revealed high genetic and virulence diversity. FEMS Microbiol. Lett. 2017, 364, fnx192. [Google Scholar] [CrossRef] [PubMed]
- Blume, V.; Luque, I.; Borge, C.; Maldonado, A.; Domínguez Rodríquez, L.; Tarradas, C.; Fernández-Garayzábal Fernández, J.F.; Vela Alonso, A.I. Genetic and virulence-phenotype characterization of serotypes 2 and 9 of Streptococcus suis swine isolates. Int. Microbiol. 2009, 12, 161–166. [Google Scholar]
- Allen, A.G.; Bolitho, S.; Lindsay, H.; Khan, S.; Bryant, C.; Norton, P.; Ward, P.; Leigh, J.; Morgan, J.; Riches, H. Generation and characterization of a defined mutant of Streptococcus suis lacking suilysin. Infect. Immun. 2001, 69, 2732–2735. [Google Scholar] [CrossRef]
- Takeuchi, D.; Akeda, Y.; Nakayama, T.; Kerdsin, A.; Sano, Y.; Kanda, T.; Hamada, S.; Dejsirilert, S.; Oishi, K. The contribution of suilysin to the pathogenesis of Streptococcus suis meningitis. J. Infect. Dis. 2014, 209, 1509–1519. [Google Scholar] [CrossRef]
- Seitz, M.; Beineke, A.; Singpiel, A.; Willenborg, J.; Dutow, P.; Goethe, R.; Valentin-Weigand, P.; Klos, A.; Baums, C. Role of capsule and suilysin in mucosal infection of complement-deficient mice with Streptococcus suis. Infect. Immun. 2014, 82, 2460–2471. [Google Scholar] [CrossRef]
- Baums, C.G.; Valentin-Weigand, P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim. Health Res. Rev. 2009, 10, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Weinert, L.A.; Chaudhuri, R.R.; Wang, J.; Peters, S.E.; Corander, J.; Jombart, T.; Baig, A.; Howell, K.J.; Vehkala, M.; Välimäki, N. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat. Commun. 2015, 6, 6740. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Tan, C.; Liu, Z.; Yang, K.; Zhou, D.; Liu, W.; Duan, Z.; Guo, R.; Chen, H.; Tian, Y. The 1910HK/RR two-component system is essential for the virulence of Streptococcus suis serotype 2. Microb. Pathog. 2017, 104, 137–145. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, Z.; Pan, Z.; Bai, Q.; Zhong, X.; Zhu, Y.; Zhang, Y.; Wu, Z.; Liu, G.; Yao, H. Streptococcus suis uptakes carbohydrate source from host glycoproteins by N-glycans degradation system for optimal survival and full virulence during infection. Pathogens 2020, 9, 387. [Google Scholar] [CrossRef]
- Merhej, V.; Georgiades, K.; Raoult, D. Postgenomic analysis of bacterial pathogens repertoire reveals genome reduction rather than virulence factors. Brief. Funct. Genom. 2013, 12, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Weiss, D.S.; Newton, K.; McBride, J.; O’Rourke, K.; Roose-Girma, M.; Lee, W.P.; Weinrauch, Y.; Monack, D.M.; Dixit, V.M. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006, 440, 228–232. [Google Scholar] [CrossRef]
- Jacobi, J. Pathophysiology of sepsis. Am. J. Health-Syst. Pharm. 2002, 59, S3–S8. [Google Scholar] [CrossRef]
- Nedeva, C.; Menassa, J.; Puthalakath, H. Sepsis: Inflammation is a necessary evil. Front. Cell Dev. Biol. 2019, 7, 108. [Google Scholar] [CrossRef]
- Hackert, N.S.; Radtke, F.A.; Exner, T.; Lorenz, H.-M.; Müller-Tidow, C.; Nigrovic, P.A.; Wabnitz, G.; Grieshaber-Bouyer, R. Human and mouse neutrophils share core transcriptional programs in both homeostatic and inflamed contexts. Nat. Commun. 2023, 14, 8133. [Google Scholar] [CrossRef]
- Lin, L.; Xu, L.; Lv, W.; Han, L.; Xiang, Y.; Fu, L.; Jin, M.; Zhou, R.; Chen, H.; Zhang, A. An NLRP3 inflammasome-triggered cytokine storm contributes to Streptococcal toxic shock-like syndrome (STSLS). PLoS Pathog. 2019, 15, e1007795. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, H.; Bian, C.; Chen, H.; Shen, Y.; Gao, X.; Ma, J.; Yao, H.; Wang, L.; Wu, Z. The antimicrobial systems of Streptococcus suis promote niche competition in pig tonsils. Virulence 2022, 13, 781–793. [Google Scholar] [CrossRef] [PubMed]
- da Silva, G.J.; Mendonça, N. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 2012, 3, 18–28. [Google Scholar] [CrossRef] [PubMed]
- MOA. Announcement of the Ministry of Agriculture and Rural People’s Republic of China No. 193. 2002. Available online: http://www.moa.gov.cn/ztzl/ncpzxzz/flfg/200709/t20070919_893091.htm (accessed on 1 September 2023).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Ma, B.; Jia, X.; Wan, Y.; Ni, S.; Chen, G.; Zong, X.; Jin, H.; Li, J.; Tan, C. Population Genomics, Virulence Traits, and Antimicrobial Resistance of Streptococcus suis Isolated in China. Microorganisms 2025, 13, 1197. https://doi.org/10.3390/microorganisms13061197
Li Y, Ma B, Jia X, Wan Y, Ni S, Chen G, Zong X, Jin H, Li J, Tan C. Population Genomics, Virulence Traits, and Antimicrobial Resistance of Streptococcus suis Isolated in China. Microorganisms. 2025; 13(6):1197. https://doi.org/10.3390/microorganisms13061197
Chicago/Turabian StyleLi, Yuying, Bin Ma, Xue Jia, Yanxi Wan, Shiting Ni, Guosheng Chen, Xin Zong, Hui Jin, Jinquan Li, and Chen Tan. 2025. "Population Genomics, Virulence Traits, and Antimicrobial Resistance of Streptococcus suis Isolated in China" Microorganisms 13, no. 6: 1197. https://doi.org/10.3390/microorganisms13061197
APA StyleLi, Y., Ma, B., Jia, X., Wan, Y., Ni, S., Chen, G., Zong, X., Jin, H., Li, J., & Tan, C. (2025). Population Genomics, Virulence Traits, and Antimicrobial Resistance of Streptococcus suis Isolated in China. Microorganisms, 13(6), 1197. https://doi.org/10.3390/microorganisms13061197






