Establishment of a Sensitive and Visual Detection Platform for Viable Salmonella in Wastewater That Combines Propidium Monoazide with Recombinase Polymerase Amplification—CRISPR/Cas12a System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Viable and Inactivated Cells
2.2. Wastewater Samples
2.3. PMA Condition Optimization
2.4. DNA Extraction, PCR and qPCR
2.5. RPA Primer Design and Optimization
2.6. Cas12a-Mediated Cleavage Analysis of the CRISPR/Cas12a System
2.7. Specificity and Sensitivity of the RPA-CRISPR/Cas12a System in Wastewater
2.7.1. Specificity Testing
2.7.2. Sensitivity Testing
2.8. Identification of Viable Salmonella in Inoculated Wastewater and Real-World Wastewater with the PMA-RPA-CRISPR/Cas12a System
2.9. Statistical Analysis
3. Results and Discussion
3.1. Optimization of PMA Treatment Concentration and Exposure Time
3.2. Development of RPA-CRISPR/Cas12a System
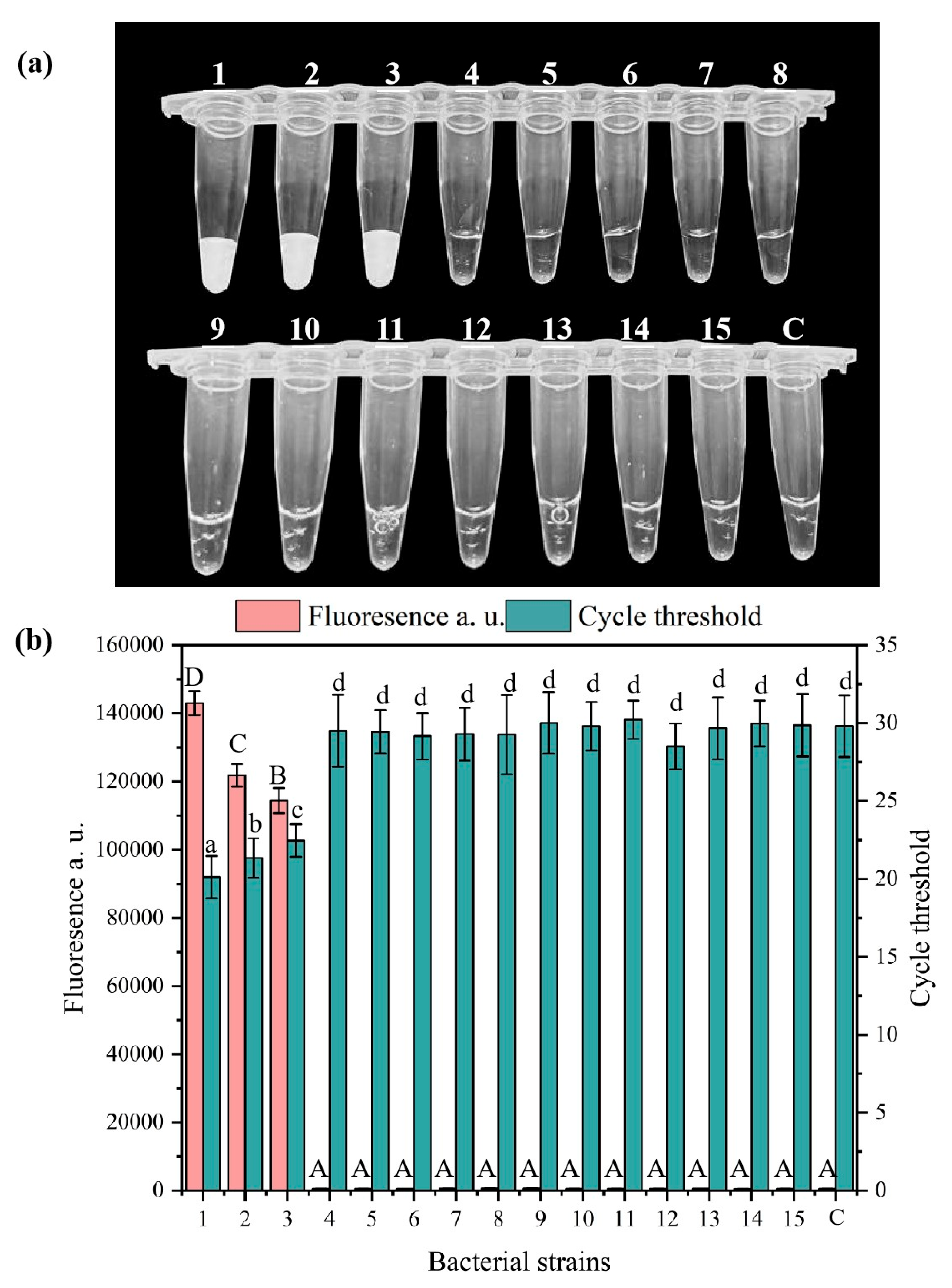
3.3. Specificity and Sensitivity Analysis of the RPA-CRISPR/Cas12a System
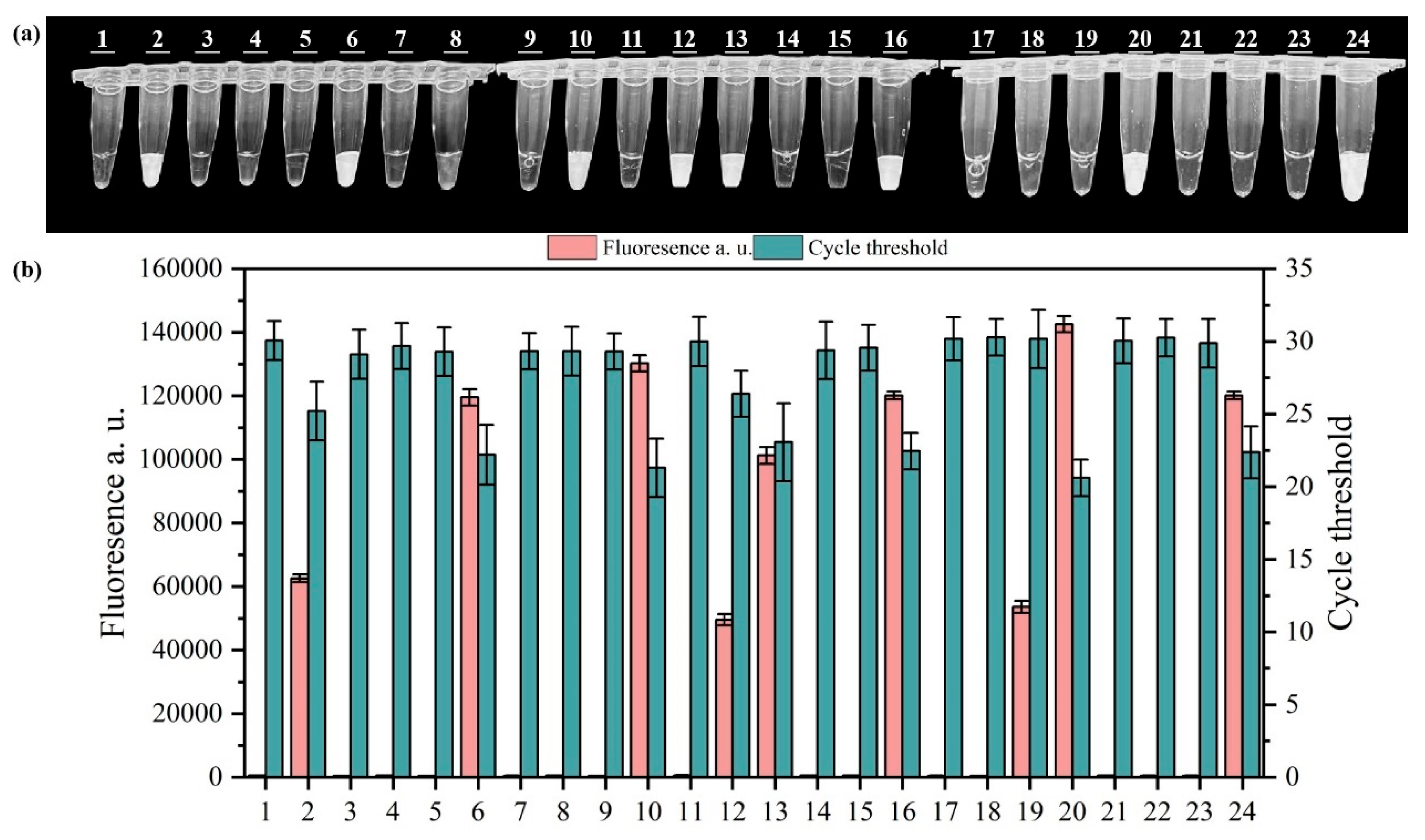
3.4. Detection of Salmonella in Wastewater Using the PMA-RPA-CRISPR/Cas12a System
4. Conclusions
5. Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, Y.; Li, J.; Zhu, L.; Jiang, L. Cooperation and competition between CRISPR- and omics-based technologies in foodborne pathogens detection: A state of the art review. Curr. Opin. Food Sci. 2022, 44, 100813. [Google Scholar] [CrossRef]
- Rabsch, W.; Tschäpe, H.; Bäumler, A.J. Non-typhoidal salmonellosis: Emerging problems. Microbes Infect. 2001, 3, 237–247. [Google Scholar] [CrossRef]
- Winfield, M.D.; Groisman, E.A. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 2003, 69, 3687–3694. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, G.; Li, X.; Xu, Z.; Lei, H.; Shen, X. Development of rapid and easy detection of Salmonella in food matrics using RPA-CRISPR/Cas12a method. LWT 2022, 162, 113443. [Google Scholar] [CrossRef]
- Zhao, Q.; Lu, D.; Zhang, G.; Zhang, D.; Shi, X. Recent improvements in enzyme-linked immunosorbent assays based on nanomaterials. Talanta 2020, 223, 121722. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Salazar, J.K. Culture-Independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food Sci. Food Saf. 2015, 15, 183–205. [Google Scholar] [CrossRef]
- Luo, X.; Wang, K.; Xue, Y.; Cao, X.; Zhou, J.; Wang, J. Digital PCR-free technologies for absolute quantitation of nucleic acids at single-molecule level. Chin. Chem. Lett. 2024, 36, 109924. [Google Scholar] [CrossRef]
- Salvo, P.; Vivaldi, F.M.; Bonini, A.; Biagini, D.; Bellagambi, F.G.; Miliani, F.M.; Francesco, F.D.; Lomonaco, T. Biosensors for Detecting Lymphocytes and Immunoglobulins. Biosensors 2020, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yang, S.; Li, S.; Liu, X.; Liu, Y.; Chen, Z.; Chen, H.; Li, S.; He, N.; Cui, H.; et al. Viral aptamer screening and aptamer-based biosensors for virus detection: A review. Int. J. Biol. Macromol. 2024, 276, 133935. [Google Scholar] [CrossRef]
- Kucherenko, I.S.; Soldatkin, O.O.; Dzyadevych, S.V.; Soldatkin, A.P. Electrochemical biosensors based on multienzyme systems: Main groups, advantages and limitations—A review. Anal. Chim. Acta 2020, 1111, 114–131. [Google Scholar] [CrossRef]
- Wang, J.; Davidson, J.L.; Kaur, S.; Dextre, A.A.; Ranjbaran, M.; Kamel, M.S.; Athalye, S.M.; Verma, M.S. Paper-Based Biosensors for the Detection of Nucleic Acids from Pathogens. Biosensors 2022, 12, 1094. [Google Scholar] [CrossRef]
- Purcarea, C.; Ruginescu, R.; Banciu, R.M.; Vasilescu, A. Extremozyme-Based Biosensors for Environmental Pollution Monitoring: Recent Developments. Biosensors 2024, 14, 143. [Google Scholar] [CrossRef] [PubMed]
- Jarque, S.; Bittner, M.; Blaha, L.; Hilscherova, K. Yeast Biosensors for Detection of Environmental Pollutants: Current State and Limitations. Trends Biotechnol. 2016, 34, 408–419. [Google Scholar] [CrossRef]
- Panferov, V.G.; Liu, J. Optical and Catalytic Properties of Nanozymes for Colorimetric Biosensors: Advantages, Limitations, and Perspectives. Adv. Opt. Mater. 2024, 12, 2401318. [Google Scholar] [CrossRef]
- Ngoc, L.T.N.; Lee, Y.-C. Current Trends in RNA Virus Detection via Nucleic Acid Isothermal Amplification-Based Platforms. Biosensors 2024, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Zhao, X. Isothermal Amplification Technologies for the Detection of Foodborne Pathogens. Food Anal. Methods 2018, 11, 1543–1560. [Google Scholar] [CrossRef]
- Padzil, F.; Mariatulqabtiah, A.R.; Tan, W.S.; Ho, K.L.; Isa, N.M.; Lau, H.Y.; Abu, J.; Chuang, K.-P. Loop-Mediated Isothermal Amplification (LAMP) as a Promising Point-of-Care Diagnostic Strategy in Avian Virus Research. Animals 2021, 12, 76. [Google Scholar] [CrossRef]
- Yan, S.; Li, C.; Lan, H.; Pan, D.; Wu, Y. Comparison of four isothermal amplification techniques: LAMP, SEA, CPA, and RPA for the identification of chicken adulteration. Food Control 2024, 159, 110302. [Google Scholar] [CrossRef]
- Ibler, A.E.M.; ElGhazaly, M.; Naylor, K.L.; Bulgakova, N.A.; F El-Khamisy, S.; Humphreys, D. Typhoid toxin exhausts the RPA response to DNA replication stress driving senescence and Salmonella infection. Nat. Commun. 2019, 10, 4040. [Google Scholar] [CrossRef]
- Kubo, S.; Niimi, H.; Kitajima, I. Improved reverse transcription-recombinase polymerase amplification assay for blood mRNA screening: Comparison with one-step RT-qPCR assay. Forensic Sci. Int. Genet. 2022, 63, 102808. [Google Scholar] [CrossRef]
- Wang, S.-W.; Gao, C.; Zheng, Y.-M.; Yi, L.; Lu, J.-C.; Huang, X.-Y.; Cai, J.-B.; Zhang, P.-F.; Cui, Y.-H.; Ke, A.-W. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol. Cancer 2022, 21, 57. [Google Scholar] [CrossRef]
- Liang, P.; Lv, B.; Chen, K.; Li, D. Sensitive aptasensing of ATP based on a PAM site-regulated CRISPR/Cas12a activation. Microchim. Acta 2024, 191, 386. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Bu, S.; Wang, C.; Wang, J.; Gao, Y.; Xu, M.; Zhang, M.; Feng, X.; Li, C.; Wan, J. A Novel CRISPR/Cas13a Biosensing Platform Comprising Dual Hairpin Probe and Traditional Lateral Flow Assays. Sens. Actuators B Chem. 2024, 423, 136752. [Google Scholar] [CrossRef]
- Zhou, B.; Yang, R.; Sohail, M.; Kong, X.; Zhang, X.; Fu, N.; Li, B. CRISPR/Cas14 provides a promising platform in facile and versatile aptasensing with improved sensitivity. Talanta 2022, 254, 124120. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, S.; Pei, X.; Li, S.; He, Y.; Tong, Y.; Liu, G. Fluorescence Signal-Readout of CRISPR/Cas Biosensors for Nucleic Acid Detection. Biosensors 2022, 12, 779. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M. Revolutionizing Point-of-Care Diagnostics via CRISPR Systems. ACS Synth. Biol. 2024, 13, 411–412. [Google Scholar] [CrossRef]
- Paul, B.; Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020, 43, 8–17. [Google Scholar] [CrossRef]
- Bae, J.-Y.; Lee, S.-y.; Oh, S.-W. Development of filtration-based RPA–CRISPR/Cas12a system for rapid, sensitive and visualized detection of Salmonella in ready-to-eat salads. Microchem. J. 2024, 206, 111527. [Google Scholar] [CrossRef]
- Li, F.; Li, F.; Chen, B.; Zhou, B.; Yu, P.; Yu, S.; Lai, W.; Xu, H. Sextuplex PCR combined with immunomagnetic separation and PMA treatment for rapid detection and specific identification of viable Salmonella spp., Salmonella enterica serovars Paratyphi B, Salmonella Typhimurium, and Salmonella Enteritidis in raw meat. Food Control 2017, 73, 587–594. [Google Scholar] [CrossRef]
- Li, H.-Z.; Yang, K.; Liao, H.; Lassen, S.B.; Su, J.-Q.; Zhang, X.; Cui, L.; Zhu, Y.-G. Active antibiotic resistome in soils unraveled by single-cell isotope probing and targeted metagenomics. Proc. Natl. Acad. Sci. USA 2022, 119, e2201473119. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xiang, P.; Ji, Y.; Chen, Z.; Lei, Z.; Huang, W.; Huang, W.; Liu, D. Response of viable bacteria to antibiotics in aerobic granular sludge: Resistance mechanisms and behaviors, bacterial communities, and driving factors. Water Res. 2023, 245, 120656. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Oh, S.-W. Point-of-Care Diagnostic System for Viable Salmonella Species via Improved Propidium Monoazide and Recombinase Polymerase Amplification Based Nucleic Acid Lateral Flow. Diagnostics 2024, 14, 831. [Google Scholar] [CrossRef]
- Bui, X.T.; Wolff, A.; Madsen, M.; Bang, D.D. Reverse transcriptase real-time PCR for detection and quantification of viable Campylobacter jejuni directly from poultry faecal samples. Res. Microbiol. 2012, 163, 64–72. [Google Scholar] [CrossRef]
- Xue, T.; Lu, Y.; Yang, H.; Hu, X.; Zhang, K.; Ren, Y.; Wu, C.; Xia, X.; Deng, R.; Wang, Y. Isothermal RNA Amplification for the Detection of Viable Pathogenic Bacteria to Estimate the Salmonella Virulence for Causing Enteritis. J. Agric. Food Chem. 2022, 70, 1670–1678. [Google Scholar] [CrossRef]
- van Prooije, T.; Ruigrok, S.; van den Berkmortel, N.; Maas, R.P.P.W.M.; Wijn, S.; van Roon-Mom, W.M.C.; van de Warrenburg, B.; Grutters, J.P.C. The potential value of disease-modifying therapy in patients with spinocerebellar ataxia type 1: An early health economic modeling study. J. Neurol. 2023, 270, 3788–3798. [Google Scholar] [CrossRef]
- Flores-Ramírez, A.; Ortega-Cuenca, J.; Cuetero-Martínez, Y.; de Los Cobos, D.; Noyola, A. Viability and removal assessment of Escherichia coli and Salmonella spp. by real-time PCR with propidium monoazide in the hygienization of sewage sludge using three anaerobic processes. Waste Manag. 2023, 161, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zheng, G.; Lin, M.; Mustapha, A. Detection of viable Escherichia coli in environmental water using combined propidium monoazide staining and quantitative PCR. Water Res. 2018, 145, 398–407. [Google Scholar] [CrossRef]
- Ni, J.; Ji, J.; Li, Y.-Y.; Kubota, K. Propidium monoazide—Polymerase chain reaction reveals viable microbial community shifts in anaerobic membrane bioreactors treating domestic sewage at low temperature. Bioresour. Technol. 2023, 387, 129564. [Google Scholar] [CrossRef]
- Chen, X.; Li, W.; Ma, Y. Real-time and visual detection of viable Salmonella in milk by a competitive annealing mediated isothermal amplification (CAMP) combined with propidium monoazide (PMA). Anal. Methods 2022, 14, 3773–3779. [Google Scholar] [CrossRef]
- Xie, G.; Yu, S.; Li, W.; Mu, D.; Aguilar, Z.P.; Xu, H. Simultaneous detection of Salmonella spp., Pseudomonas aeruginosa, Bacillus cereus, and Escherichia coli O157:H7 in environmental water using PMA combined with mPCR. J. Microbiol. 2020, 58, 668–674. [Google Scholar] [CrossRef] [PubMed]





| Primer Set | Sequence (5′–3′) |
|---|---|
| F1 | ATGTCGTGGAAAGTAACGTTTAGCTGCTG |
| F2 | CTGGATCAGCCGAAGAAAGCTTTGCCTGTG |
| F3 | GTGGGGAAGGTTAAGGAGGGTGATAAGTTG |
| R1 | TATAACACAGTTTATCCGTATGGCTGGGCG |
| R2 | TGGTATCAGATAAAACCTCCGCTATAACAC |
| R3 | TGTCTCCTGTATTGAGGCGCTTAACCAGC |
| Primer Set | Sequence (5′–3′) |
|---|---|
| crRNA1 | GGGTAATTTCTACTAAGTGTAGATAGCCGGTAAACTACACGATG |
| crRNA2 | GGGTAATTTCTACTAAGTGTAGATAAGAGGCGCCTTGCGCTAAA |
| crRNA3 | GGGTAATTTCTACTAAGTGTAGATAAAGAAATAGCACGTCAGCA |
| crRNA4 | GGGTAATTTCTACTAAGTGTAGATGCCGTACTGACTGGTTGATG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Sui, X.; Xu, Y.; Zheng, X.; Tan, L. Establishment of a Sensitive and Visual Detection Platform for Viable Salmonella in Wastewater That Combines Propidium Monoazide with Recombinase Polymerase Amplification—CRISPR/Cas12a System. Microorganisms 2025, 13, 1166. https://doi.org/10.3390/microorganisms13051166
Liang J, Sui X, Xu Y, Zheng X, Tan L. Establishment of a Sensitive and Visual Detection Platform for Viable Salmonella in Wastewater That Combines Propidium Monoazide with Recombinase Polymerase Amplification—CRISPR/Cas12a System. Microorganisms. 2025; 13(5):1166. https://doi.org/10.3390/microorganisms13051166
Chicago/Turabian StyleLiang, Jiayin, Xintian Sui, Yan Xu, Xiangqun Zheng, and Lu Tan. 2025. "Establishment of a Sensitive and Visual Detection Platform for Viable Salmonella in Wastewater That Combines Propidium Monoazide with Recombinase Polymerase Amplification—CRISPR/Cas12a System" Microorganisms 13, no. 5: 1166. https://doi.org/10.3390/microorganisms13051166
APA StyleLiang, J., Sui, X., Xu, Y., Zheng, X., & Tan, L. (2025). Establishment of a Sensitive and Visual Detection Platform for Viable Salmonella in Wastewater That Combines Propidium Monoazide with Recombinase Polymerase Amplification—CRISPR/Cas12a System. Microorganisms, 13(5), 1166. https://doi.org/10.3390/microorganisms13051166






