Bioprotection with Saccharomyces cerevisiae: A Promising Strategy
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains, Storage, and Culture Media
2.2. Molecular Profiling and Identification of S. cerevisiae Strains
2.2.1. Genomic DNA Extraction
2.2.2. InterDelta-PCR Fingerprinting
2.3. Killing Acticity Assay and Measurment
2.4. Antagonism of S. cerevisiae Against Spoilage Yeast in Synthetic Must
2.5. Measurement of Yeast Growth
2.6. Killer Toxin Production and Quantification
2.7. Well Plate Assay of Sc54Kt and Arbitrary Unit
2.8. Sensitivity of B. bruxellensis and H. uvarum Strains to Sc54Kt Toxin in Must
2.9. Analytical Determinations
2.10. Statistical Analysis
3. Results and Discussion
3.1. Evaluation of Killing Activity
3.2. Biopreservative Efficacy of Sc54 in Co-Culture with B. bruxellensis and H. uvarum in SM
3.3. Effect of Killer Toxin Sc54Kt on B. bruxellensis and H. uvarum Culturability and Growth
3.3.1. Killing Activity Assay of Sc54Kt
3.3.2. Spectrum of Action and Antimicrobial Properties of Killer Toxin Sc54Kt in Synthetic Must
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alexandre, H.; Puyo, M.; Tourdot-Maréchal, R. Bioprotection in Winemaking. In New Advances in Saccharomyces [Working Title]; IntechOpen: London, UK, 2023. [Google Scholar]
- Escribano-Viana, R.; González-Arenzana, L.; Garijo, P.; Fernández, L.; López, R.; Santamaría, P.; Gutiérrez, A.R. Bioprotective Effect of a Torulaspora delbrueckii/Lachancea thermotolerans-Mixed Inoculum in Red Winemaking. Fermentation 2022, 8, 337. [Google Scholar] [CrossRef]
- Rubio-Bretón, P.; Gonzalo-Diago, A.; Iribarren, M.; Garde-Cerdán, T.; Pérez-Álvarez, E.P. Bioprotection as a Tool to Free Additives Winemaking: Effect on Sensorial, Anthocyanic and Aromatic Profile of Young Red Wines. LWT 2018, 98, 458–464. [Google Scholar] [CrossRef]
- Tzamourani, A.; Evangelou, A.; Ntourtoglou, G.; Lytra, G.; Paraskevopoulos, I.; Dimopoulou, M. Effect of Non-Saccharomyces Species Monocultures on Alcoholic Fermentation Behavior and Aromatic Profile of Assyrtiko Wine. Appl. Sci. 2024, 14, 1522. [Google Scholar] [CrossRef]
- Napa-Almeyda, C.A.; Criado, C.; Mayta-Hancco, J.; Silva-Jaimes, M.; Condezo-Hoyos, L.; Pozo-Bayón, M.Á. Non-Saccharomyces Yeast Strains, Aromatic Compounds and Sensory Analysis of Italy and Negra Criolla Pisco from the Moquegua Region of Peru. Fermentation 2023, 9, 757. [Google Scholar] [CrossRef]
- Yao, M.; Wang, F.; Arpentin, G. Bioprotection as a Tool to Produce Natural Wine: Impact on Physicochemical and Sensory Analysis. BIO Web Conf. 2023, 56, 02019. [Google Scholar] [CrossRef]
- SantˈAna, A.S.; Lemos Junior, W.J.F. Microbial Synergies and Their Impact on Economic and Quality Innovation in Sustainable Winemaking: Yeast and Lactic Acid Bacteria Interconnections. Food Biosci. 2024, 62, 105238. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, Y.; Li, Y.; Lou, Y.; Feng, X.; Yang, B. Comparison of Phenolic Profiles of Albino Bilberry (Vaccinium Myrtillus L.) Wines Fermented by non-Saccharomyces Yeasts. Food Biosci. 2023, 55, 102980. [Google Scholar] [CrossRef]
- Bordet, F.; Joran, A.; Klein, G.; Roullier-Gall, C.; Alexandre, H. Yeast–Yeast Interactions: Mechanisms, Methodologies and Impact on Composition. Microorganisms 2020, 8, 600. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Henschke, P.A. The ‘Buttery’ Attribute of Wine—Diacetyl—Desirability, Spoilage and Beyond. Int. J. Food Microbiol. 2004, 96, 235–252. [Google Scholar] [CrossRef]
- Liu, S. Practical Implications of Lactate and Pyruvate Metabolism by Lactic Acid Bacteria in Food and Beverage Fermentations. Int. J. Food Microbiol. 2003, 83, 115–131. [Google Scholar] [CrossRef]
- Barbe, J.-C.; Pelonnier-Magimel, E.; Windhotz, S.; Masneuf Pomarède, I. Sensory Characterisation of Wines without Added Sulfites via Specific and Adapted Sensory Profile. OENO One 2020, 54, 671–685. [Google Scholar] [CrossRef]
- Simonin, S.; Honoré-Chedozeau, C.; Monnin, L.; David-Vaizant, V.; Bach, B.; Alexandre, H.; Chatelet, B.; Tourdot-Marechal, R. Bioprotection on Chardonnay Grape: Limits and Impacts of Settling Parameters. Aust. J. Grape Wine Res. 2022, 2022, 1489094. [Google Scholar] [CrossRef]
- Windholtz, S.; Dutilh, L.; Lucas, M.; Maupeu, J.; Vallet-Courbin, A.; Farris, L.; Coulon, J.; Masneuf-Pomarède, I. Population Dynamics and Yeast Diversity in Early Winemaking Stages without Sulfites Revealed by Three Complementary Approaches. Appl. Sci. 2021, 11, 2494. [Google Scholar] [CrossRef]
- Reiter, T.; Montpetit, R.; Byer, S.; Frias, I.; Leon, E.; Viano, R.; Mcloughlin, M.; Halligan, T.; Hernandez, D.; Figueroa-Balderas, R.; et al. Transcriptomics Provides a Genetic Signature of Vineyard Site and Offers Insight into Vintage-Independent Inoculated Fermentation Outcomes. mSystems 2021, 6, e00033-21. [Google Scholar] [CrossRef] [PubMed]
- Windholtz, S.; Redon, P.; Lacampagne, S.; Farris, L.; Lytra, G.; Cameleyre, M.; Barbe, J.-C.; Coulon, J.; Thibon, C.; Masneuf-Pomarède, I. Non-Saccharomyces Yeasts as Bioprotection in the Composition of Red Wine and in the Reduction of Sulfur Dioxide. LWT 2021, 149, 111781. [Google Scholar] [CrossRef]
- Hamm, D.; Muñoz González, B. Use of Other Species in Winemaking, and Their Interaction with Saccharomyces cerevisiae. In New Advances in Saccharomyces; Morata, A., Loira, I., González, C., Escott, C., Eds.; IntechOpen: London, UK, 2023; ISBN 978-0-85466-128-2. [Google Scholar]
- Comuzzo, P.; Del Fresno, J.M.; Voce, S.; Loira, I.; Morata, A. Emerging Biotechnologies and Non-Thermal Technologies for Winemaking in a Context of Global Warming. Front. Microbiol. 2023, 14, 1273940. [Google Scholar] [CrossRef] [PubMed]
- Giménez, P.; Just-Borras, A.; Pons, P.; Gombau, J.; Heras, J.M.; Sieczkowski, N.; Canals, J.M.; Zamora, F. Biotechnological Tools for Reducing the Use of Sulfur Dioxide in White Grape Must and Preventing Enzymatic Browning: Glutathione; Inactivated Dry Yeasts Rich in Glutathione; and Bioprotection with Metschnikowia pulcherrima. Eur. Food Res. Technol. 2023, 249, 1491–1501. [Google Scholar] [CrossRef]
- Puyo, M.; Simonin, S.; Klein, G.; David-Vaizant, V.; Quijada-Morín, N.; Alexandre, H.; Tourdot-Maréchal, R. Use of Oenological Tannins to Protect the Colour of Rosé Wine in a Bioprotection Strategy with Metschnikowia pulcherrima. Foods 2023, 12, 735. [Google Scholar] [CrossRef]
- Viola, E.; Naselli, V.; Prestianni, R.; Pirrone, A.; Porrello, A.; Amato, F.; Savastano, R.; Maggio, A.; Carusi, M.; Seminerio, V.; et al. The Impact of a Saccharomyces cerevisiae Bio-Protective Strain during Cold Static Clarification on Catarratto Wine. AIMS Microbiol. 2025, 11, 40–58. [Google Scholar] [CrossRef]
- Wang, C.; Mas, A.; Esteve-Zarzoso, B. The Interaction between Saccharomyces cerevisiae and non-Saccharomyces Yeast during Alcoholic Fermentation Is Species and Strain Specific. Front. Microbiol. 2016, 7, 502. [Google Scholar] [CrossRef]
- El Dana, F.; Hayar, S.; Colosio, M.-C. Selection of Three Indigenous Lebanese Yeast Saccharomyces cerevisiae with Physiological Traits from Grape Varieties in Western Semi-Desert and Pedoclimatic Conditions in the Bekaa Valley. Fermentation 2021, 7, 280. [Google Scholar] [CrossRef]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of Yeasts by RFLP Analysis of the 5.8S rRNA Gene and the Two Ribosomal Internal Transcribed Spacers. Int. J. Syst. Evol. Microbiol. 1999, 49, 329–337. [Google Scholar] [CrossRef]
- Lopes, C.A.; Sangorrín, M.P. Optimization of Killer Assays for Yeast Selection Protocols. Rev. Argent. Microbiol. 2010, 42, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Harju, S.; Fedosyuk, H.; Peterson, K.R. Rapid Isolation of Yeast Genomic DNA: Bust n’ Grab. BMC Biotechnol. 2004, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.-L.; Karst, F. Optimisation of Interdelta Analysis for Saccharomyces cerevisiae Strain Characterisation. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- De Celis, M.; Ruiz, J.; Martín-Santamaría, M.; Alonso, A.; Marquina, D.; Navascués, E.; Gómez-Flechoso, M.Á.; Belda, I.; Santos, A. Diversity of Saccharomyces cerevisiae Yeasts Associated to Spontaneous and Inoculated Fermenting Grapes from Spanish Vineyards. Lett. Appl. Microbiol. 2019, 68, 580–588. [Google Scholar] [CrossRef]
- Cappello, M.S.; Bleve, G.; Grieco, F.; Dellaglio, F.; Zacheo, G. Characterization of Saccharomyces cerevisiae Strains Isolated from Must of Grape Grown in Experimental Vineyard. J. Appl. Microbiol. 2004, 97, 1274–1280. [Google Scholar] [CrossRef]
- Belloch, C.; Barrio, E.; García, M.D.; Querol, A. Phylogenetic Reconstruction of the Yeast Genus Kluyveromyces: Restriction Map Analysis of the 5.8S rRNA Gene and the Two Ribosomal Internal Transcribed Spacers. Syst. Appl. Microbiol. 1998, 21, 266–273. [Google Scholar] [CrossRef]
- Bely, M.; Sablayrolles, J.-M.; Barre, P. Automatic Detection of Assimilable Nitrogen Deficiencies during Alcoholic Fermentation in Oenological Conditions. J. Ferment. Bioeng. 1990, 70, 246–252. [Google Scholar] [CrossRef]
- Evers, M.S.; Roullier-Gall, C.; Morge, C.; Sparrow, C.; Gobert, A.; Vichi, S.; Alexandre, H. Thiamine and Biotin: Relevance in the Production of Volatile and Non-Volatile Compounds during Saccharomyces cerevisiae Alcoholic Fermentation in Synthetic Grape Must. Foods 2023, 12, 972. [Google Scholar] [CrossRef]
- Puyo, M.; Mas, P.; Roullier-Gall, C.; Romanet, R.; Lebleux, M.; Klein, G.; Alexandre, H.; Tourdot-Maréchal, R. Bioprotection Efficiency of Metschnikowia Strains in Synthetic Must: Comparative Study and Metabolomic Investigation of the Mechanisms Involved. Foods 2023, 12, 3927. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Francisco, D.; Chambon, C.; Hébraud, M.; Arneborg, N.; Almeida, M.G.; Caldeira, J.; Albergaria, H. Identification of Novel GAPDH-Derived Antimicrobial Peptides Secreted by Saccharomyces cerevisiae and Involved in Wine Microbial Interactions. Appl. Microbiol. Biotechnol. 2014, 98, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; San Mauro, M.; Bravo, E.; Marquina, D. PMKT2, a New Killer Toxin from Pichia membranifaciens, and Its Promising Biotechnological Properties for Control of the Spoilage Yeast Brettanomyces bruxellensis. Microbiology 2009, 155, 624–634. [Google Scholar] [CrossRef]
- Büyüksırıt-Bedir, T.; Kuleaşan, H. Purification and Characterization of a Metschnikowia pulcherrima Killer Toxin with Antagonistic Activity against Pathogenic Microorganisms. Arch. Microbiol. 2022, 204, 337. [Google Scholar] [CrossRef]
- Büyüksırıt Bedir, T.; Kuleaşan, H. A Natural Approach, the Use of Killer Toxin Produced by Metschnikowia pulcherrima in Fresh Ground Beef Patties for Shelf Life Extention. Int. J. Food Microbiol. 2021, 345, 109154. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kielkopf, C.L.; Bauer, W.; Urbatsch, I.L. Bradford Assay for Determining Protein Concentration. Cold Spring Harb. Protoc. 2020, 5. [Google Scholar] [CrossRef]
- Kielkopf, C.L.; Bauer, W.; Urbatsch, I.L. Methods for Measuring the Concentrations of Proteins. Cold Spring Harb. Protoc. 2020, 5. [Google Scholar] [CrossRef]
- Kumar Sharma, P.; Chand, D. Purification and Characterization of Thermostable Cellulase Free Xylanase from Pseudomonas Sp. XPB-6. Adv. Microbiol. 2012, 2, 17–25. [Google Scholar] [CrossRef][Green Version]
- Carboni, G.; Fancello, F.; Zara, G.; Zara, S.; Ruiu, L.; Marova, I.; Pinna, G.; Budroni, M.; Mannazzu, I. Production of a Lyophilized Ready-to-Use Yeast Killer Toxin with Possible Applications in the Wine and Food Industries. Int. J. Food Microbiol. 2020, 335, 108883. [Google Scholar] [CrossRef]
- Comitini, F.; Ingeniis De, J.; Pepe, L.; Mannazzu, I.; Ciani, M. Pichia anomala and Kluyveromyces wickerhamii Killer Toxins as New Tools against Dekkera/Brettanomyces Spoilage Yeasts. FEMS Microbiol. Lett. 2004, 238, 235–240. [Google Scholar] [CrossRef]
- Romano, P.; Suzzi, G.; Comi, G.; Zironi, R. Higher Alcohol and Acetic Acid Production by Apiculate Wine Yeasts. J. Appl. Bacteriol. 1992, 73, 126–130. [Google Scholar] [CrossRef]
- Aragno, J.; Fernandez-Valle, P.; Thiriet, A.; Grondin, C.; Legras, J.-L.; Camarasa, C.; Bloem, A. Two-Stage Screening of Metschnikowia spp. Bioprotective Properties: From Grape Juice to Fermented Must by Saccharomyces cerevisiae. Microorganisms 2024, 12, 1659. [Google Scholar] [CrossRef]
- Li, Y.-Q.; Hu, K.; Xu, Y.-H.; Mei, W.-C.; Tao, Y.-S. Biomass Suppression of Hanseniaspora uvarum by Killer Saccharomyces cerevisiae Highly Increased Fruity Esters in Mixed Culture Fermentation. LWT 2020, 132, 109839. [Google Scholar] [CrossRef]
- Hu, K.; Zhao, H.; Edwards, N.; Peyer, L.; Tao, Y.; Arneborg, N. The Effects of Cell-Cell Contact between Pichia kluyveri and Saccharomyces cerevisiae on Amino Acids and Volatiles in Mixed Culture Alcoholic Fermentations. Food Microbiol. 2022, 103, 103960. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Sabir, F.; Diniz, M.; Carvalho, L.; Albergaria, H.; Prista, C. Biocontrol of Brettanomyces/Dekkera bruxellensis in Alcoholic Fermentations Using Saccharomycin-Overproducing Saccharomyces cerevisiae Strains. Appl. Microbiol. Biotechnol. 2019, 103, 3073–3083. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Tufariello, M.; Capozzi, V.; Spano, G.; Mita, G.; Grieco, F. The Oenological Potential of Hanseniaspora uvarum in Simultaneous and Sequential Co-Fermentation with Saccharomyces cerevisiae for Industrial Wine Production. Front. Microbiol. 2016, 7, 670. [Google Scholar] [CrossRef]
- Vaquero, C.; Escott, C.; Heras, J.M.; Carrau, F.; Morata, A. Co-Inoculations of Lachancea thermotolerans with Different Hanseniaspora spp.: Acidification, Aroma, Biocompatibility, and Effects of Nutrients in Wine. Food Res. Int. 2022, 161, 111891. [Google Scholar] [CrossRef]
- Cibrario, A.; Miot-Sertier, C.; Paulin, M.; Bullier, B.; Riquier, L.; Perello, M.-C.; De Revel, G.; Albertin, W.; Masneuf-Pomarède, I.; Ballestra, P.; et al. Brettanomyces bruxellensis Phenotypic Diversity, Tolerance to Wine Stress and Wine Spoilage Ability. Food Microbiol. 2020, 87, 103379. [Google Scholar] [CrossRef]
- Kemsawasd, V.; Viana, T.; Ardö, Y.; Arneborg, N. Influence of Nitrogen Sources on Growth and Fermentation Performance of Different Wine Yeast Species during Alcoholic Fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 10191–10207. [Google Scholar] [CrossRef]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in Alcoholic Fermentation Processes: Role of Physiological Fitness and Microbial Interactions. Appl. Microbiol. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef]
- Branco, P.; Coutinho, R.; Malfeito-Ferreira, M.; Prista, C.; Albergaria, H. Wine Spoilage Control: Impact of Saccharomycin on Brettanomyces bruxellensis and Its Conjugated Effect with Sulfur Dioxide. Microorganisms 2021, 9, 2528. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Li, X.; Sheng, X.; Li, T.; Tang, W.; Yu, Z.; Wang, Y. Effect of Hanseniaspora uvarum–Saccharomyces cerevisiae Mixed Fermentation on Aroma Characteristics of Rosa Roxburghii Tratt, Blueberry, and Plum Wines. Molecules 2022, 27, 8097. [Google Scholar] [CrossRef]
- Wang, C.; Mas, A.; Esteve-Zarzoso, B. Interaction between Hanseniaspora uvarum and Saccharomyces cerevisiae during Alcoholic Fermentation. Int. J. Food Microbiol. 2015, 206, 67–74. [Google Scholar] [CrossRef]
- Li, L.; Yuan, C.; Zhang, L.; Chu, R.; Yu, Q.; Cai, J.; Yang, T.; Zhang, M. The Impact of Simultaneous Inoculation with Torulaspora delbrueckii and Hanseniaspora uvarum Combined with Saccharomyces cerevisiae on Chemical and Sensory Quality of Sauvignon Blanc Wines. Front. Microbiol. 2024, 15, 1413650. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; Agarbati, A.; Canonico, L.; Galli, E.; Ciani, M. Purification and Characterization of WA18, a New Mycocin Produced by Wickerhamomyces anomalus Active in Wine Against Brettanomyces bruxellensis Spoilage Yeasts. Microorganisms 2020, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.L.; Mazzucco, M.B.; Lopes, C.A.; Ganga, M.A.; Sangorrín, M.P. Purification and Characterization of Saccharomyces eubayanus Killer Toxin: Biocontrol Effectiveness against Wine Spoilage Yeasts. Int. J. Food Microbiol. 2020, 331, 108714. [Google Scholar] [CrossRef]
- De Ullivarri, M.F.; Mendoza, L.M.; Raya, R.R. Killer Activity of Saccharomyces cerevisiae Strains: Partial Characterization and Strategies to Improve the Biocontrol Efficacy in Winemaking. Antonie Van. Leeuwenhoek 2014, 106, 865–878. [Google Scholar] [CrossRef]
- Branco, P.; Francisco, D.; Monteiro, M.; Almeida, M.G.; Caldeira, J.; Arneborg, N.; Prista, C.; Albergaria, H. Antimicrobial Properties and Death-Inducing Mechanisms of Saccharomycin, a Biocide Secreted by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 159–171. [Google Scholar] [CrossRef]
- Albergaria, H.; Francisco, D.; Gori, K.; Arneborg, N.; Gírio, F. Saccharomyces cerevisiae CCMI 885 Secretes Peptides That Inhibit the Growth of Some non-Saccharomyces Wine-Related Strains. Appl. Microbiol. Biotechnol. 2010, 86, 965–972. [Google Scholar] [CrossRef]
- Pereznevado, F.; Albergaria, H.; Hogg, T.; Girio, F. Cellular Death of Two non-Saccharomyces Wine-Related Yeasts during Mixed Fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 2006, 108, S0168160506000419. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; Mannazzu, I.; Ciani, M. Tetrapisispora phaffii Killer Toxin Is a Highly Specific β-Glucanase That Disrupts the Integrity of the Yeast Cell Wall. Microb. Cell Factories 2009, 8, 55. [Google Scholar] [CrossRef]
- Bussey, H.; Sherman, D.; Somers, J.M. Action of Yeast Killer Factor: A Resistant Mutant with Sensitive Spheroplasts. J. Bacteriol. 1973, 113, 1193–1197. [Google Scholar] [CrossRef] [PubMed]
- Kurzweilová, H.; Sigler, K. Factors Affecting the Susceptibility of Sensitive Yeast Cells to Killer Toxin K1. Folia Microbiol. 1993, 38, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Breinig, F.; Tipper, D.J.; Schmitt, M.J. Kre1p, the Plasma Membrane Receptor for the Yeast K1 Viral Toxin. Cell 2002, 108, 395–405. [Google Scholar] [CrossRef]
- Boone, C.; Sommer, S.S.; Hensel, A.; Bussey, H. Yeast KRE Genes Provide Evidence for a Pathway of Cell Wall Beta-Glucan Assembly. J. Cell Biol. 1990, 110, 1833–1843. [Google Scholar] [CrossRef]
- De La Peña, P.; Barros, F.; Gascón, S.; Lazo, P.S.; Ramos, S. Effect of Yeast Killer Toxin on Sensitive Cells of Saccharomyces cerevisiae. J. Biol. Chem. 1981, 256, 10420–10425. [Google Scholar] [CrossRef]
- Skipper, N.; Bussey, H. Mode of Action of Yeast Toxins: Energy Requirement for Saccharomyces cerevisiae Killer Toxin. J. Bacteriol. 1977, 129, 668–677. [Google Scholar] [CrossRef]
- Dignard, D.; Whiteway, M.; Germain, D.; Tessier, D.; Thomas, D.Y. Expression in Yeast of a cDNA Copy of the K2 Killer Toxin Gene. Molec. Gen. Genet. 1991, 227, 127–136. [Google Scholar] [CrossRef]
- Lukša, J.; Podoliankaitė, M.; Vepštaitė, I.; Strazdaitė-Žielienė, Ž.; Urbonavičius, J.; Servienė, E. Yeast β-1,6-Glucan Is a Primary Target for the Saccharomyces cerevisiae K2 Toxin. Eukaryot. Cell 2015, 14, 406–414. [Google Scholar] [CrossRef]
- Novotna, D.; Flegelova, H.; Janderova, B. Different Action of Killer Toxins K1 and K2 on the Plasma Membrane and the Cell Wall of Saccharomyces cerevisiae. FEMS Yeast Res. 2004, 4, 803–813. [Google Scholar] [CrossRef]
- Flegelová, H.; Novotná, D.; Vojtíšková, K.; Janderová, B. Isolation and Characterization of Saccharomyces cerevisiae Mutants with a Different Degree of Resistance to Killer Toxins K1 and K2. FEMS Yeast Res. 2002, 2, 73–79. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmitt, M.J. Cloning and Expression of a cDNA Copy of the Viral K28 Killer Toxin Gene in Yeast. Molec. Gen. Genet. 1995, 246, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A New Wine Saccharomyces cerevisiae Killer Toxin (Klus), Encoded by a Double-Stranded RNA Virus, with Broad Antifungal Activity Is Evolutionarily Related to a Chromosomal Host Gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
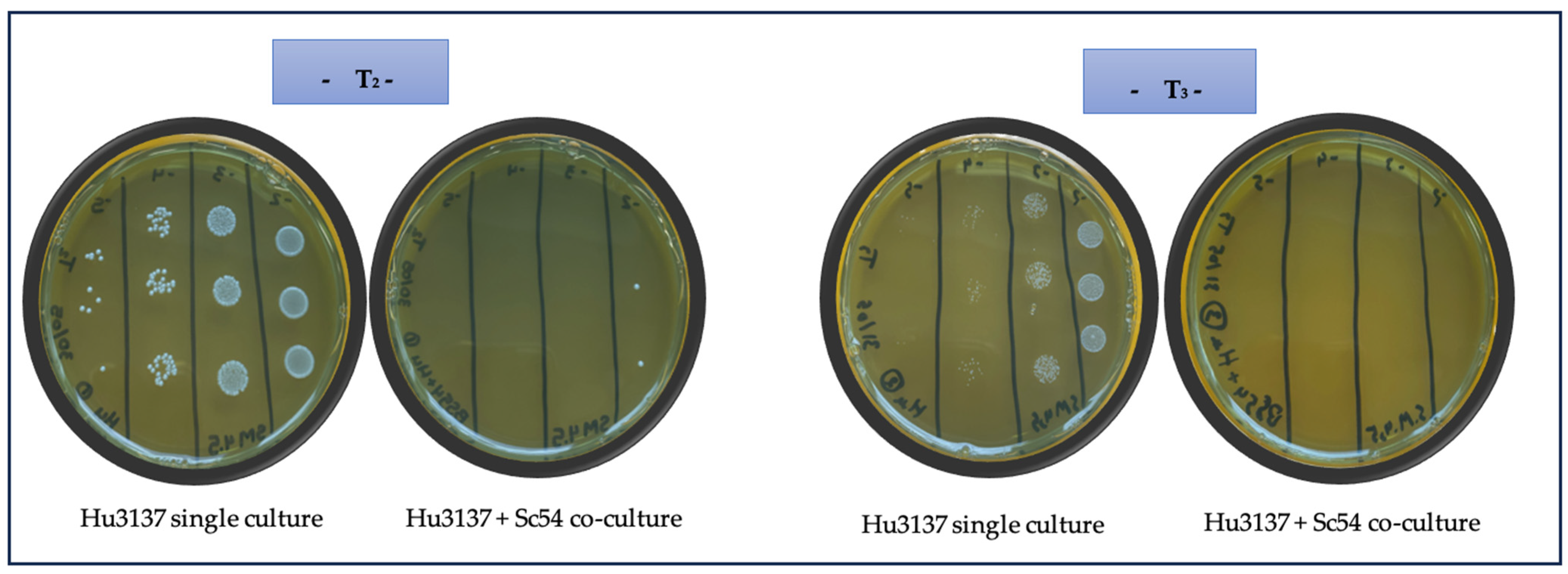
 and
and  ethanol, is displayed as striped bars for Sc54 in single culture and solid bars for co-culture conditions.
ethanol, is displayed as striped bars for Sc54 in single culture and solid bars for co-culture conditions.
 and
and  ethanol, is displayed as striped bars for Sc54 in single culture and solid bars for co-culture conditions.
ethanol, is displayed as striped bars for Sc54 in single culture and solid bars for co-culture conditions.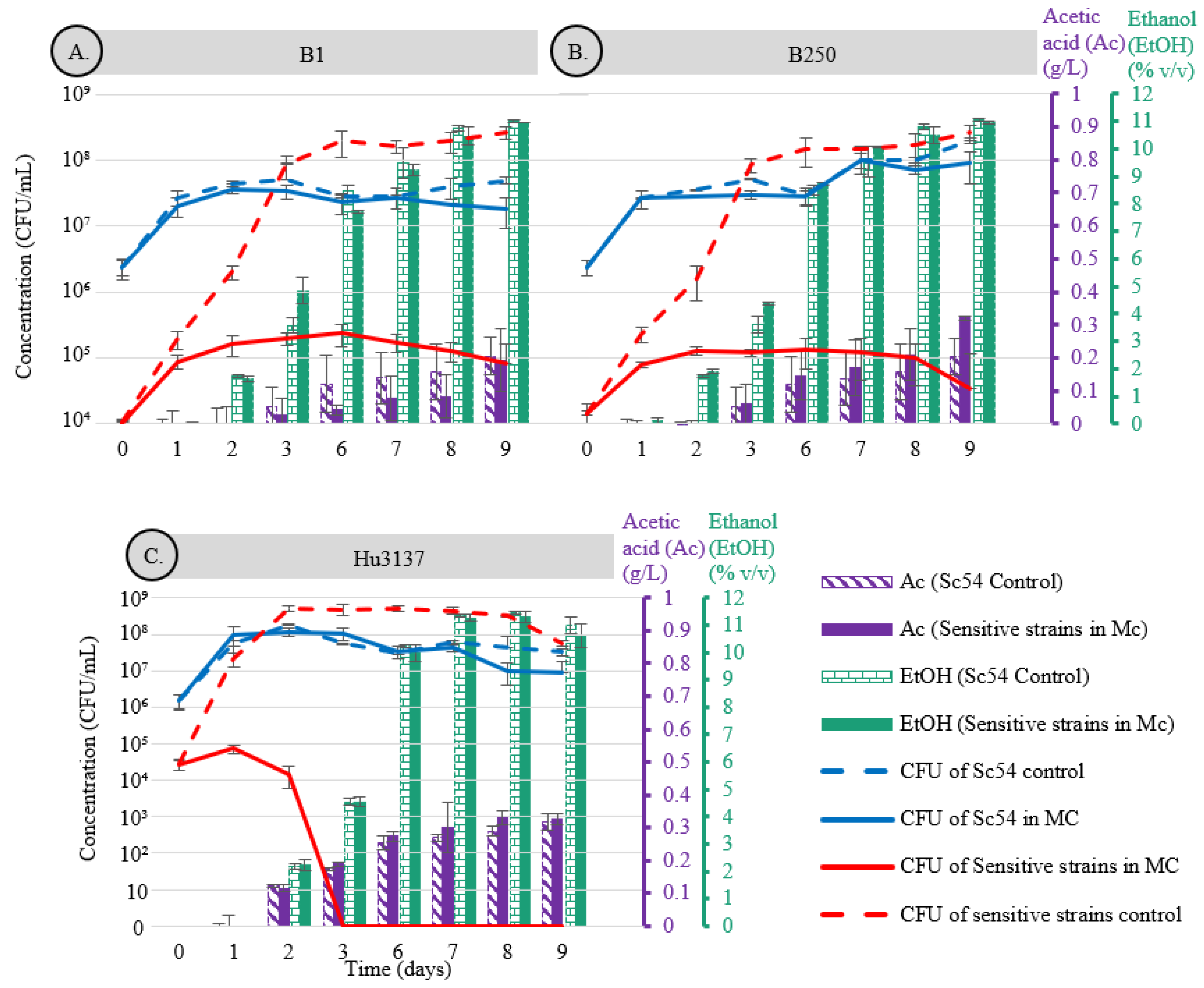
| S. cerevisiae Codes | Killing Activity Agar Medium | ||
|---|---|---|---|
| B1 | B250 | Hu3137 | |
| Sc54 | ++ | ++ | ++ |
| S342 | ++ | + | + |
| S340 | + | + | +/− |
| S334 | + | − | +/− |
| VL2 | ++ | + | Nd |
| Sensitive Strain | Killer Strain | µmax (h−1) | Final Population (CFU/mL) (9 Days) |
|---|---|---|---|
| B1 1 | Control | 0.126 ± 0.001 a | 2.72 × 108 ± 0.11 × 108 a |
| Sc54 2 | 0.042 ± 0.003 b | 8.10 × 104 ± 0.82 × 104 b | |
| B250 1 | Control | 0.123 ± 0.002 a | 2.7 × 108 ± 0.06 × 108 a |
| Sc54 2 | 0.026 ± 0.01 c | 3.44 × 104 ± 0.87 × 104 b | |
| Hu3137 1 | Control | 0.204 ± 0.001 a | 5.8 × 107 ± 1.2 × 107 a |
| Sc54 2 | −0.01 ± 0.012 b | 0 b |
| Killer Toxin | Halo Diameter (mm) | |||||
|---|---|---|---|---|---|---|
| H. uvarum Hu3137 | B. bruxellensis B1 | B. bruxellensis B250 | ||||
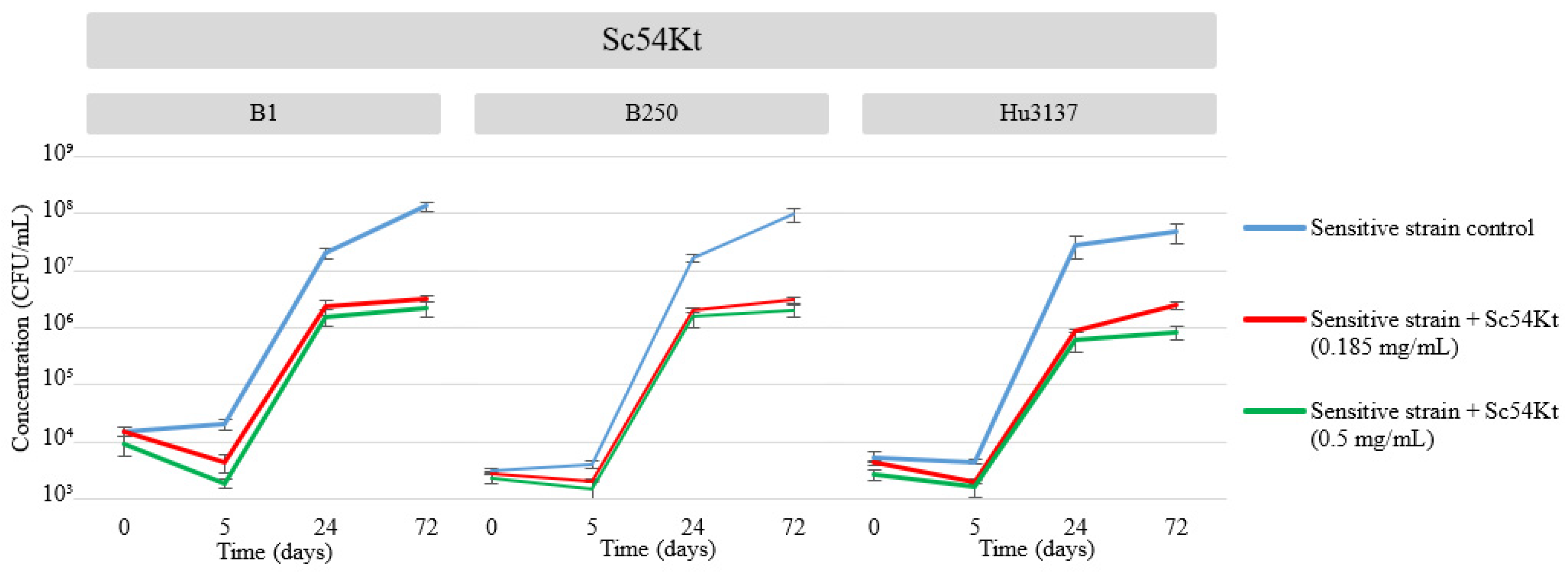
| 0.5 mg/mL | 0.185 mg/mL | 0.5 mg/mL | 0.185 mg/mL | 0.5 mg/mL | 0.185 mg/mL | |
| Sc54Kt | 13 | 7 | 11 | 5 | 10 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Dana, F.; David, V.; Tourdot-Maréchal, R.; Hayar, S.; Colosio, M.-C.; Alexandre, H. Bioprotection with Saccharomyces cerevisiae: A Promising Strategy. Microorganisms 2025, 13, 1163. https://doi.org/10.3390/microorganisms13051163
El Dana F, David V, Tourdot-Maréchal R, Hayar S, Colosio M-C, Alexandre H. Bioprotection with Saccharomyces cerevisiae: A Promising Strategy. Microorganisms. 2025; 13(5):1163. https://doi.org/10.3390/microorganisms13051163
Chicago/Turabian StyleEl Dana, Fatima, Vanessa David, Raphaëlle Tourdot-Maréchal, Salem Hayar, Marie-Charlotte Colosio, and Hervé Alexandre. 2025. "Bioprotection with Saccharomyces cerevisiae: A Promising Strategy" Microorganisms 13, no. 5: 1163. https://doi.org/10.3390/microorganisms13051163
APA StyleEl Dana, F., David, V., Tourdot-Maréchal, R., Hayar, S., Colosio, M.-C., & Alexandre, H. (2025). Bioprotection with Saccharomyces cerevisiae: A Promising Strategy. Microorganisms, 13(5), 1163. https://doi.org/10.3390/microorganisms13051163







