Selection of Probiotics for Honey Bees: The In Vitro Inhibition of Paenibacillus larvae, Melissococcus plutonius, and Serratia marcescens Strain Sicaria by Host-Specific Lactobacilli and Bifidobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Isolation of Bifidobacterium and Lactobacillus Strains
2.3. Isolation of Bacterial Honey Bee Pathogens
2.4. Classification of Isolates
2.5. In Vitro Inhibition of Honey Bee Bacterial Pathogens
2.6. Phylogenetic Analyses
3. Results
3.1. Classification of Bacterial Strains
3.2. In Vitro Inhibition of Honey Bee Bacterial Pathogens
3.3. Phylogenetic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alberoni, D.; Gaggìa, F.; Baffoni, L.; Modesto, M.M.; Biavati, B.; Di Gioia, D. Bifidobacterium Xylocopae sp. nov. and Bifidobacterium Aemilianum sp. nov., from the Carpenter Bee (Xylocopa Violacea) Digestive Tract. Syst. Appl. Microbiol. 2019, 42, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, J.; Zheng, H. Characterization of Bifidobacterium Apousia sp. nov., Bifidobacterium Choladohabitans sp. nov., and Bifidobacterium Polysaccharolyticum sp. nov., Three Novel Species of the Genus Bifidobacterium from Honeybee Gut. Syst. Appl. Microbiol. 2021, 44, 126247. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Kwong, W.K.; Moran, N.A. Frischella Perrara gen. nov., sp. nov., a Gammaproteobacterium Isolated from the Gut of the Honeybee, Apis Mellifera. Int. J. Syst. Evol. Microbiol. 2013, 63, 3646–3651. [Google Scholar] [CrossRef] [PubMed]
- Killer, J.; Dubná, S.; Sedláček, I.; Švec, P. Lactobacillus Apis sp. nov., from the Stomach of Honeybees (Apis mellifera), Having an in vitro Inhibitory Effect on the Causative Agents of American and European Foulbrood. Int. J. Syst. Evol. Microbiol. 2014, 64, 152–157. [Google Scholar] [CrossRef]
- Killer, J.; Kopečný, J.; Mrázek, J.; Havlík, J.; Koppová, I.; Benada, O.; Rada, V.; Kofroňová, O. Bombiscardovia Coagulans gen. nov., sp. nov., a New Member of the Family Bifidobacteriaceae Isolated from the Digestive Tract of Bumblebees. Syst. Appl. Microbiol. 2010, 33, 359–366. [Google Scholar] [CrossRef]
- Killer, J.; Votavová, A.; Valterová, I.; Vlková, E.; Rada, V.; Hroncová, Z. Lactobacillus Bombi sp. nov., from the Digestive Tract of Laboratory-Reared Bumblebee Queens (Bombus Terrestris). Int. J. Syst. Evol. Microbiol. 2014, 64, 2611–2617. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Cultivation and Characterization of the Gut Symbionts of Honey Bees and Bumble Bees: Description of Snodgrassella Alvi gen. nov., sp. nov., a Member of the Family Neisseriaceae of the Betaproteobacteria, and Gilliamella Apicola gen. nov., sp. nov., a Member of Orbaceae Fam. nov., Orbales Ord. nov., a Sister Taxon to the Order “Enterobacteriales” of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013, 63, 2008–2018. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Alsterfjord, M.; Nilson, B.; Butler, È.; Vásquez, A. Lactobacillus Apinorum sp. nov., Lactobacillus Mellifer sp. nov., Lactobacillus Mellis sp. nov., Lactobacillus Melliventris sp. nov., Lactobacillus Kimbladii sp. nov., Lactobacillus Helsingborgensis sp. nov. and Lactobacillus Kullabergensis sp. nov., Isolated from the Honey Stomach of the Honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol. 2014, 64, 3109–3119. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Modesto, M.; Pascarelli, S. Bifidobacterium Mellis sp. nov., Isolated from the Honey Stomach of the Honey Bee Apis mellifera. Int. J. Syst. Evol. Microbiol. 2023, 73, 3. [Google Scholar] [CrossRef]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling Metabolic Functions of Bacteria in the Honey Bee Gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Gage, A.; Smith, T.E.; Blake, K.J.; Kwong, W.K.; Riddington, I.M.; Moran, N.A. Host-Microbiome Metabolism of a Plant Toxin in Bees. Elife 2022, 11, e82595. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Perreau, J.; Elijah Powell, J.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of Labor in Honey Bee Gut Microbiota for Plant Polysaccharide Digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef] [PubMed]
- Leska, A.; Nowak, A.; Miśkiewicz, K.; Rosicka-Kaczmarek, J. Binding and Detoxification of Insecticides by Potentially Probiotic Lactic Acid Bacteria Isolated from Honeybee (Apis mellifera L.) Environment—An In Vitro Study. Cells 2022, 11, 3743. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Niu, J.; Zhu, Y.; Li, Z.; Ye, L.; Cao, H.; Shi, T.; Yu, L. Apilactobacillus kunkeei Alleviated Toxicity of Acetamiprid in Honeybee. Insects 2022, 13, 1167. [Google Scholar] [CrossRef]
- Nowak, A.; Szczuka, D.; Górczyńska, A.; Motyl, I.; Kręgiel, D. Characterization of Apis Mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection-A Review. Cells 2021, 10, 701. [Google Scholar] [CrossRef]
- Smriti; Rana, A.; Singh, G.; Gupta, G. Prospects of Probiotics in Beekeeping: A Review for Sustainable Approach to Boost Honeybee Health. Arch. Microbiol. 2024, 206, 205. [Google Scholar] [CrossRef]
- Garrido, P.M.; Porrini, M.P.; Alberoni, D.; Baffoni, L.; Scott, D.; Mifsud, D.; Eguaras, M.J.; Di Gioia, D. Beneficial Bacteria and Plant Extracts Promote Honey Bee Health and Reduce Nosema Ceranae Infection. Probiotics Antimicrob. Proteins 2024, 16, 259–274. [Google Scholar] [CrossRef]
- Janashia, I.; Alaux, C. Specific Immune Stimulation by Endogenous Bacteria in Honey Bees (Hymenoptera: Apidae). J. Econ. Entomol. 2016, 109, 1474–1477. [Google Scholar] [CrossRef]
- Ye, M.; Li, X.; Yang, F.; Zhou, B. Beneficial Bacteria as Biocontrol Agents for American Foulbrood Disease in Honey Bees (Apis mellifera). J. Insect Sci. 2023, 23, 6. [Google Scholar] [CrossRef]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee Gut Microbiota Promotes Host Weight Gain via Bacterial Metabolism and Hormonal Signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Alberoni, D.; Baffoni, L.; Gaggìa, F.; Ryan, P.M.; Murphy, K. Impact of Beneficial Bacteria Supplementation on the Gut Microbiota, Colony Development and Productivity of Apis mellifera L. Benef. Microbes 2018, 9, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Ricigliano, V.A.; Mott, B.M.; Copeland, D.C.; Floyd, A.S.; Maes, P. The Queen’s Gut Refines with Age: Longevity Phenotypes in a Social Insect Model. Microbiome 2018, 6, 108. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lei, L.; Chen, W.; Chi, X.; Han, K.; Wang, Y.; Ma, L.; Liu, Z.; Xu, B. The Comparison of Antioxidant Performance, Immune Performance, IIS Activity and Gut Microbiota Composition between Queen and Worker Bees Revealed the Mechanism of Different Lifespan of Female Casts in the Honeybee. Insects 2022, 13, 772. [Google Scholar] [CrossRef]
- Janashia, I.; Choiset, Y.; Jozefiak, D.; Déniel, F.; Coton, E.; Moosavi-Movahedi, A.A.; Chanishvili, N.; Haertlé, T. Beneficial Protective Role of Endogenous Lactic Acid Bacteria against Mycotic Contamination of Honeybee Beebread. Probiotics Antimicrob. Proteins 2018, 10, 638–646. [Google Scholar] [CrossRef]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Gibbons, S.; Chernyshova, A.M.; Al, K.F.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Lactobacillus spp. Attenuate Antibiotic-Induced Immune and Microbiota Dysregulation in Honey Bees. Commun. Biol. 2020, 3, 534. [Google Scholar] [CrossRef]
- Powell, J.E.; Carver, Z.; Leonard, S.P.; Moran, N.A. Field-Realistic Tylosin Exposure Impacts Honey Bee Microbiota and Pathogen Susceptibility, Which Is Ameliorated by Native Gut Probiotics. Microbiol. Spectr. 2021, 9, e0010321. [Google Scholar] [CrossRef]
- Wu, J.; Lang, H.; Mu, X.; Zhang, Z.; Su, Q.; Hu, X.; Zheng, H. Honey Bee Genetics Shape the Strain-Level Structure of Gut Microbiota in Social Transmission. Microbiome 2021, 9, 225. [Google Scholar] [CrossRef]
- Zhang, Z.; Mu, X.; Cao, Q.; Shi, Y.; Hu, X.; Zheng, H. Honeybee Gut Lactobacillus Modulates Host Learning and Memory Behaviors via Regulating Tryptophan Metabolism. Nat. Commun. 2022, 13, 2037. [Google Scholar] [CrossRef]
- Vernier, C.L.; Nguyen, L.A.; Gernat, T.; Ahmed, A.C.; Chen, Z.; Robinson, G.E. Gut Microbiota Contribute to Variations in Honey Bee Foraging Intensity. ISME J. 2024, 18, wrae030. [Google Scholar] [CrossRef]
- Forsgren, E.; Olofsson, T.C.; Vásquez, A.; Fries, I. Novel Lactic Acid Bacteria Inhibiting Paenibacillus Larvae in Honey Bee Larvae Novel Lactic Acid Bacteria Inhibiting Paenibacillus Larvae in Honey Bee Larvae. Apidologie 2010, 41, 99–108. [Google Scholar] [CrossRef]
- Iorizzo, M.; Ganassi, S.; Albanese, G.; Letizia, F.; Testa, B.; Tedino, C.; Petrarca, S.; Mutinelli, F.; Mazzeo, A.; De Cristofaro, A. Antimicrobial Activity from Putative Probiotic Lactic Acid Bacteria for the Biological Control of American and European Foulbrood Diseases. Vet. Sci. 2022, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.; Duan, H.; Wang, J.; Zhang, W.; Guo, J.; Zhang, X.; Hu, X.; Zheng, H. Specific Strains of Honeybee Gut Lactobacillus Stimulate Host Immune System to Protect against Pathogenic Hafnia Alvei. Microbiol. Spectr. 2022, 23, e0189621. [Google Scholar] [CrossRef]
- Truong, A.T.; Kang, J.E.; Yoo, M.S.; Nguyen, T.T.; Youn, S.Y.; Yoon, S.S.; Cho, Y.S. Probiotic Candidates for Controlling Paenibacillus Larvae, a Causative Agent of American Foulbrood Disease in Honey Bee. BMC Microbiol. 2023, 23, 150. [Google Scholar] [CrossRef]
- Wu, M.; Sugimura, Y.; Takaya, N.; Takamatsu, D.; Kobayashi, M.; Taylor, D.M.; Yoshiyama, M. Characterization of Bifidobacteria in the Digestive Tract of the Japanese Honeybee, Apis Cerana Japonica. J. Invertebr. Pathol. 2013, 112, 88–93. [Google Scholar] [CrossRef]
- Zeid, A.A.A.; Khattaby, A.M.; El-Khair, I.A.A.; Gouda, H.I.A. Detection Bioactive Metabolites of Fructobacillus Fructosus strain HI-1 Isolated from Honey Bee’s Digestive Tract Against Paenibacillus Larvae. Probiotics Antimicrob. Proteins 2022, 14, 476–485. [Google Scholar] [CrossRef]
- Arredondo, D.; Castelli, L.; Porrini, M.P.; Garrido, P.M.; Eguaras, M.J.; Zunino, P.; Antúnez, K. Lactobacillus Kunkeei Strains Decreased the Infection by Honey Bee Pathogens Paenibacillus Larvae and Nosema Ceranae. Benef. Microbes 2018, 9, 279–290. [Google Scholar] [CrossRef]
- Baffoni, L.; Gaggìa, F.; Alberoni, D.; Cabbri, R.; Nanetti, A.; Biavati, B.; Di Gioia, D. Effect of Dietary Supplementation of Bifidobacterium and Lactobacillus Strains in Apis Mellifera L. Against Nosema ceranae. Benef. Microbes 2016, 7, 45–51. [Google Scholar] [CrossRef]
- Iorizzo, M.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics 2020, 9, 262. [Google Scholar] [CrossRef]
- Tejerina, M.R.; Cabana, M.J.; Benitez-Ahrendts, M.R. Strains of Lactobacillus spp. Reduce Chalkbrood in Apis mellifera. J. Invertebr. Pathol. 2021, 178, 107521. [Google Scholar] [CrossRef]
- Brar, G.; Ngor, L.; McFrederick, Q.S.; Torson, A.S.; Rajamohan, A.; Rinehart, J.; Singh, P.; Bowsher, J.H. High Abundance of Lactobacilli in the Gut Microbiome of Honey Bees During Winter. Sci. Rep. 2025, 15, 7409. [Google Scholar] [CrossRef]
- Reid, G.; Gadir, A.A.; Dhir, R. Probiotics: Reiterating What They Are and What They Are Not. Front. Microbiol. 2019, 10, 424. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Salminen, S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 2013, 36, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Vásquez, A.; Olofsson, T.C.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Odham, G. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Killer, J.; Kopečný, J.; Mrázek, J.; Rada, V.; Dubná, S.; Marounek, M. Bifidobacteria in the Digestive Tract of Bumblebees. Anaerobe 2010, 16, 165–170. [Google Scholar] [CrossRef]
- Matiašovic, J.; Bzdil, J.; Papežíková, I.; Čejková, D.; Vasina, E.; Bizos, J.; Navrátil, S.; Šedivá, M.; Klaudiny, J.; Pikula, J. Genomic Analysis of Paenibacillus larvae Isolates from the Czech Republic and the Neighbouring Regions of Slovakia. Res. Vet. Sci. 2023, 158, 34–40. [Google Scholar] [CrossRef]
- Thebeau, J.M.; Liebe, D.; Masood, F.; Kozii, I.V.; Klein, C.D.; Zabrodski, M.W.; Moshynskyy, I.; Sobchishin, L.; Wilson, G.; Guarna, M.M.; et al. Article Investigation of Melissococcus Plutonius Isolates from 3 Outbreaks of European Foulbrood Disease in Commercial Beekeeping Operations in Western Canada. Can. Vet. J. 2022, 63, 935–942. [Google Scholar]
- Burritt, N.L.; Foss, N.J.; Neeno-Eckwall, E.C.; Church, J.O.; Hilger, A.M.; Hildebrand, J.A.; Warshauer, D.M.; Perna, N.T.; Burritt, J.B. Sepsis and Hemocyte Loss in Honey Bees (Apis Mellifera) Infected with Serratia Marcescens Strain Sicaria. PLoS ONE 2016, 11, e0167752. [Google Scholar] [CrossRef]
- Forster, R.J.; Teather, R.M.; Gong, J.; Deng, S.J. 16S rDNA Analysis of Butyrivibrio Fibrisolvens: Phylogenetic Position and Relation to Butyrate-Producing Anaerobic Bacteria from the Rumen of White-Tailed Deer. Lett. Appl. Microbiol. 1996, 23, 218–222. [Google Scholar] [CrossRef]
- Galkiewicz, J.P.; Kellogg, C.A. Cross-Kingdom Amplification Using Bacteria-Specific Primers: Complications for Studies of Coral Microbial Ecology. Appl Environ Microbiol 2008, 74, 7828–7831. [Google Scholar] [CrossRef]
- Killer, J.; Mekadim, C.; Pechar, R.; Bunešová, V.; Mrázek, J.; Vlková, E. Gene Encoding the CTP Synthetase as an Appropriate Molecular Tool for Identification and Phylogenetic Study of the Family Bifidobacteriaceae. Microbiologyopen 2018, 7, e00579. [Google Scholar] [CrossRef]
- Cheng, L.; Kiewiet, M.B.G.; Logtenberg, M.J.; Groeneveld, A.; Nauta, A.; Schols, H.A.; Walvoort, M.T.C.; Harmsen, H.J.M.; de Vos, P. Effects of Different Human Milk Oligosaccharides on Growth of Bifidobacteria in Monoculture and Co-Culture with Faecalibacterium Prausnitzii. Front. Microbiol. 2020, 11, 569700. [Google Scholar] [CrossRef] [PubMed]
- Killer, J.; Havlík, J.; Bunešová, V.; Vlková, E.; Benada, O. Pseudoscardovia radai sp. nov., A Representative of the Family Bifidobacteriaceae Isolated from the Digestive Tract of a Wild Pig (Sus Scrofa Scrofa). Int. J. Syst. Evol. Microbiol. 2014, 64, 2932–2938. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, K.M.; Tamarit, D.; Javelind, E.; Olofsson, T.C.; Andersson, S.G.E.; Vásquez, A. Extensive Intra-Phylotype Diversity in Lactobacilli and Bifidobacteria from the Honeybee Gut. BMC Genom. 2015, 16, 284. [Google Scholar] [CrossRef]
- Heyndrickx, M.; Vandemeulebroecke, K.; Hoste, B. Reclassification of Paenibacillus (Formerly Bacillus) Pulvifaciens (Nakamura 1984) Ash et al. 1994, a Later Subjective Synonym of Paenibacillus (Formerly Bacillus) larvae (White 1906) Ash et al. 1994, as a Subspecies of P. Larvae, with Emended Descriptions of P. Larvae as P. Larvae subs P. Larvae and P. Larvae subsp. Pulvifaciens. Int. J. Syst. Bacteriol. 1996, 46, 270–279. [Google Scholar] [CrossRef]
- Fijan, S. Microorganisms with Claimed Probiotic Properties: An Overview of Recent Literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef]
- Hejazi, A.; Falkiner, F.R. Serratia marcescens. J. Med. Microbiol. 1997, 46, 903–912. [Google Scholar] [CrossRef]
- Raymann, K.; Coon, K.L.; Shaffer, Z.; Salisbury, S.; Moran, N.A. Pathogenicity of Serratia marcescens Strains in Honey Bees. mBio 2018, 9, e01649-18, Erratum in mBio 2019, 10, e02855-18. [Google Scholar] [CrossRef]
- Ansari, M.J.; Al-Ghamdi, A.; Nuru, A.; Ahmed, A.M.; Ayaad, T.H.; Al-Qarni, A.; Alattal, Y.; Al-Waili, N. Survey and Molecular Detection of Melissococcus plutonius, the Causative Agent of European Foulbrood in Honeybees in Saudi Arabia. Saudi J. Biol. Sci. 2017, 24, 1327–1335. [Google Scholar] [CrossRef]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Al, K.F.; Chernyshova, A.M.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Novel Probiotic Approach to Counter Paenibacillus larvae Infection in Honey Bees. ISME J. 2020, 14, 476–491. [Google Scholar] [CrossRef]
- Butler, È.; Alsterfjord, M.; Olofsson, T.C.; Karlsson, C.; Malmström, J.; Vásquez, A. Proteins of Novel Lactic Acid Bacteria from Apis mellifera mellifera: An Insight into the Production of Known Extra-Cellular Proteins during Microbial Stress. BMC Microbiol. 2013, 13, 235. [Google Scholar] [CrossRef] [PubMed]
- Masood, F.; Thebeau, J.M.; Cloet, A.; Kozii, I.V.; Zabrodski, M.W.; Biganski, S.; Wood, S.C. Evaluating approved and alternative treatments against an oxytetracycline-resistant bacterium responsible for European foulbrood disease in honey bees. Sci. Rep. 2022, 12, 5906. [Google Scholar] [CrossRef] [PubMed]
- Daisley, B.A.; Pitek, A.P.; Mallory, E.; Chernyshova, A.M.; Allen-Vercoe, E.; Reid, G.; Thompson, G.J. Disentangling the microbial ecological factors impacting honey bee susceptibility to Paenibacillus larvae infection. Trends Microbiol. 2023, 31, 521–534. [Google Scholar] [CrossRef]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; Ganassi, S.; Ianiro, M.; Letizia, F.; De Cristofaro, A. Antimicrobial activity against Paenibacillus larvae and functional properties of Lactiplantibacillus plantarum strains: Potential benefits for honeybee health. Antibiotics 2020, 9, 442. [Google Scholar] [CrossRef]
- Borges, D.; Guzman-Novoa, E.; Goodwin, P.H. Effects of prebiotics and probiotics on honey bees (Apis mellifera) infected with the microsporidian parasite Nosema ceranae. Microorganisms 2021, 9, 481. [Google Scholar] [CrossRef]
- Mallory, E.; Freeze, G.; Daisley, B.A.; Allen-Vercoe, E. Revisiting the role of pathogen diversity and microbial interactions in honeybee susceptibility and treatment of Melissococcus plutonius infection. Front. Vet. Sci. 2024, 11, 1495010. [Google Scholar] [CrossRef]
- Toutiaee, S.; Mojgani, N.; Harzandi, N.; Moharrami, M.; Mokhberosafa, L. In vitro probiotic and safety attributes of Bacillus spp. isolated from beebread, honey samples and digestive tract of honeybees Apis mellifera. Lett. Appl. Microbiol. 2022, 74, 656–665. [Google Scholar] [CrossRef]

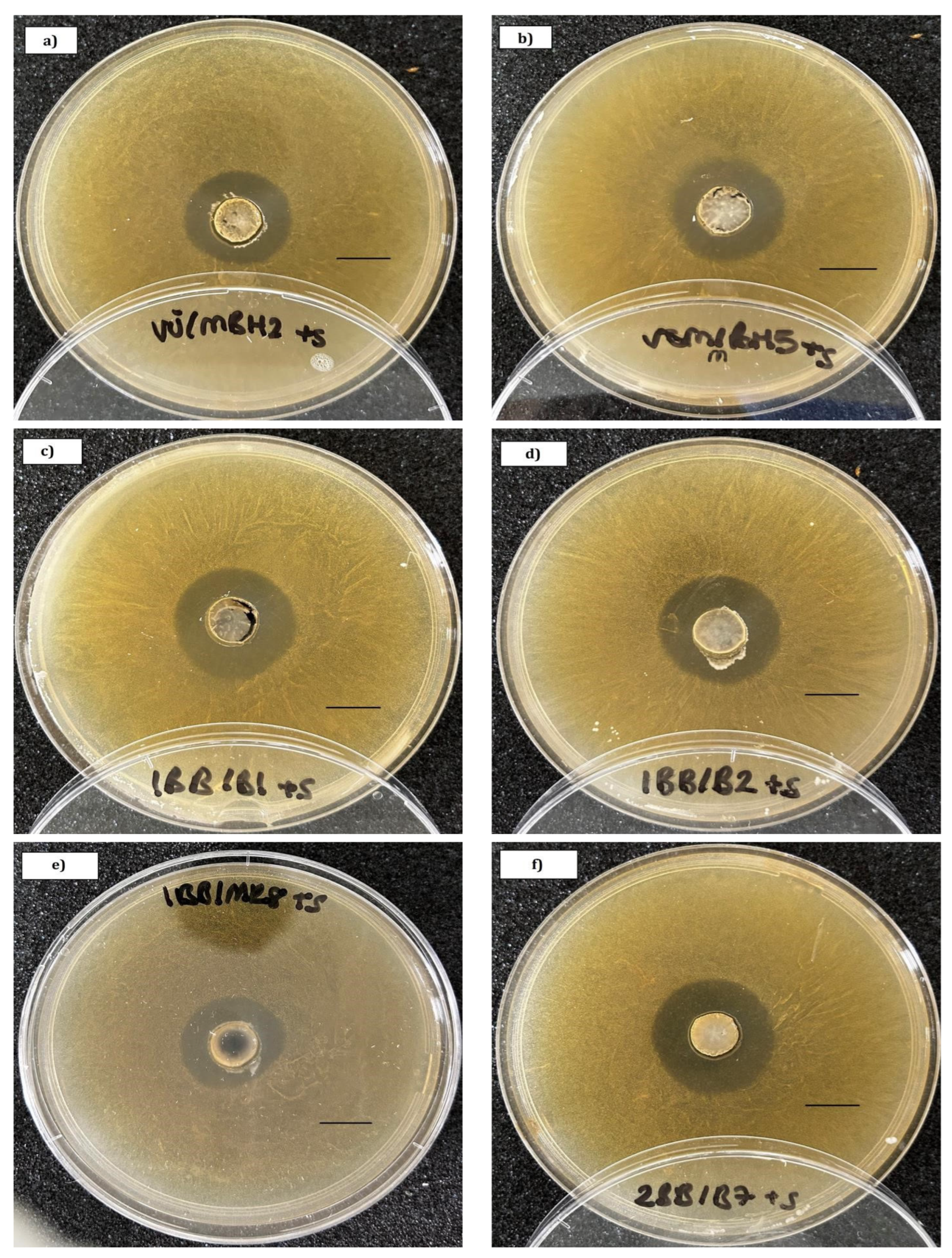
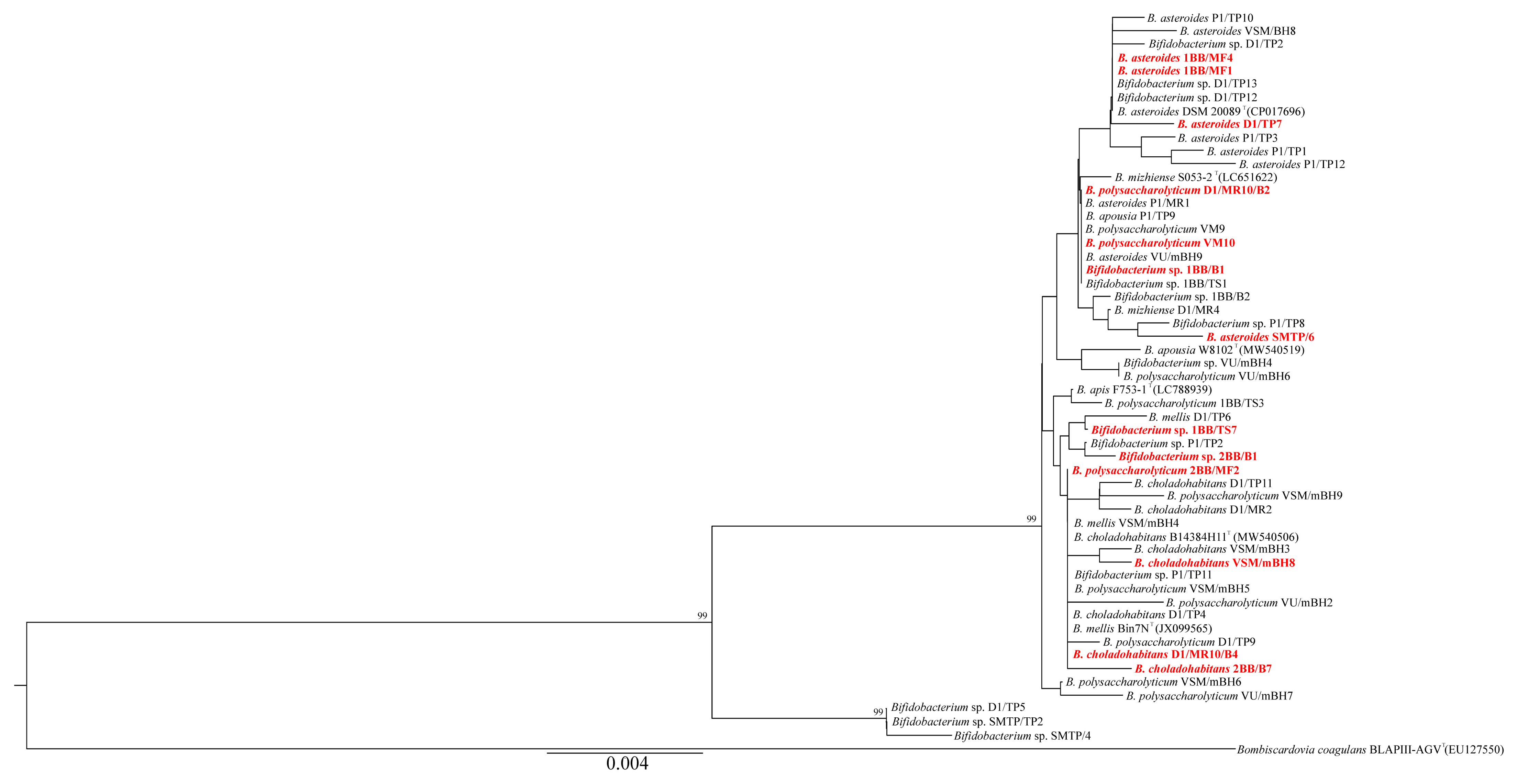
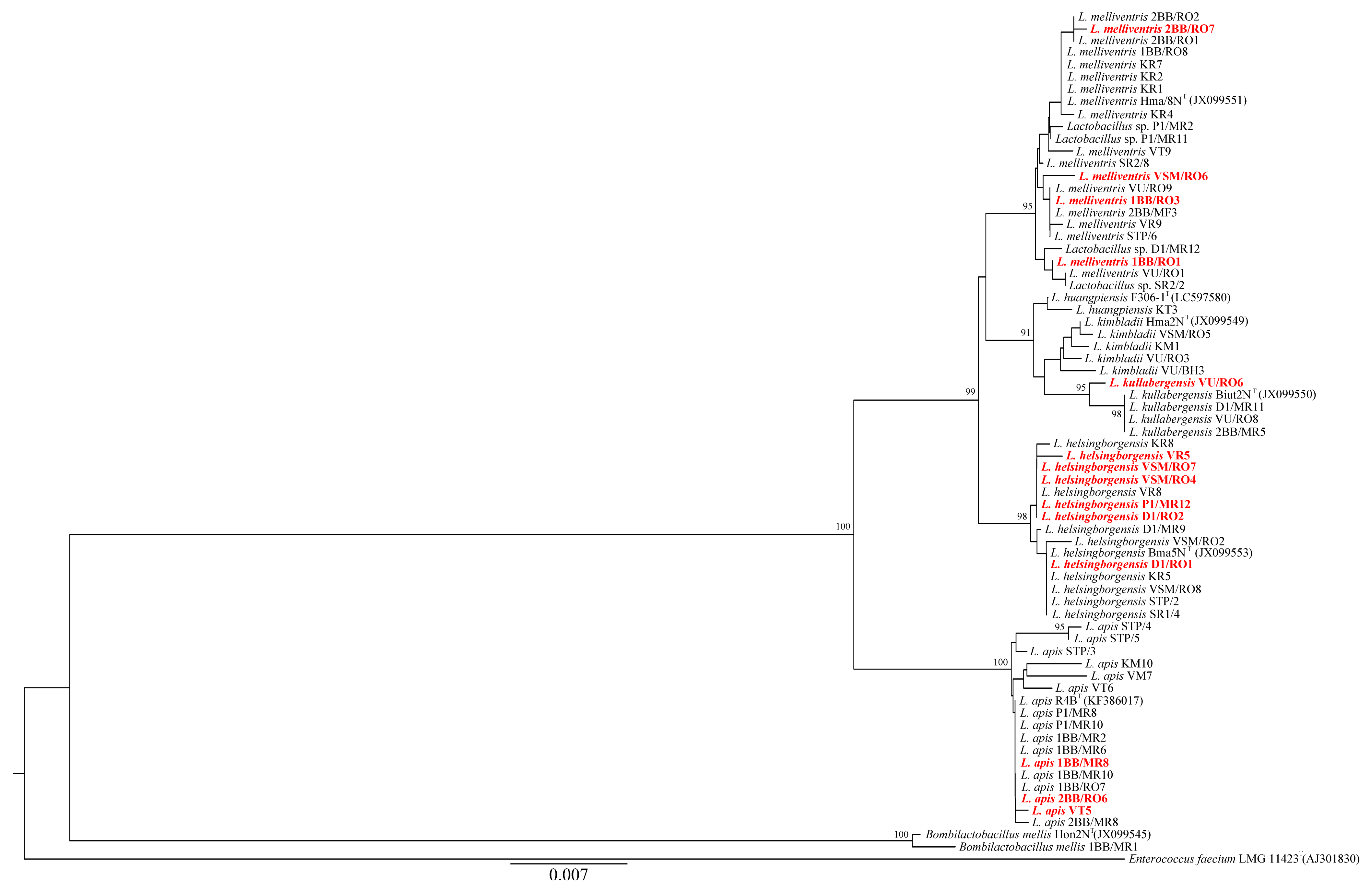
| Strain | Classification | PL2 | PL41 | PL48 | PL52 | MP | Ss1 | GenBank a.n. |
|---|---|---|---|---|---|---|---|---|
| D1/TP2 | Bifidobacterium apousia | - | 21 | - | - | - | - | PP754609 |
| D1/TP4 | Bifidobacterium choladohabitans | - | - | - | - | - | - | PP754610 |
| D1/TP5 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | - | PP754611 |
| D1/TP6 | Bifidobacterium mellis | - | - | - | - | - | - | PP754612 |
| D1/TP7 | Bifidobacterium asteroides | - | 20 | 38 | 38 | - | - | PP754613 |
| D1/TP9 | Bifidobacterium asteroides | - | 16 | 24 | - | - | - | PP754614 |
| D1/TP11 | Bifidobacterium choladohabitans | - | 33 | - | - | - | - | PP754615 |
| D1/TP12 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | - | PP754616 |
| D1/TP13 | Bifidobacterium sp. (p.n.sp.) | - | 36 | - | - | - | - | PP754617 |
| D1/MR2 | Bifidobacterium choladohabitans | 18 | 16 | 20 | 22 | - | - | PP754618 |
| D1/MR4 | Bifidobacterium mizhiense | - | - | - | - | - | - | PP754619 |
| D1/MR9. | Lactobacillus helsingborgensis | - | - | - | 36 | - | - | PP754620 |
| D1/MR10/B2 | Bifidobacterium polysaccharolyticum | 33 | 34 | 42 | PP754621 | |||
| D1/MR10/B4 | Bifidobacterium choladohabitans | 19 | 25 | 21 | 19 | - | - | PP754622 |
| D1/MR11 | Lactobacillus kullabergensis | - | - | - | - | - | - | PP754623 |
| D1/MR12 | Lactobacillus sp. (p.n.sp.) | - | - | - | - | - | - | PP754624 |
| D1/RO1 | Lactobacillus helsingborgensis | 21 | 36 | - | 22 | - | - | PP754625 |
| D1/RO2 | Lactobacillus helsingborgensis | 32 | 22 | 32 | 34 | - | - | PP754626 |
| KM1 | Lactobacillus kimbladii | - | - | - | - | - | - | PP754627 |
| KM10 | Lactobacillus apis | - | - | - | - | - | - | PP754628 |
| KR1 | Lactobacillus melliventris | - | - | 36 | 22 | - | - | PP754629 |
| KR2 | Lactobacillus melliventris | - | - | - | - | - | - | PP754630 |
| KR4 | Lactobacillus melliventris | - | - | - | - | - | - | PP754631 |
| KR5 | Lactobacillus helsingborgensis | 20 | 18 | - | 24 | 26 | - | PP754632 |
| KR7 | Lactobacillus melliventris | - | - | - | - | - | - | PP754633 |
| KR8 | Lactobacillus helsingborgensis | - | - | 21 | 17 | - | - | PP754634 |
| KT3 | Lactobacillus huangpiensis | - | - | - | - | - | - | PP754635 |
| P1/MR1 | Bifidobacterium asteroides | 18 | 19 | 18 | 18 | - | 21 | PP754636 |
| P1/MR2 | Lactobacillus melliventris | - | 17 | - | 17 | - | - | PP754637 |
| P1/MR8 | Lactobacillus apis | - | 24 | 18 | 17 | - | 18 | PP754638 |
| P1/MR10 | Lactobacillus apis | - | 31 | - | - | 17 | - | PP754639 |
| P1/MR11 | Lactobacillus melliventris | - | - | - | - | - | - | PP754640 |
| P1/MR12 | Lactobacillus helsingborgensis | - | 44 | 34 | 40 | - | - | PP754641 |
| P1/TP1 | Bifidobacterium asteroides | - | - | 21 | - | - | - | PP754642 |
| P1/TP2 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | - | PP754643 |
| P1/TP3 | Bifidobacterium asteroides | 17 | 18 | 18 | - | - | - | PP754644 |
| P1/TP8 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | - | PP754645 |
| P1/TP9 | Bifidobacterium apousia | - | - | - | 48 | - | - | PP754646 |
| P1/TP10 | Bifidobacterium asteroides | - | 46 | - | 26 | - | - | PP754647 |
| P1/TP11 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | - | PP754648 |
| P1/TP12 | Bifidobacterium asteroides | - | - | - | - | - | - | PP754649 |
| VM7 | Lactobacillus apis | - | - | 18 | - | - | - | PP754650 |
| VM9 | Bifidobacterium polysaccharolyticum | 19 | - | 25 | 20 | - | - | PP754651 |
| VM10 | Bifidobacterium polysaccharolyticum | 20 | 28 | 21 | 17 | - | - | PP754652 |
| VR5 | Lactobacillus helsingborgensis | 22 | 22 | 24 | 26 | 22 | - | PP754653 |
| VR8 | Lactobacillus helsingborgensis | - | - | - | - | - | - | PP754654 |
| VR9 | Lactobacillus melliventris | - | - | - | - | - | - | PP754655 |
| VT5 | Lactobacillus apis | 26 | 34 | 26 | 24 | - | - | PP754656 |
| VT6 | Lactobacillus apis | - | - | - | - | - | - | PP754657 |
| VT9 | Lactobacillus melliventris | - | - | - | - | - | - | PP754658 |
| VSM/BH8 | Bifidobacterium asteroides | - | - | - | - | - | - | PP754659 |
| VSM/mBH3 | Bifidobacterium choladohabitans | - | - | - | - | - | - | PP754660 |
| VSM/mBH4 | Bifidobacterium mellis | - | - | - | 30 | - | 20 | PP754661 |
| VSM/mBH5 | Bifidobacterium polysaccharolyticum | - | - | - | - | - | 24 | PP754662 |
| VSM/mBH6 | Bifidobacterium polysaccharolyticum | - | - | - | - | - | - | PP754663 |
| VSM/mBH8 | Bifidobacterium choladohabitans | - | - | 28 | 21 | - | 21 | PP754664 |
| VSM/mBH9 | Bifidobacterium polysaccharolyticum | - | - | - | - | - | 23 | PP754665 |
| VSM/RO2 | Lactobacillus helsingborgensis | - | - | - | - | - | - | PP754666 |
| VSM/RO4 | Lactobacillus helsingborgensis | 48 | 36 | 52 | > 70 | - | - | PP754667 |
| VSM/RO5 | Lactobacillus kimbladii | - | - | - | - | - | - | PP754668 |
| VSM/RO6 | Lactobacillus melliventris | 18 | 18 | 24 | 21 | - | - | PP754669 |
| VSM/RO7 | Lactobacillus helsingborgensis | - | - | 21 | 27 | - | 23 | PP754670 |
| VSM/RO8 | Lactobacillus helsingborgensis | - | - | - | - | - | - | PP754671 |
| VU/BH3 | Lactobacillus kimbladii | - | - | - | - | - | - | PP754672 |
| VU/mBH2 | Bifidobacterium polysaccharolyticum | - | - | - | - | - | 21 | PP754673 |
| VU/mBH4 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | 20 | PP754674 |
| VU/mBH6 | Bifidobacterium polysaccharolyticum | - | - | - | - | - | 18 | PP754675 |
| VU/mBH7 | Bifidobacterium polysaccharolyticum | - | - | - | - | - | - | PP754676 |
| VU/mBH9 | Bifidobacterium asteroides | - | - | - | - | - | - | PP754677 |
| VU/RO1 | Lactobacillus melliventris | - | - | - | - | - | - | PP754678 |
| VU/RO3 | Lactobacillus kimbladii | - | - | - | - | - | - | PP754679 |
| VU/RO6 | Lactobacillus kullabergensis | 36 | 26 | 31 | 51 | - | - | PP754680 |
| VU/RO8 | L. kullabergensis | - | - | - | - | - | 18 | PP754681 |
| VU/RO9 | L. melliventris | - | - | 20 | 36 | - | - | PP754682 |
| 1BB/B1 | Bifidobacterium sp. (p.n.sp.) | >70 | 33 | 26 | - | - | 25 | PP754683 |
| 1BB/B2 | Bifidobacterium sp. (p.n.sp.) | 24 | - | 23 | 22 | - | 25 | PP754684 |
| 1BB/MF1 | Bifidobacterium asteroides | 18 | - | 25 | 51 | - | 19 | PP754685 |
| 1BB/MF4 | Bifidobacterium asteroides | 24 | 24 | 18 | - | - | - | PP754686 |
| 1BB/MR1 | Bombilactobacillus mellis | - | - | - | - | - | - | PP754687 |
| 1BB/MR2 | Lactobacillus apis | - | - | 27 | - | - | - | PP754688 |
| 1BB/MR6 | Lactobacillus apis | - | - | 17 | 17 | - | 23 | PP754689 |
| 1BB/MR8 | Lactobacillus apis | >70 | 33 | 30 | - | - | 22 | PP754690 |
| 1BB/MR10 | Lactobacillus apis | - | - | 16 | 16 | - | 23 | PP754691 |
| 1BB/RO1 | Lactobacillus melliventris | 39 | 27 | - | 24 | - | - | PP754692 |
| 1BB/RO3 | Lactobacillus melliventris | >70 | 26 | 26 | - | - | - | PP754693 |
| 1BB/RO7 | Lactobacillus apis | - | - | - | - | - | 18 | PP754694 |
| 1BB/RO8 | Lactobacillus melliventris | - | - | - | - | - | - | PP754695 |
| 1BB/TS1 | Bifidobacterium sp. (p.n.sp.) | - | - | - | 16 | - | - | PP754696 |
| 1BB/TS3 | Bifidobacterium polysaccharolyticum | - | - | - | 20 | - | - | PP754697 |
| 1BB/TS7 | Bifidobacterium sp. (p.n.sp.) | 22 | 18 | 29 | 19 | - | - | PP754698 |
| 2BB/B1 | Bifidobacterium sp. (p.n.sp.) | 27 | 46 | - | 16 | - | 19 | PP754699 |
| 2BB/B7 | Bifidobacterium choladohabitans | 22 | - | 22 | 20 | - | 25 | PP754700 |
| 2BB/MF2 | Bifidobacterium polysaccharolyticum | - | - | 22 | 26 | - | - | PP754701 |
| 2BB/MF3 | Lactobacillus melliventris | - | - | - | 20 | - | - | PP754702 |
| 2BB/MR5 | Lactobacillus kullabergensis | - | - | - | - | - | - | PP754703 |
| 2BB/MR8 | Lactobacillus apis | - | - | 20 | 18 | - | - | PP754704 |
| 2BB/RO1 | Lactobacillus melliventris | - | - | - | - | - | - | PP754705 |
| 2BB/RO2 | Lactobacillus melliventris | - | - | - | - | - | - | PP754706 |
| 2BB/RO6 | Lactobacillus apis | - | - | 22 | 29 | - | - | PP754707 |
| 2BB/RO7 | Lactobacillus melliventris | - | - | 25 | 25 | - | - | PP754708 |
| SMTP/2 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | - | PP754709 |
| SMTP/4 | Bifidobacterium sp. (p.n.sp.) | - | - | - | - | - | - | PP754710 |
| SMTP/6 | Bifidobacterium asteroides | 24 | 21 | 17 | 18 | - | - | PP754711 |
| STP/2 | Lactobacillus helsingborgensis | - | - | - | - | - | - | PP754712 |
| STP/3 | Lactobacillus apis | - | - | - | - | - | - | PP754713 |
| STP/4 | Lactobacillus apis | - | - | - | - | - | - | PP754714 |
| STP/5 | Lactobacillus apis | - | - | - | - | - | - | PP754715 |
| STP/6 | Lactobacillus melliventris | - | - | - | - | - | - | PP754716 |
| SR1/4 | Lactobacillus helsingborgensis | - | - | - | - | - | - | PP754717 |
| SR2/2 | Lactobacillus sp. (p.n.sp.) | - | - | - | 16 | - | - | PP754718 |
| SR2/8 | Lactobacillus melliventris | - | - | - | - | - | - | PP754719 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dengiz, B.; Killer, J.; Havlík, J.; Dobeš, P.; Hyršl, P. Selection of Probiotics for Honey Bees: The In Vitro Inhibition of Paenibacillus larvae, Melissococcus plutonius, and Serratia marcescens Strain Sicaria by Host-Specific Lactobacilli and Bifidobacteria. Microorganisms 2025, 13, 1159. https://doi.org/10.3390/microorganisms13051159
Dengiz B, Killer J, Havlík J, Dobeš P, Hyršl P. Selection of Probiotics for Honey Bees: The In Vitro Inhibition of Paenibacillus larvae, Melissococcus plutonius, and Serratia marcescens Strain Sicaria by Host-Specific Lactobacilli and Bifidobacteria. Microorganisms. 2025; 13(5):1159. https://doi.org/10.3390/microorganisms13051159
Chicago/Turabian StyleDengiz, Buse, Jiří Killer, Jaroslav Havlík, Pavel Dobeš, and Pavel Hyršl. 2025. "Selection of Probiotics for Honey Bees: The In Vitro Inhibition of Paenibacillus larvae, Melissococcus plutonius, and Serratia marcescens Strain Sicaria by Host-Specific Lactobacilli and Bifidobacteria" Microorganisms 13, no. 5: 1159. https://doi.org/10.3390/microorganisms13051159
APA StyleDengiz, B., Killer, J., Havlík, J., Dobeš, P., & Hyršl, P. (2025). Selection of Probiotics for Honey Bees: The In Vitro Inhibition of Paenibacillus larvae, Melissococcus plutonius, and Serratia marcescens Strain Sicaria by Host-Specific Lactobacilli and Bifidobacteria. Microorganisms, 13(5), 1159. https://doi.org/10.3390/microorganisms13051159






