Hemolysin-like Protein of ‘Candidatus Phytoplasma Mali’ Is an NTPase and Binds Arabidopsis thaliana Toc33
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of HLPPM19 DNA
2.2. Yeast Two-Hybrid (Y2H) Screening
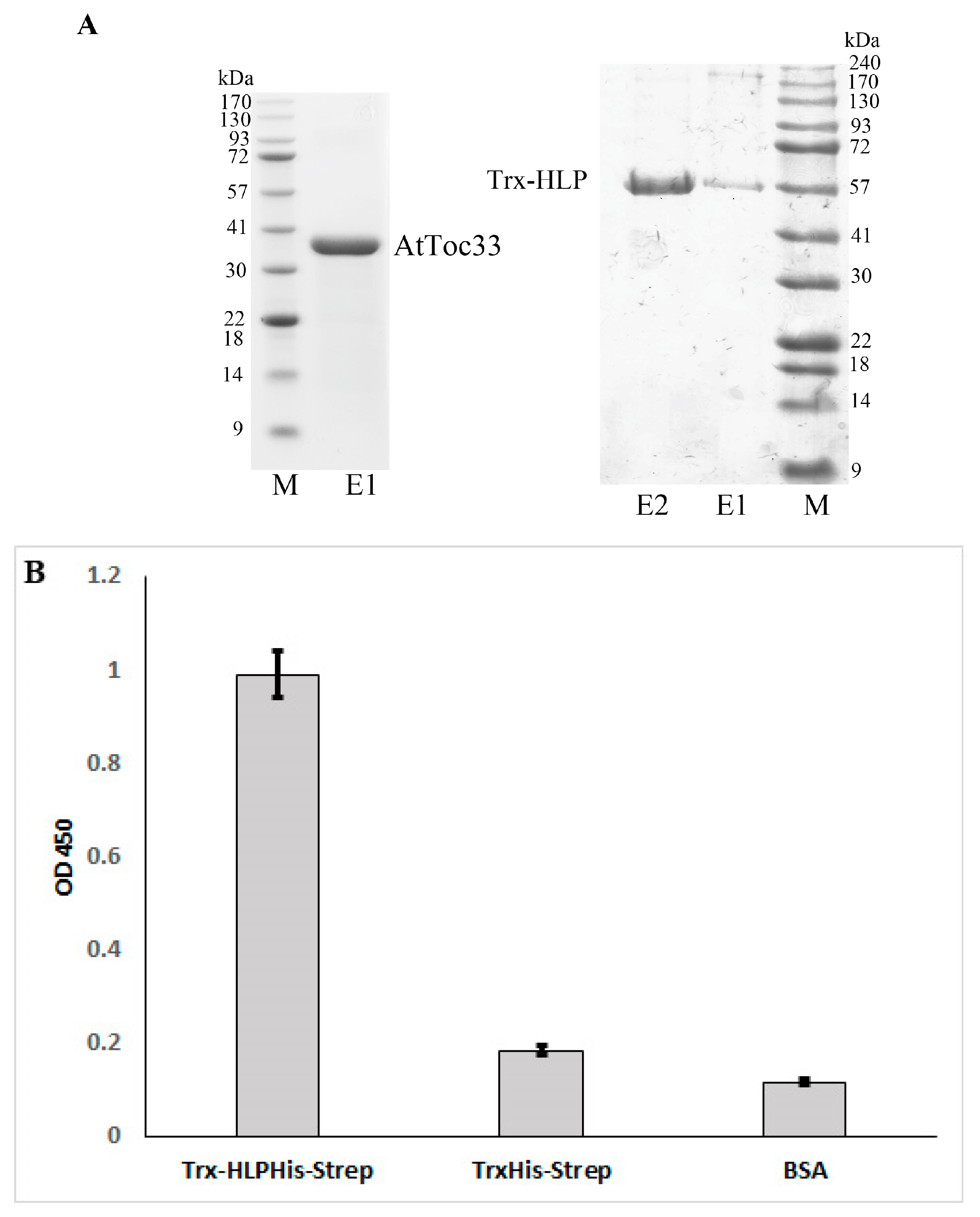
2.3. Recombinant Expression and Purification of HLPPM19 and AtToc33
2.4. ELISA
2.5. GTP-Binding Assay
2.6. Immuno-Fluorescence Histochemical Staining
2.7. Nucleotidase Activity Assay
2.8. Thin Layer Chromatography (TLC) Analysis
3. Results
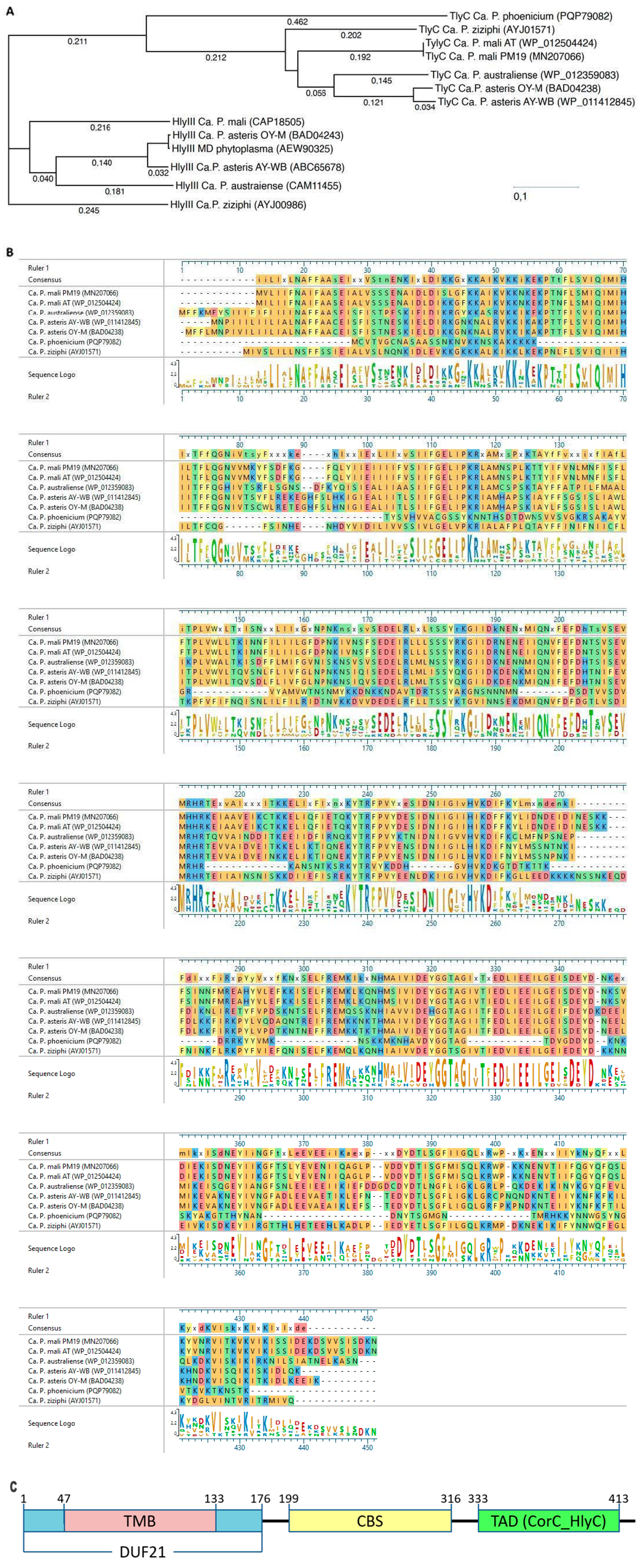
3.1. Sequence Analysis of HLPPM19
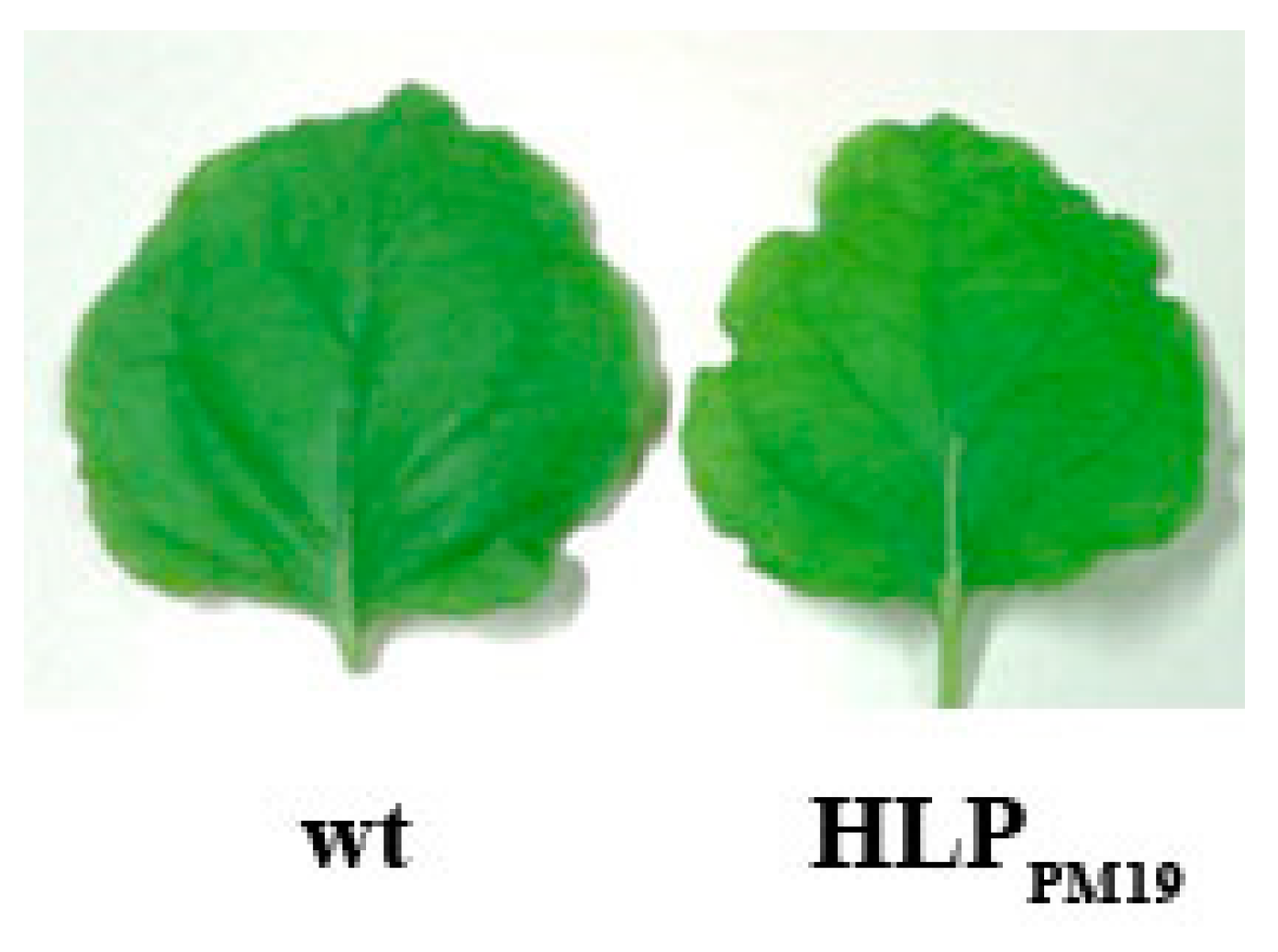
3.2. Recombinant Expression of HLPPM19 Does Not Change the Plant Phenotype
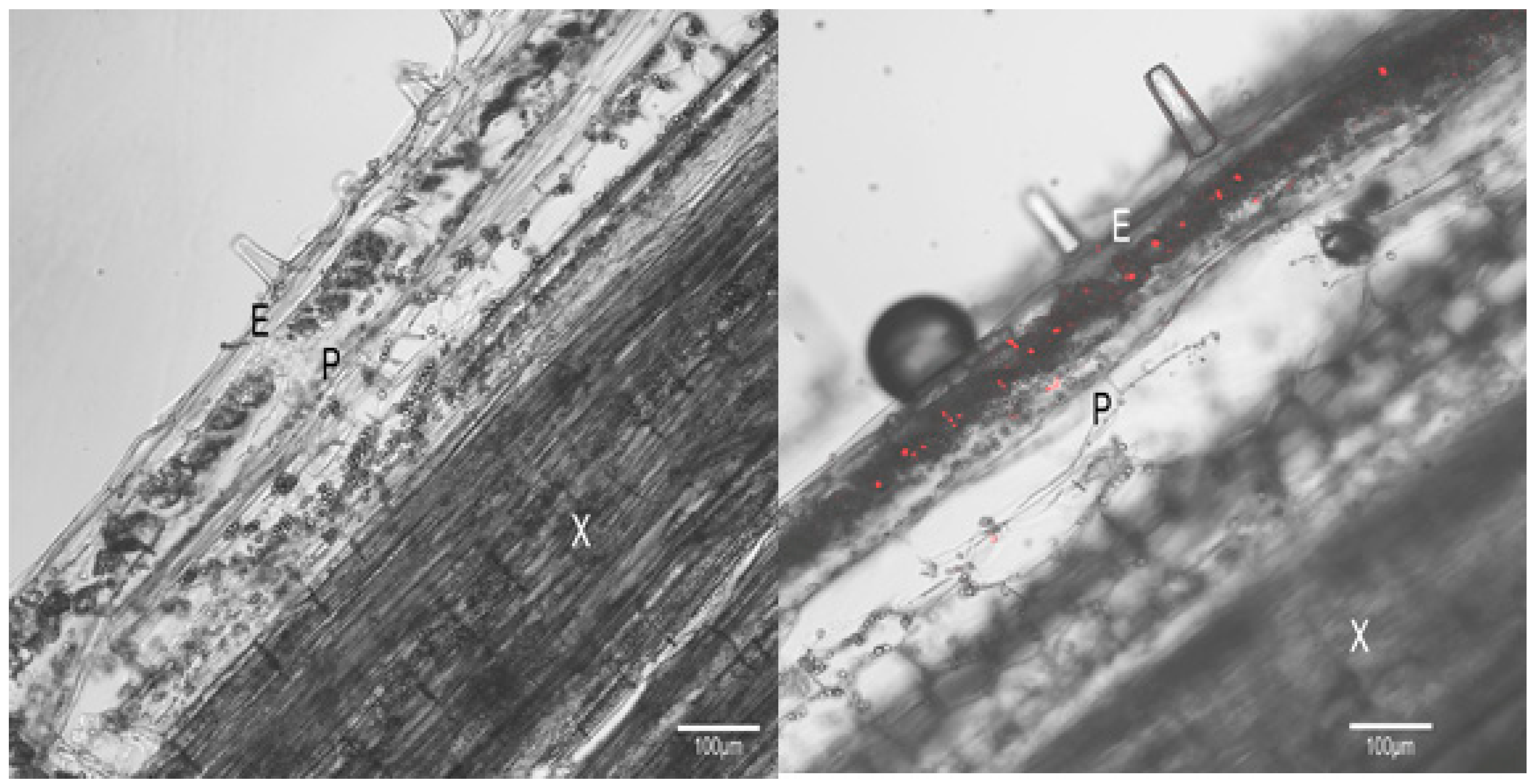
3.3. HLP Is Located on the Phytoplasma Membrane
3.4. HLPPM19 Interacts with AtToc33 in Y2H Assay
3.5. Amino Acids 133 to 198 of HLPPM19 Are Important for Interaction with AtToc33 in Y2H
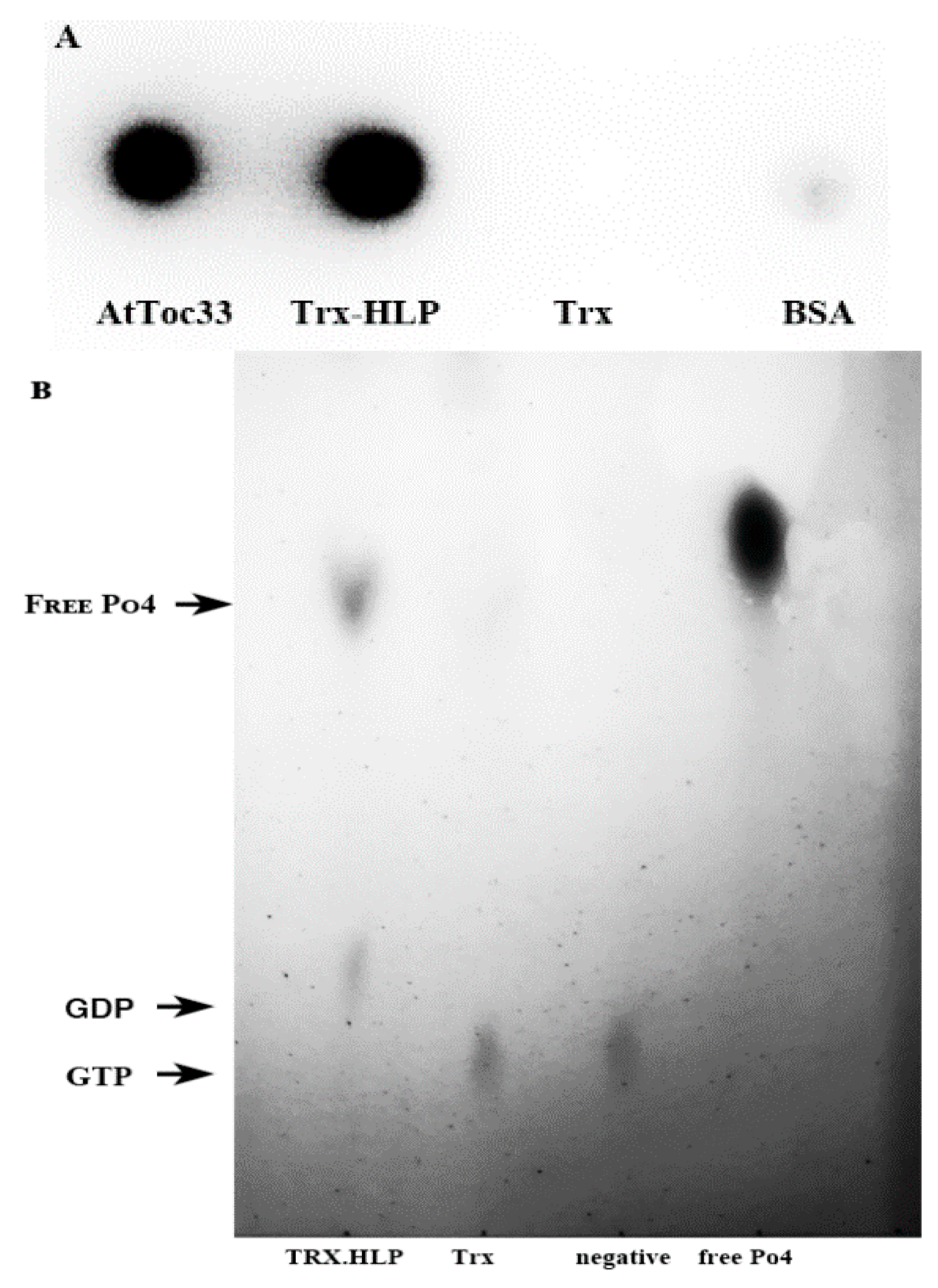
3.6. HLPPM19-ΔTM Binds AtToc33 in ELISA
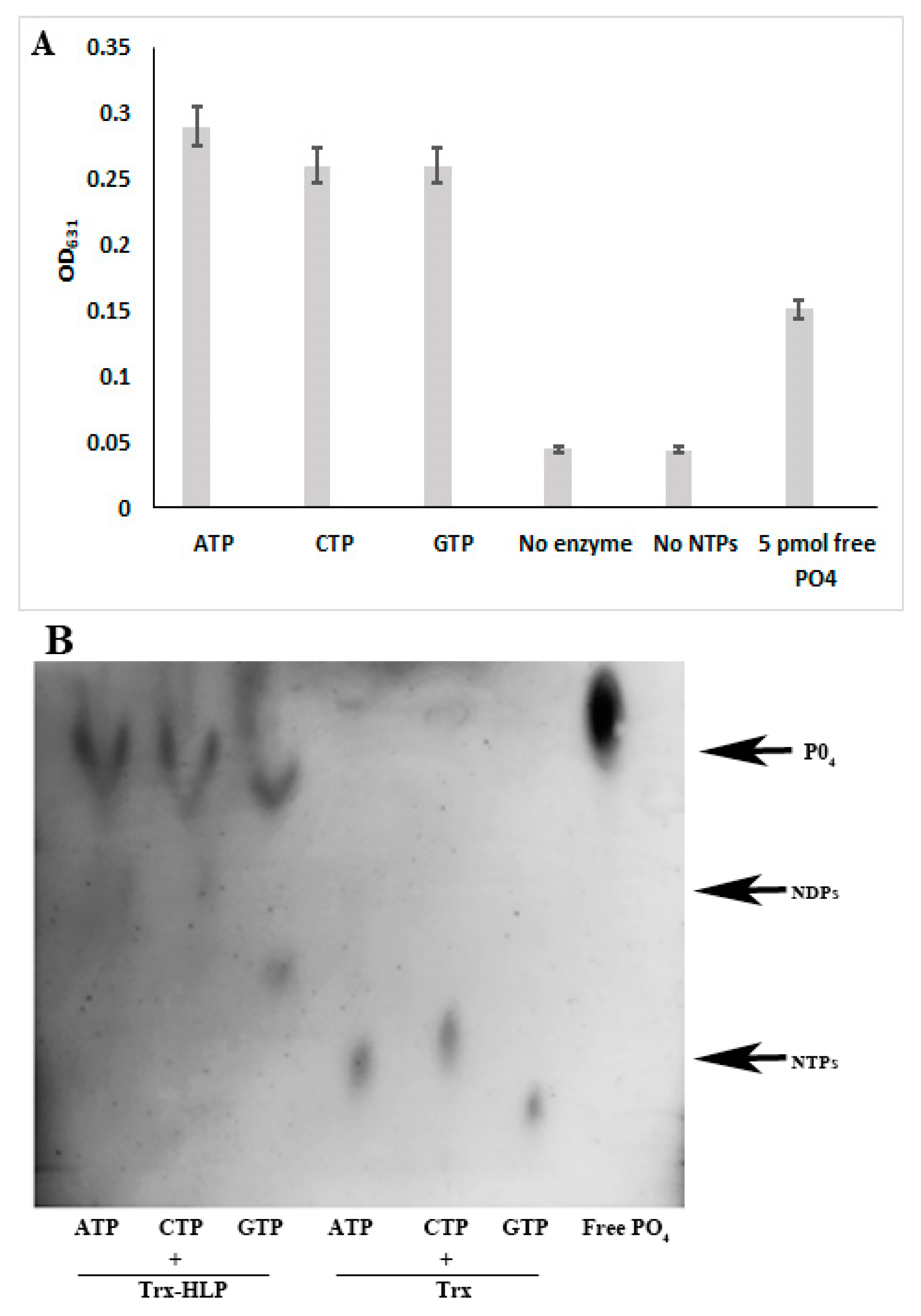
3.7. HLPPM19 Binds and Hydrolyzes GTP
3.8. HLPPM19 Functions as Nucleosidase
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, R.; Bai, B.; Li, D.; Wang, J.; Huang, W.; Wu, Y. Phytoplasma: A plant pathogen that cannot be ignored in agricultural production—Research progress and outlook. Mol. Plant Pathol. 2024, 25, e13437. [Google Scholar] [CrossRef] [PubMed]
- Oshima, K.; Kakizawa, S.; Nishigawa, H.; Jung, H.Y.; Wei, W.; Suzuki, S.; Arashida, R.; Nakata, D.; Miyata, S.; Ugaki, M.; et al. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 2004, 36, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, J.; Ewing, A.; Miller, S.A.; Jancso, R.A.; Shevchenko, D.V.; Tsukerman, K.; Walunas, T.; Lapidus, A.; Campbell, J.W.; et al. Living with genome instability: The adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 2006, 188, 3682–3696. [Google Scholar] [CrossRef] [PubMed]
- Tran-Nguyen, L.T.T.; Kube, M.; Schneider, B.; Reinhardt, R.; Gibb, K.S. Comparative genome analysis of ‘Candidatus Phytoplasma australiense’ (subgroup tuf-Australia I; rp-A) and ‘Ca. Phytoplasma asteris’ strains OY-M and AY-WB. J. Bacteriol. 2008, 190, 3979–3991. [Google Scholar] [CrossRef]
- Quaglino, F.; Kube, M.; Jawhari, M.; Abou-Jawdah, Y.; Siewert, C.; Choueiri, E.; Sobh, H.; Casati, P.; Tedeschi, R.; Lova, M.M.; et al. ‘Candidatus Phytoplasma phoenicium’ associated with almond witches’ broom disease: From draft genome to genetic diversity among strain populations. BMC Microbiol. 2015, 15, 148. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Jiao, Q. Comparative genome analysis of jujube witches’-broom phytoplasma, an obligate pathogen that causes jujube witches’-broom disease. BMC Genom. 2018, 19, 689. [Google Scholar] [CrossRef]
- Ji, X.; Gai, Y.; Lu, B.; Zheng, C.; Mu, Z. Shotgun proteomic analysis of mulberry dwarf phytoplasma. Proteome Sci. 2010, 8, 20. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Stilwagen, S.; Ivanova, N.; D’Souza, M.; Bernal, A.; Lykidis, A.; Kapatral, V.; Anderson, I.; Larsen, N.; Los, T.; et al. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl. Acad. Sci. USA 2002, 99, 12403–12408. [Google Scholar] [CrossRef]
- Kube, M.; Schneider, B.; Kuhl, H.; Dandekar, T.; Heitmann, K.; Migdoll, A.M.; Reinhardt, R.; Seemüller, E. The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali’. BMC Genom. 2008, 9, 306. [Google Scholar] [CrossRef]
- Kemp, B.E. Bateman domains and adenosine derivatives form a binding contract. J. Clin. Investig. 2004, 113, 182–184. [Google Scholar] [CrossRef]
- Ignoul, S.; Eggermont, J. CBS domains: Structure, function, and pathology in human proteins. Am. J. Physiol.-Cell Physiol. 2005, 289, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- Jarausch, B.; Schwind, N.; Fuchs, A.; Jarausch, W. Characteristics of the spread of apple proliferation by its vector Cacopsylla picta. Phytopathology 2011, 101, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Strohmayer, A.; Moser, M.; Si-Ammour, A.; Krczal, G.; Boonrod, K. ‘Candidatus Phytoplasma mali’ genome encodes a protein that functions as a E3 Ubiquitin Ligase and could inhibit plant basal defense. Mol. Plant-Microbe Interact. 2019, 32, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Weibel, P.; Hiltbrunner, A.; Brand, L.; Kessler, F. Dimerization of Toc-GTPases at the chloroplast protein import machinery. J. Biol. Chem. 2003, 278, 37321–37329. [Google Scholar] [CrossRef]
- Munteanu, B.; Braun, M.; Boonrod, K. Improvement of PCR reaction conditions for site-directed mutagenesis of big plasmids. J. Zhejiang Univ. Sci. B 2012, 13, 244–247. [Google Scholar] [CrossRef]
- Carson, S.D. Ammonium molybdate-stannous chloride determination of orthophosphate in the presence of Triton X-100. Anal Biochem. 1976, 75, 472–477. [Google Scholar] [CrossRef]
- Forestan, C.; Varotto, S. Auxin Immunolocalization in Plant Tissues. In Plant Organogenesis: Methods in Molecular Biology; De Smet, I., Ed.; Humana Press: Totowa, NJ, USA, 2013. [Google Scholar]
- Murphy, J.; Riley, J.P. A Single-Solution Method for the Determination of Soluble Phosphate in Sea Water. J. Mar. Biol. Assoc. U. K. 1958, 37, 9–14. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar]
- Truernit, E.; Sauer, N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 1995, 196, 564–570. [Google Scholar] [CrossRef]
- Konnerth, A.; Krczal, G.; Boonrod, K. Immunodominant membrane proteins of phytoplasmas. Microbiology 2016, 162, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Hiltbrunner, A.; Bauer, J.; Vidi, P.A.; Infanger, S.; Weibel, P.; Hohwy, M.; Kessler, F. Targeting of an abundant cytosolic form of the protein import receptor at Toc159 to the outer chloroplast membrane. J. Cell Biol. 2001, 154, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Kessler, F.; Blobel, G.; Patel, H.A.; Schnell, D.J. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 1994, 266, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, P.; Soll, J. Toc, Tic, and chloroplast protein import. Biochim. Biophys. Acta 2002, 1509, 177–189. [Google Scholar] [CrossRef]
- Schnell, D.J.; Kessler, F.; Blobel, G. Isolation of components of the chloroplast protein import machinery. Science 1994, 266, 1007–1012. [Google Scholar] [CrossRef]
- Christensen, N.M.; Axelsen, K.B.; Nicolaisen, M.; Schulz, A. Phytoplasmas and their interactions with hosts. Trends Plant Sci. 2005, 10, 526–535. [Google Scholar] [CrossRef]
- Rudzińska-Langwald, A.; Kaminska, M. Changes of ultrastructure and cytoplasmic free calcium in Gladiolus x hybridus Van Houtte roots infected by aster yellows phytoplasma. Acta Soc. Bot. Pol. 2011, 72, 269–282. [Google Scholar] [CrossRef]
- Esau, K. Comparative Structure of Companion Cells and Phloem Parenchyma Cells in Mimosa pudica L. Ann. Bot. 1973, 37, 625–632. [Google Scholar] [CrossRef]
- Paramonova, N.V.; Krasavina, M.S.; Sokolova, S.V. Ultrastructure of chloroplasts in phloem companion cells and mesophyll cells as related to the stimulation of sink activity by cytokinins. Russ. J. Plant Physiol. 2002, 49, 187–195. [Google Scholar] [CrossRef]
- Cayla, T.; Batailler, B.; Le Hir, R.; Revers, F.; Anstead, J.A.; Thompson, G.A.; Grandjean, O.; Dinant, S. Live imaging of companion cells and sieve elements in Arabidopsis leaves. PLoS ONE 2015, 25, e0118122. [Google Scholar] [CrossRef]
- Dhanoa, P.K.; Richardson, L.G.; Smith, M.D.; Gidda, S.K.; Henderson, M.P.; Andrews, D.W.; Mullen, R.T. Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 2010, 4, e10098. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, H.; Combe, J.; Patel, R.; Agne, B.; Martin, M.; Kessler, F.; Jarvis, P. Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J. 2010, 63, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, P.; Chen, L.J.; Li, H.; Peto, C.A.; Fankhauser, C.; Chory, J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 1998, 2, 100–103. [Google Scholar] [CrossRef]
- Kubis, S.; Baldwin, A.; Patel, R.; Razzaq, A.; Dupree, P.; Lilley, K.; Kurth, J.; Leister, D.; Jarvis, P. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 2003, 15, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coggill, P.; Finn, R.D. DUFs: Families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1148–1152. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, 281–288. [Google Scholar] [CrossRef]
- Tedeschi, R.; Alma, A. Transmission of apple proliferation phytoplasma by Cacopsylla melanoneura (Homoptera: Psyllidae). J. Econ. Entomol. 2004, 97, 8–13. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boonrod, K.; Konnerth, A.; Braun, M.; Krczal, G. Hemolysin-like Protein of ‘Candidatus Phytoplasma Mali’ Is an NTPase and Binds Arabidopsis thaliana Toc33. Microorganisms 2025, 13, 1150. https://doi.org/10.3390/microorganisms13051150
Boonrod K, Konnerth A, Braun M, Krczal G. Hemolysin-like Protein of ‘Candidatus Phytoplasma Mali’ Is an NTPase and Binds Arabidopsis thaliana Toc33. Microorganisms. 2025; 13(5):1150. https://doi.org/10.3390/microorganisms13051150
Chicago/Turabian StyleBoonrod, Kajohn, Alisa Konnerth, Mario Braun, and Gabi Krczal. 2025. "Hemolysin-like Protein of ‘Candidatus Phytoplasma Mali’ Is an NTPase and Binds Arabidopsis thaliana Toc33" Microorganisms 13, no. 5: 1150. https://doi.org/10.3390/microorganisms13051150
APA StyleBoonrod, K., Konnerth, A., Braun, M., & Krczal, G. (2025). Hemolysin-like Protein of ‘Candidatus Phytoplasma Mali’ Is an NTPase and Binds Arabidopsis thaliana Toc33. Microorganisms, 13(5), 1150. https://doi.org/10.3390/microorganisms13051150






