Characterization of Sex-Based Differences in Gut Microbiota That Correlate with Suppression of Lupus in Female BWF1 Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Animal Facility Descriptions
2.2. Animal Handling and Fecal Sample Collection
2.3. Cecal (Microbiota) Transplant Preparation and Processing
2.4. Cecal Microbiota Transfer
2.5. Measurement of Proteinuria (Kidney Disease Score Assessment)
2.6. Microbiota Composition Analysis and Statistical Analysis
2.7. Statistical Analyses
3. Results
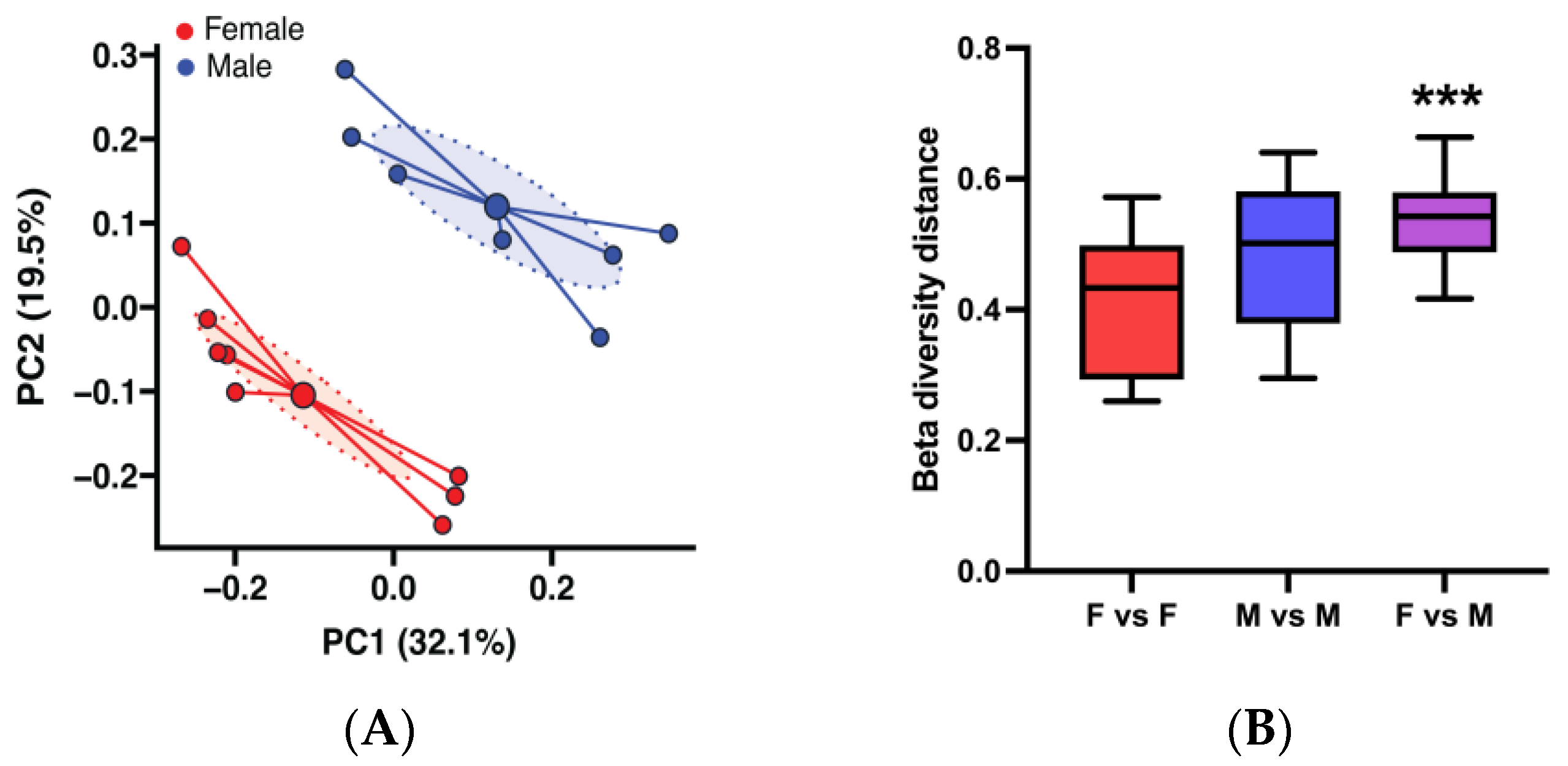
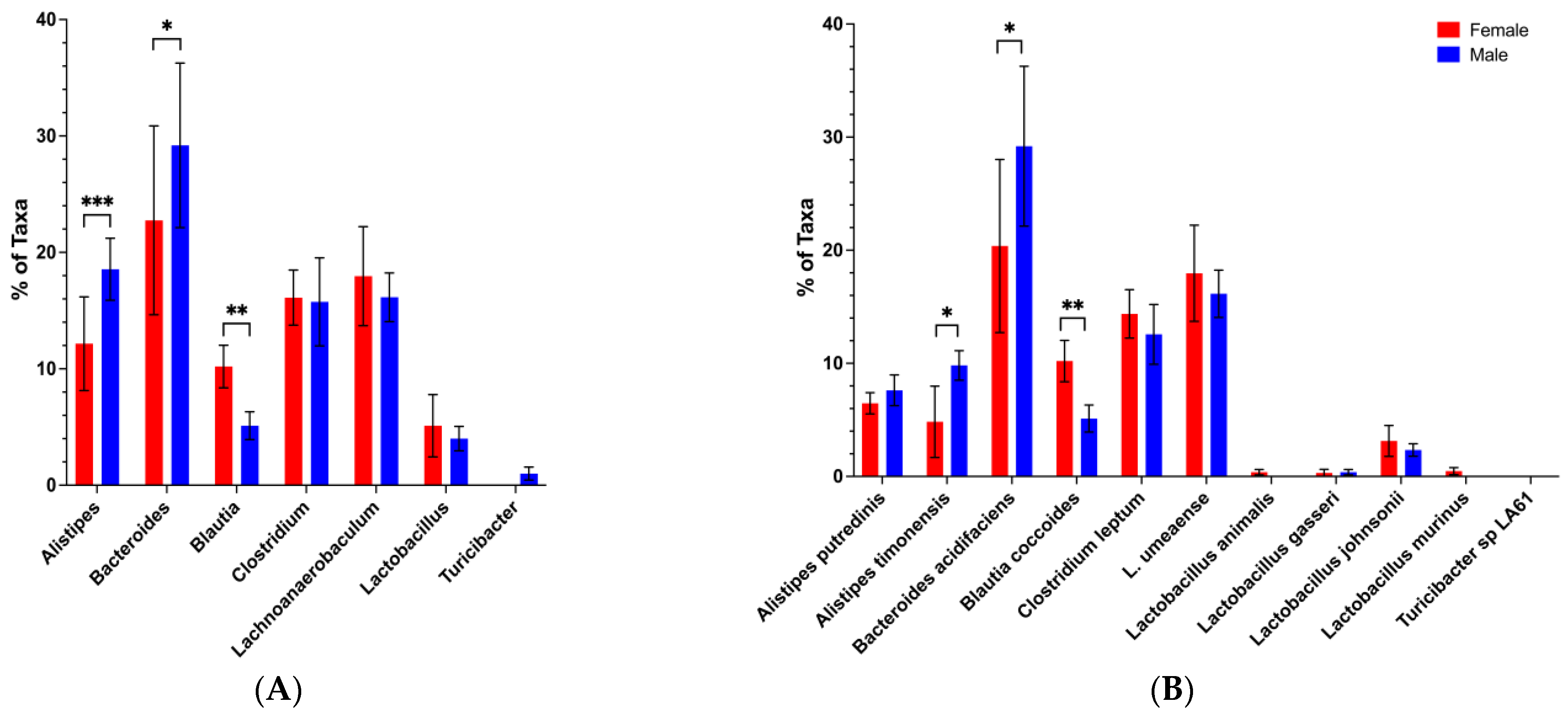
3.1. Sex-Based Differences in Gut Microbiota Composition and Function in the BWF1 Mouse Model of Lupus
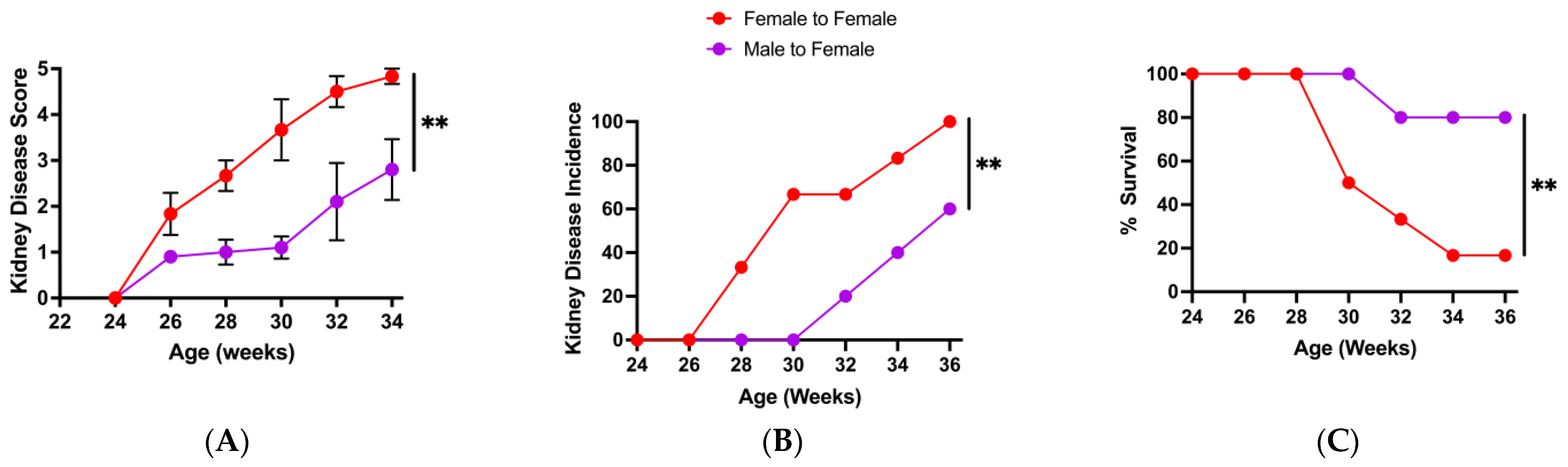
3.2. Change in Microbiota Composition Is Associated with Loss of Protective Male Microbiota Transfer Phenotype
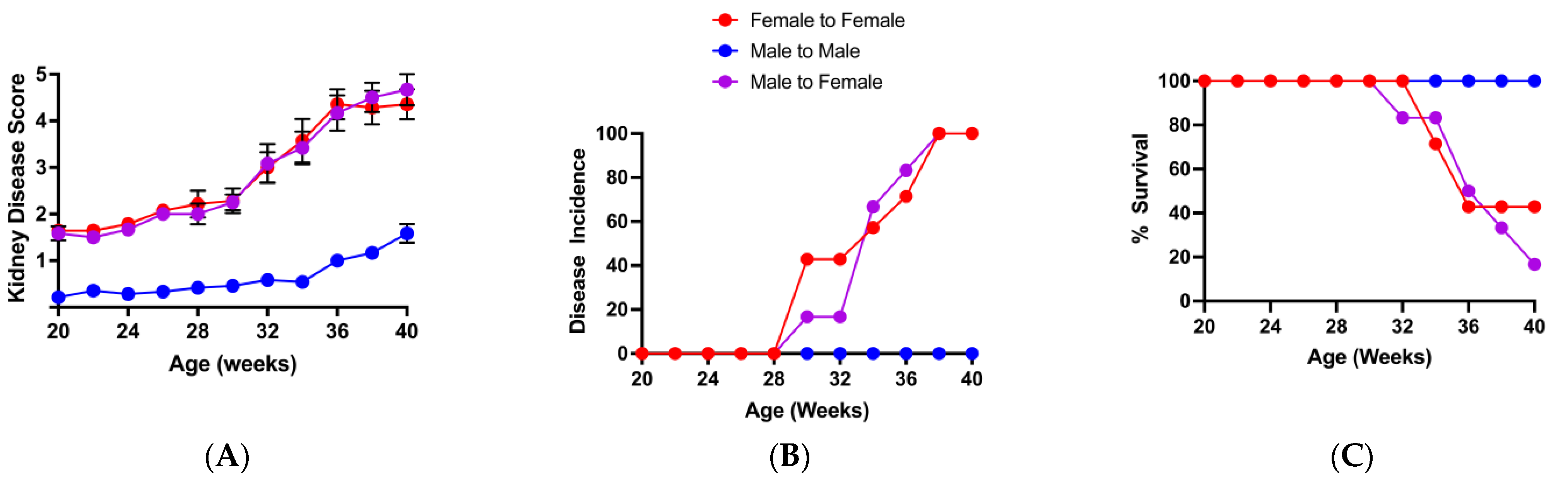
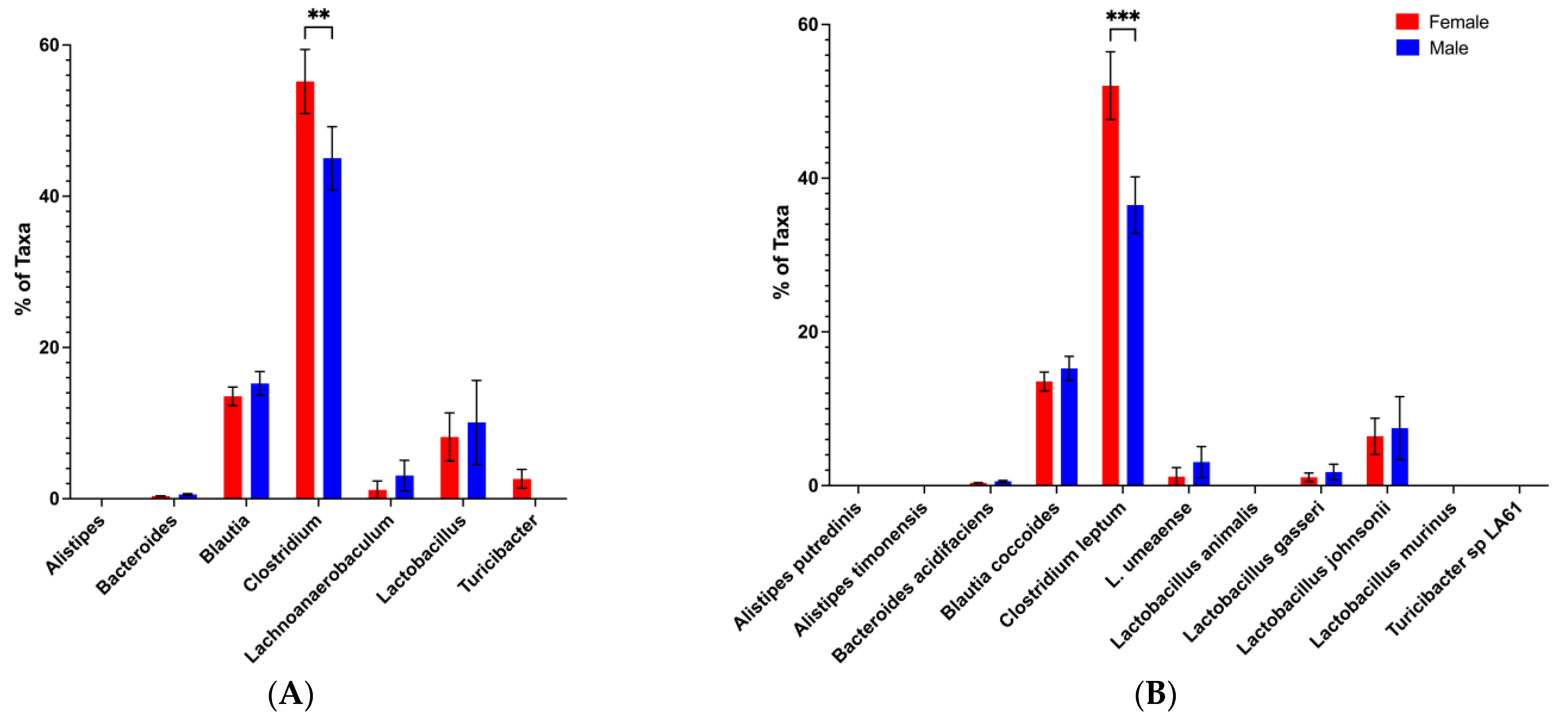
3.3. Loss of the Potential Male Microbiota Protective Phenotype Is Associated with Genus and Species-Level Changes in Male BWF1 Microbiota
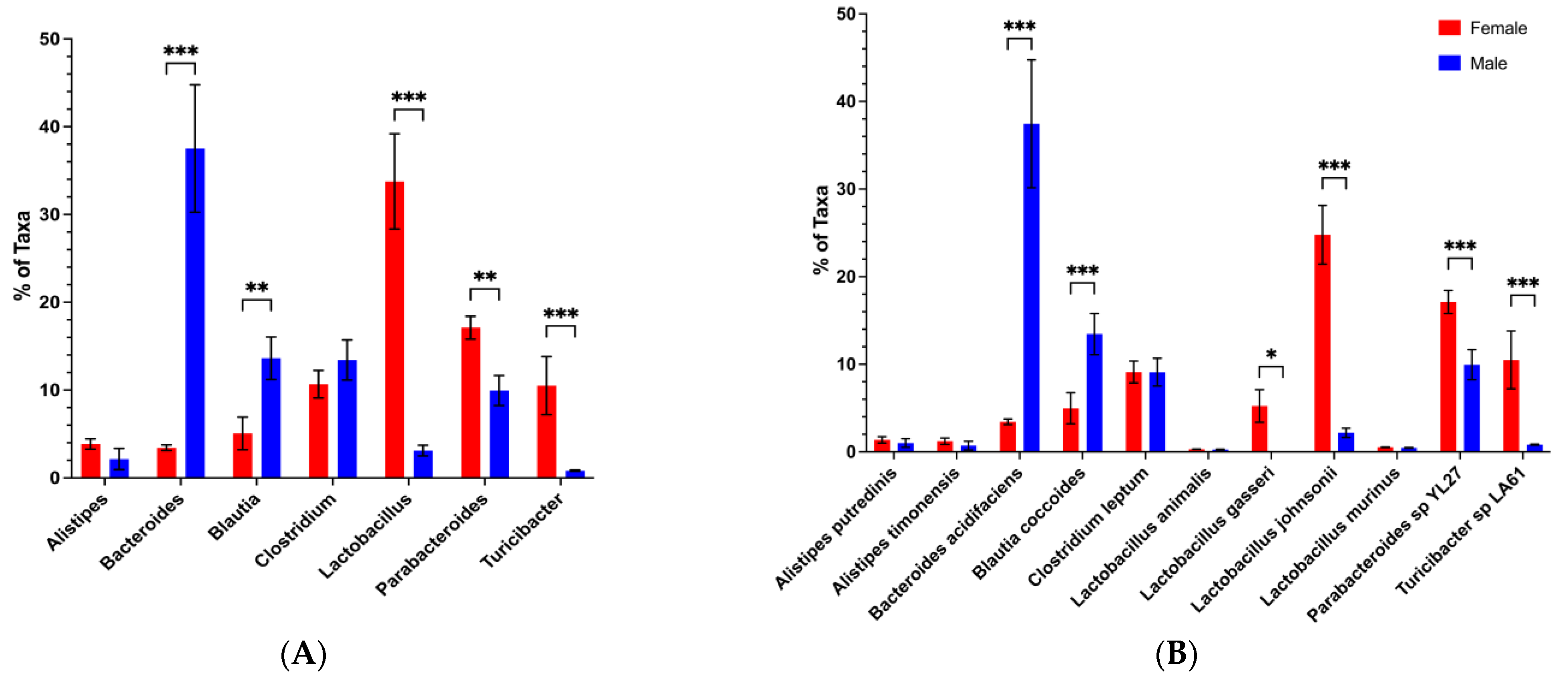
3.4. Restoration of Potential Male Microbiota Protective Phenotype Is Associated with High Levels of Bacteroides and Low Levels of Clostridium
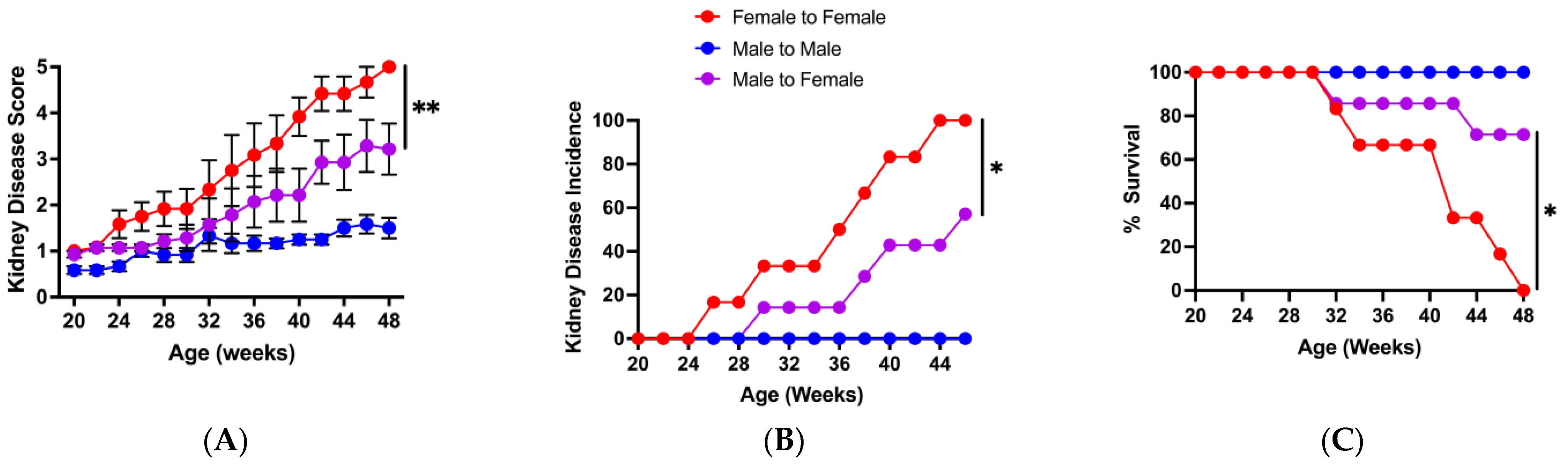
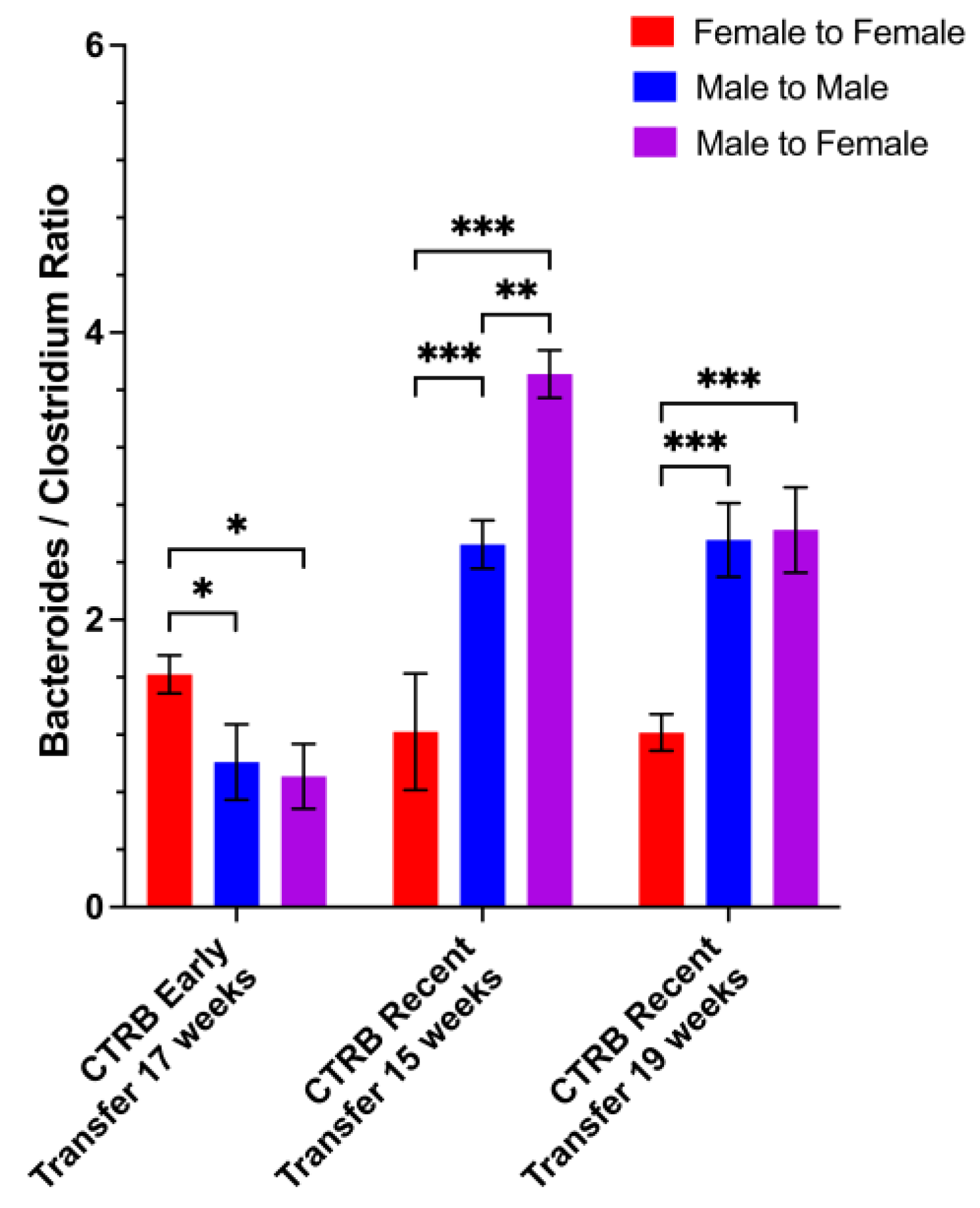
3.5. Protection from Disease by Male-to-Female Microbiota Transfer Correlates with Elevated Bacteroides and Decreased Clostridium Abundances in the Recipients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SLE | Systemic lupus erythematosus |
| BWF1 | NZWxNZBF1 |
| NOD | Non-obese diabetes |
| Tregs | Regulatory T cells |
| SNF1 | SWRxNZB F1 |
| CTRB | Clinical and Translational Building |
| PCoA | Principal coordinate analysis |
| PERMANOVA | Permutational multivariate analysis of variance |
| ANOVA | Analysis of variance |
| F | Female |
| M | Male |
| DSS | Dextran sodium sulfate |
References
- De Luca, F.; Shoenfeld, Y. The microbiome in autoimmune diseases. Clin. Exp. Immunol. 2019, 195, 74–85. [Google Scholar] [CrossRef]
- Wang, T.; Sternes, P.R.; Guo, X.K.; Zhao, H.; Xu, C.; Xu, H. Autoimmune diseases exhibit shared alterations in the gut microbiota. Rheumatology 2024, 63, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yuan, W.; Yang, C.; Wang, Z.; Zhang, J.; Xu, D.; Sun, X.; Sun, W. Emerging role of gut microbiota in autoimmune diseases. Front. Immunol. 2024, 15, 1365554. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Yang, K.; He, Q.; Zhu, X.; Long, Z.; Wu, Y.; Chen, J.; Li, Y.; Zeng, J.; Cui, G.; et al. Efficacy and safety of gut microbiota-based therapies in autoimmune and rheumatic diseases: A systematic review and meta-analysis of 80 randomized controlled trials. BMC Med. 2024, 22, 110. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, Y.; Xia, X. Autoimmune diseases and gut microbiota: A bibliometric and visual analysis from 2004 to 2022. Clin. Exp. Med. 2023, 23, 2813–2827. [Google Scholar] [CrossRef]
- d’Afflitto, M.; Upadhyaya, A.; Green, A.; Peiris, M. Association Between Sex Hormone Levels and Gut Microbiota Composition and Diversity-A Systematic Review. J. Clin. Gastroenterol. 2022, 56, 384–392. [Google Scholar] [CrossRef]
- Duan, X.; Nie, Y.; Xie, X.; Zhang, Q.; Zhu, C.; Zhu, H.; Chen, R.; Xu, J.; Zhang, J.; Yang, C.; et al. Sex differences and testosterone interfere with the structure of the gut microbiota through the bile acid signaling pathway. Front. Microbiol. 2024, 15, 1421608. [Google Scholar] [CrossRef]
- Hokanson, K.C.; Hernandez, C.; Deitzler, G.E.; Gaston, J.E.; David, M.M. Sex shapes gut-microbiota-brain communication and disease. Trends Microbiol. 2024, 32, 151–161. [Google Scholar] [CrossRef]
- Katrib, M.; Haddad, R.; Hamdan, Z.; Rida, M.A. The dynamic relationship of gut microbiota with sex hormones in systemic lupus erythematosus. Rheumatologia 2023, 61, 130–136. [Google Scholar] [CrossRef]
- Gleicher, N.; Barad, D.H. Gender as risk factor for autoimmune diseases. J. Autoimmun. 2007, 28, 1–6. [Google Scholar] [CrossRef]
- Libert, C.; Dejager, L.; Pinheiro, I. The X chromosome in immune functions: When a chromosome makes the difference. Nat. Rev. Immunol. 2010, 10, 594–604. [Google Scholar] [CrossRef]
- Helyer, B.; Howie, J. Renal disease associated with positive lupus erythematosus tests in a crossbred strain of mice. Nature 1963, 197, 197. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Harder, J.; Ma, J.; Yin, X.; Zhang, X.; Kosiewicz, M.M. Using Multiple Analytical Platforms to Investigate the Androgen Depletion Effects on Fecal Metabolites in a Mouse Model of Systemic Lupus Erythematosus. J. Proteome Res. 2020, 19, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Dubois, E.L.; Horowitz, R.E.; Demopoulos, H.B.; Teplitz, R. NZB/NZW mice as a model of systemic lupus erythematosus. JAMA 1966, 195, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Burnet, F.M.; Holmes, M.C. The natural history of the NZB/NZW F1 hybrid mouse: A laboratory model of systemic lupus erythematosus. Australas. Ann. Med. 1965, 14, 185–191. [Google Scholar] [CrossRef]
- Azzouz, D.; Omarbekova, A.; Heguy, A.; Schwudke, D.; Gisch, N.; Rovin, B.H.; Caricchio, R.; Buyon, J.P.; Alekseyenko, A.V.; Silverman, G.J. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019, 78, 947–956. [Google Scholar] [CrossRef]
- Greiling, T.M.; Dehner, C.; Chen, X.; Hughes, K.; Iniguez, A.J.; Boccitto, M.; Ruiz, D.Z.; Renfroe, S.C.; Vieira, S.M.; Ruff, W.E.; et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 2018, 10, eaan2306. [Google Scholar] [CrossRef]
- López, P.; de Paz, B.; Rodríguez-Carrio, J.; Hevia, A.; Sánchez, B.; Margolles, A.; Suárez, A. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 2016, 6, 24072. [Google Scholar] [CrossRef]
- Luo, X.M.; Edwards, M.R.; Mu, Q.; Yu, Y.; Vieson, M.D.; Reilly, C.M.; Ahmed, S.A.; Bankole, A.A. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl. Environ. Microbiol. 2018, 84, e02288-17. [Google Scholar] [CrossRef]
- Wang, A.; Zhao, J.; Qin, Y.; Zhang, Y.; Xing, Y.; Wang, Y.; Yu, Z.; Yan, J.; Han, M.; Yuan, J.; et al. Alterations of the gut microbiota in the lupus nephritis: A systematic review. Ren. Fail. 2023, 45, 2285877. [Google Scholar] [CrossRef]
- Xiang, S.; Qu, Y.; Qian, S.; Wang, R.; Wang, Y.; Jin, Y.; Li, J.; Ding, X. Association between systemic lupus erythematosus and disruption of gut microbiota: A meta-analysis. Lupus Sci. Med. 2022, 9, e000599. [Google Scholar] [CrossRef]
- Zegarra-Ruiz, D.F.; El Beidaq, A.; Iniguez, A.J.; Lubrano Di Ricco, M.; Manfredo Vieira, S.; Ruff, W.E.; Mubiru, D.; Fine, R.L.; Sterpka, J.; Greiling, T.M.; et al. A Diet-Sensitive Commensal Lactobacillus Strain Mediates TLR7-Dependent Systemic Autoimmunity. Cell Host Microbe 2019, 25, 113–127.e6. [Google Scholar] [CrossRef]
- Stoll, M.L.; Gavalchin, J. Systemic lupus erythematosus—Messages from experimental models. Rheumatology 2000, 39, 18–27. [Google Scholar] [CrossRef]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Lian, F.P.; Zhang, F.; Zhao, C.M.; Wang, X.X.; Bu, Y.J.; Cen, X.; Zhao, G.F.; Zhang, S.X.; Chen, J.W. Gut microbiota regulation of T lymphocyte subsets during systemic lupus erythematosus. BMC Immunol. 2024, 25, 41. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Zhang, H.; Liao, X.; Lin, K.; Liu, H.; Edwards, M.R.; Ahmed, S.A.; Yuan, R.; Li, L.; Cecere, T.E.; et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 2017, 5, 73. [Google Scholar] [CrossRef]
- Perl, A.; Hanczko, R.; Lai, Z.W.; Oaks, Z.; Kelly, R.; Borsuk, R.; Asara, J.M.; Phillips, P.E. Comprehensive metabolome analyses reveal N-acetylcysteine-responsive accumulation of kynurenine in systemic lupus erythematosus: Implications for activation of the mechanistic target of rapamycin. Metabolomics 2015, 11, 1157–1174. [Google Scholar] [CrossRef]
- Garcia-Gomez, E.; Gonzalez-Pedrajo, B.; Camacho-Arroyo, I. Role of sex steroid hormones in bacterial-host interactions. Biomed. Res. Int. 2013, 2013, 928290. [Google Scholar] [CrossRef]
- Haro, C.; Rangel-Zuniga, O.A.; Alcala-Diaz, J.F.; Gomez-Delgado, F.; Perez-Martinez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Landa, B.B.; Navas-Cortes, J.A.; Tena-Sempere, M.; et al. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS ONE 2016, 11, e0154090. [Google Scholar] [CrossRef]
- Kim, Y.S.; Unno, T.; Kim, B.Y.; Park, M.S. Sex Differences in Gut Microbiota. World J. Mens Health 2020, 38, 48–60. [Google Scholar] [CrossRef]
- Markle, J.G.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef]
- Johnson, B.M.; Gaudreau, M.C.; Gudi, R.; Brown, R.; Gilkeson, G.; Vasu, C. Gut microbiota differently contributes to intestinal immune phenotype and systemic autoimmune progression in female and male lupus-prone mice. J. Autoimmun. 2020, 108, 102420. [Google Scholar] [CrossRef]
- Harder, J.W.; Ma, J.; Alard, P.; Sokoloski, K.J.; Mathiowitz, E.; Furtado, S.; Egilmez, N.K.; Kosiewicz, M.M. Male microbiota-associated metabolite restores macrophage efferocytosis in female lupus-prone mice via activation of PPARγ/LXR signaling pathways. J. Leukoc. Biol. 2023, 113, 41–57. [Google Scholar] [CrossRef]
- Lawenius, L.; Cowardin, C.; Grahnemo, L.; Scheffler, J.M.; Horkeby, K.; Engdahl, C.; Wu, J.; Vandenput, L.; Koskela, A.; Tuukkanen, J.; et al. Transplantation of gut microbiota from old mice into young healthy mice reduces lean mass but not bone mass. Gut Microbes 2023, 15, 2236755. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.K.A.; Raposo, P.J.F.; Madsen, K.L.; Fenrich, K.K.; Kabarchuk, G.; Fouad, K. What Makes a Successful Donor? Fecal Transplant from Anxious-Like Rats Does Not Prevent Spinal Cord Injury-Induced Dysbiosis. Biology 2021, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Barcena, C.; Valdes-Mas, R.; Mayoral, P.; Garabaya, C.; Durand, S.; Rodriguez, F.; Fernandez-Garcia, M.T.; Salazar, N.; Nogacka, A.M.; Garatachea, N.; et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 2019, 25, 1234–1242. [Google Scholar] [CrossRef]
- Willing, B.P.; Vacharaksa, A.; Croxen, M.; Thanachayanont, T.; Finlay, B.B. Altering host resistance to infections through microbial transplantation. PLoS ONE 2011, 6, e26988. [Google Scholar] [CrossRef]
- Schriefer, A.E.; Cliften, P.F.; Hibberd, M.C.; Sawyer, C.; Brown-Kennerly, V.; Burcea, L.; Klotz, E.; Crosby, S.D.; Gordon, J.I.; Head, R.D. A multi-amplicon 16S rRNA sequencing and analysis method for improved taxonomic profiling of bacterial communities. J. Microbiol. Methods 2018, 154, 6–13. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Vazquez-Baeza, Y.; Gonzalez, A.; Smarr, L.; McDonald, D.; Morton, J.T.; Navas-Molina, J.A.; Knight, R. Bringing the Dynamic Microbiome to Life with Animations. Cell Host Microbe 2017, 21, 7–10. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025; Available online: https://www.R-project.org (accessed on 22 April 2025).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 22 April 2025).
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots; R Package Version 0.6.0. 2023. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 22 April 2025).
- Berg, R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996, 4, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Takeda, K. Host-microbiota interactions in rheumatoid arthritis. Exp. Mol. Med. 2019, 51, 1–6. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Lee, J.Y.; Mannaa, M.; Kim, Y.; Kim, J.; Kim, G.T.; Seo, Y.S. Comparative Analysis of Fecal Microbiota Composition Between Rheumatoid Arthritis and Osteoarthritis Patients. Genes 2019, 10, 748. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, K.M.; Nagy, T.; Guo, T.L. Chronic oral exposure to glycated whey proteins increases survival of aged male NOD mice with autoimmune prostatitis by regulating the gut microbiome and anti-inflammatory responses. Food Funct. 2020, 11, 153–162. [Google Scholar] [CrossRef]
- Rong, J.; Liu, S.; Hu, C.; Liu, C. Single probiotic supplement suppresses colitis-associated colorectal tumorigenesis by modulating inflammatory development and microbial homeostasis. J. Gastroenterol. Hepatol. 2019, 34, 1182–1192. [Google Scholar] [CrossRef]
- Petrov, V.A.; Laczny, C.C.; Wilmes, P. Bacteroides acidifaciens: Linking dietary fiber to liver health. Cell Metab. 2024, 36, 1908–1910. [Google Scholar] [CrossRef]
- Zheng, C.; Zhong, Y.; Xie, J.; Wang, Z.; Zhang, W.; Pi, Y.; Zhang, W.; Liu, L.; Luo, J.; Xu, W. Bacteroides acidifaciens and its derived extracellular vesicles improve DSS-induced colitis. Front. Microbiol. 2023, 14, 1304232. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Lee, Y.S.; Kim, Y.; Lee, S.H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.S.; Lim, H.S.; Kim, M.S.; et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-D.; Jia, X.-M.; Xu, J.; Zhao, L.; Ji, J.-Y.; Wu, B.-X.; Ma, Y.; Li, H.; Zuo, X.; Pan, W.-Y.; et al. The gut microbiota of non-treated patients with SLE defines an autoimmunogenic and proinflammatory profile. Arthritis Rheumatol. 2020, 73, 232–243. [Google Scholar] [CrossRef]
- Grizotte-Lake, M.; Zhong, G.; Duncan, K.; Kirkwood, J.; Iyer, N.; Smolenski, I.; Isoherranen, N.; Vaishnava, S. Commensals Suppress Intestinal Epithelial Cell Retinoic Acid Synthesis to Regulate Interleukin-22 Activity and Prevent Microbial Dysbiosis. Immunity 2018, 49, 1103–1115.e6. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Grainger, J.R.; Spencer, S.P.; Belkaid, Y. The role of retinoic acid in tolerance and immunity. Immunity 2011, 35, 13–22. [Google Scholar] [CrossRef]
- Li, W.; Deng, C.; Yang, H.; Wang, G. The Regulatory T Cell in Active Systemic Lupus Erythematosus Patients: A Systemic Review and Meta-Analysis. Front. Immunol. 2019, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, H.; Xu, S.; Zhuang, Y.; An, J.; Su, C.; Xia, Y.; Chen, J.; Xu, Z.Z.; Liu, Q.; et al. Alteration in gut microbiota is associated with dysregulation of cytokines and glucocorticoid therapy in systemic lupus erythematosus. Gut Microbes 2020, 11, 1758–1773. [Google Scholar] [CrossRef]
- Yang, P.; Xu, R.; Chen, F.; Chen, S.; Khan, A.; Li, L.; Zhang, X.; Wang, Y.; Xu, Z.; Shen, H. Fungal gut microbiota dysbiosis in systemic lupus erythematosus. Front. Microbiol. 2023, 14, 1149311. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harder, J.W.; Ma, J.; Collins, J.; Alard, P.; Jala, V.R.; Bodduluri, H.; Kosiewicz, M.M. Characterization of Sex-Based Differences in Gut Microbiota That Correlate with Suppression of Lupus in Female BWF1 Mice. Microorganisms 2025, 13, 1023. https://doi.org/10.3390/microorganisms13051023
Harder JW, Ma J, Collins J, Alard P, Jala VR, Bodduluri H, Kosiewicz MM. Characterization of Sex-Based Differences in Gut Microbiota That Correlate with Suppression of Lupus in Female BWF1 Mice. Microorganisms. 2025; 13(5):1023. https://doi.org/10.3390/microorganisms13051023
Chicago/Turabian StyleHarder, James W., Jing Ma, James Collins, Pascale Alard, Venkatakrishna R. Jala, Haribabu Bodduluri, and Michele M. Kosiewicz. 2025. "Characterization of Sex-Based Differences in Gut Microbiota That Correlate with Suppression of Lupus in Female BWF1 Mice" Microorganisms 13, no. 5: 1023. https://doi.org/10.3390/microorganisms13051023
APA StyleHarder, J. W., Ma, J., Collins, J., Alard, P., Jala, V. R., Bodduluri, H., & Kosiewicz, M. M. (2025). Characterization of Sex-Based Differences in Gut Microbiota That Correlate with Suppression of Lupus in Female BWF1 Mice. Microorganisms, 13(5), 1023. https://doi.org/10.3390/microorganisms13051023





