Identifying Root-Associated Endophytic Fungi and Bacteria in Festuca and Lolium Grasses from a Site in Lithuania
Abstract
1. Introduction
2. Materials and Methods
2.1. Root Sampling and Sterilization
2.2. Microscopic Evaluation and Estimation of the Abundance of Endophytic Fungi in the Grass Roots
2.3. Isolation of the Fungi
2.4. Isolation of the Bacteria
2.5. DNA Extraction from the Fungi and Bacteria
2.6. Standard DNA Amplification and Sequencing
2.7. Morphological Characterization of the Endophytic Fungi
2.8. Statistical Analysis
2.9. Photography
3. Results
3.1. The Frequency of Colonization by Endophytic Fungi in Festuca and Lolium Species and Their Hybrids
3.2. Diversity and Abundance of the Endophytic Fungi
3.3. Diversity and Abundance of the Endophytic Bacteria
3.4. The Fungal and Bacterial Taxon Richness Across Plant Species
4. Discussion
4.1. The Distribution of Fungal Root-Derived Endophytes in Festuca/Lolium Grasses
4.2. Fungal Endophytes Across Grass Growth Types and Habitats
4.3. The Distribution of Bacterial Root-Derived Endophytes in Festuca/Lolium Grasses
4.4. Bacterial Endophytes Across Grass Growth Types and Habitats
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Porras-Alfaro, A.; Herrera, J.; Sinsabaugh, R.L.; Odenbach, K.J.; Lowrey, T.; Natvig, D.O. Novel root fungal consortium associated with a dominant desert grass. Appl. Environ. Microbiol. 2008, 74, 2805–2813. [Google Scholar] [CrossRef] [PubMed]
- Mandyam, K.; Loughin, T.; Jumpponen, A. Isolation and morphological and metabolic characterization of common endophytes in annually burned tallgrass prairie. Mycologia 2010, 102, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Glynou, K.; Ali, T.; Buch, A.-K.; Haghi Kia, S.; Ploch, S.; Xia, X.; Çelik, A.; Thines, M.; Maciá-Vicente, J.G. The local environment determines the assembly of root endophytic fungi at a continental scale. Environ. Microbiol. 2016, 18, 2418–2434. [Google Scholar] [CrossRef] [PubMed]
- Knapp, D.G.; Imrefi, I.; Boldpurev, E.; Csíkos, S.; Akhmetova, G.; Berek-Nagy, P.J.; Otgonsuren, B.; Kovács, G.M. Root-colonizing endophytic fungi of the dominant grass Stipa krylovii from a mongolian steppe grassland. Front. Microbiol. 2019, 10, 2565. [Google Scholar] [CrossRef]
- Knapp, D.G.; Németh, J.B.; Barry, K.; Hainaut, M.; Henrissat, B.; Johnson, J.; Kuo, A.; Lim, J.H.P.; Lipzen, A.; Nolan, M.; et al. Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci. Rep. 2018, 8, 6321. [Google Scholar] [CrossRef]
- Toju, H.; Yamamoto, S.; Sato, H.; Tanabe, A.S.; Gilbert, G.S.; Kadowaki, K. Community composition of root-associated fungi in a Quercus-dominated temperate forest: “Codominance” of mycorrhizal and root-endophytic fungi. Ecol. Evol. 2013, 3, 1281–1293. [Google Scholar] [CrossRef]
- Mandyam, K.; Jumpponen, A. Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud. Mycol. 2005, 53, 173–189. [Google Scholar] [CrossRef]
- Knapp, D.G.; Pintye, A.; Kovács, G.M. The dark side is not fastidious—Dark septate endophytic fungi of native and invasive plants of semiarid sandy areas. PLoS ONE 2012, 7, e32570. [Google Scholar] [CrossRef]
- Andrade-Linares, D.; Franken, P. Fungal endophytes in plant roots: Taxonomy, colonization patterns, and functions. In Symbiotic Endophytes; Springer: Berlin/Heidelberg, Germany, 2013; pp. 311–334. [Google Scholar] [CrossRef]
- Pašakinskienė, I.; Stakelienė, V.; Matijošiūtė, S.; Martūnas, J.; Rimkevičius, M.; Būdienė, J.; Aučina, A.; Skridaila, A. Growth-Promoting Effects of Grass Root-Derived Fungi Cadophora fastigiata, Paraphoma fimeti and Plectosphaerella cucumerina on Spring Barley (Hordeum vulgare) and Italian Ryegrass (Lolium multiflorum). Microorganisms 2025, 13, 25. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth promoting Rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Aloo, B.N.; Nyongesa, B.O.; Were, J.O.; Were, B.A.; Tumuhairwe, J.B. Rhizobacterial biomolecules for sustainable crop production and environmental management: Plausible functions and molecular mechanism. In Microbial Biomolecules Emerging Approach in Agriculture, Pharmaceuticals and Environment Management, 1st ed.; Kumar, A., Bilal, M., Ferreira, L.F.R., Madhuree, K., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 1–30. [Google Scholar]
- Addy, H.D.; Piercey, M.M.; Currah, R.S. Microfungal endophytes in roots. Can. J. Bot. 2005, 83, 1–13. [Google Scholar] [CrossRef]
- Wearn, J.A.; Sutton, B.C.; Morley, N.J.; Gange, A.C. Species and organ specificity of fungal endophytes in herbaceous grassland plants. J. Ecol. 2012, 100, 1085–1092. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L. The epichloae, symbionts of the grass subfamily Poöideae. Ann. Mo. Bot. Gard. 2010, 97, 646–665. [Google Scholar] [CrossRef]
- Tadych, M.; Bergen, M.S.; White, J.F. Epichloë spp. associated with grasses: New insights on life cycles, dissemination and evolution. Mycologia 2014, 106, 181–201. [Google Scholar] [CrossRef]
- Saikkonen, K.; Ahlholm, J.; Helander, M.; Lehtimäki, S.; Niemeläinen, O. Endophytic fungi in wild and cultivated grasses in Finland. Ecography 2000, 23, 360–366. [Google Scholar] [CrossRef]
- Leyronas, C.; Raynal, G. Presence of Neotyphodium-like endophytes in European grasses. Ann. Appl. Biol. 2001, 139, 119–127. [Google Scholar] [CrossRef]
- Müller, C.B.; Krauss, J. Symbiosis between grasses and asexual fungal endophytes. Curr. Opin. Plant Biol. 2005, 8, 450–456. [Google Scholar] [CrossRef]
- Soto-Barajas, M.; Azquez-De-Aldana, B.; Alvarez, A.; Zabalgogeazcoa, I. Sympatric Epichloë species and chemotypic profiles in natural populations of Lolium perenne. Fungal Ecol. 2019, 39, 231–241. [Google Scholar] [CrossRef]
- Krauss, J.; Vikuk, V.; Young, C.A.; Krischke, M.; Mueller, M.J.; Baerenfaller, K. Epichloë endophyte infection rates and alkaloid content in commercially available grass seed mixtures in Europe. Microorganisms 2020, 8, 498. [Google Scholar] [CrossRef]
- Hume, D.E.; Stewart, A.V.; Simpson, W.R.; Johnson, R.D. Epichloë fungal endophytes play a fundamental role in New Zealand grasslands. J. R. Soc. N. Z. 2020, 50, 279–298. [Google Scholar] [CrossRef]
- Garces, K.R.; Sage, H.E.; Christian, N.; Emery, S.M. Epichloë increases root fungal endophyte richness and alters root fungal endophyte composition in a changing world. J. Fungi 2022, 8, 1142. [Google Scholar] [CrossRef] [PubMed]
- Pašakinskienė, I.; Stakelienė, V.; Matijošiūtė, S.; Martūnas, J. Diversity of Endophytic Fungi and Bacteria Inhabiting the Roots of the Woodland Grass, Festuca gigantea (Poaceae). Diversity 2024, 16, 453. [Google Scholar] [CrossRef]
- Del los Santos, M.C.; Taulé, C.; Mareque, C.; Baracochea, M.; Battistoni, F. Identification and characterization of the part of the bacterial community associated with field-grown tall fescue (Festuca arundinacea) cv. SFRO Don Tomás in Uruguay. Ann. Microbiol. 2016, 66, 329–342. [Google Scholar] [CrossRef]
- Przemieniecki, S.W.; Damszel, M.; Kurowski, T.P.; Mastalerz, J.; Kotlarz, K. Identification, ecological evaluation and phylogenetic analysis of non-symbiotic endophytic fungi colonizing timothy grass and perennial ryegrass grown in adjacent plots. Grass Forage Sci. 2019, 74, 42–52. [Google Scholar] [CrossRef]
- Coombs, J.T.; Franco, C.M.M. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 2003, 69, 5603–5608. [Google Scholar] [CrossRef]
- Groenewald, J.Z.; Nakashima, C.; Nishikawa, J.; Shin, H.-D.; Park, J.-H.; Jama, A.N.; Groenewald, M.; Braun, U.; Crous, P.W. Species concepts in Cercospora: Spotting the weeds among the roses. Stud. Mycol. 2013, 75, 115–170. [Google Scholar] [CrossRef]
- Toju, H.; Kurokawa, H.; Kenta, T. Factors influencing leaf- and root-associated communities of bacteria and fungi across 33 plant orders in a grassland. Front. Microbiol. 2019, 10, 241. [Google Scholar] [CrossRef]
- Baluška, F.; Mancuso, S. Root apex transition zone as oscillatory zone. Front. Plant Sci. 2013, 4, 354. [Google Scholar] [CrossRef]
- Baskin, T.I. Patterns of root growth acclimation: Constant processes, changing boundaries. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 65–73. [Google Scholar] [CrossRef]
- Kiheri, H.; Heinonsalo, J.; Timonen, S. Staining and microscopy of mycorrhizal fungal colonization in preserved ericoid plant roots. J. Berry Res. 2017, 4, 231–237. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Sung, G.-H.; Sung, J.-M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Huo, J.; Wan, L.; Pan, L.; Jiang, N.; Fu, J.; Wei, S.; He, L. Differences and biocontrol potential of haustorial endophytic fungi from Taxillus chinensis on different host plants. BMC Microbiol. 2023, 23, 128. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Sánchez Márquez, S.; Bills, G.; Zabalgogeazcoa, I. The endophytic mycobiota of the grass Dactylis glomerata. Fungal Divers. 2007, 27, 171–195. [Google Scholar]
- Rashmi, M.; Kushveer, J.S.; Sarma, V.V. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 2019, 10, 798–1079. [Google Scholar] [CrossRef]
- Wirsel, S.G.R.; Leibinger, W.; Ernst, M.; Mendgen, K. Genetic diversity of fungi closely associated with common reed. N. Phytol. 2001, 149, 589–598. [Google Scholar] [CrossRef]
- Gao, Y.; Ren, G.-C.; Wanasinghe, D.; Xu, J.-C.; Farias, A.; Gui, H. Two new species and a new record of Microdochium from grasses in Yunnan province, South-West China. J. Fungi 2022, 8, 1297. [Google Scholar] [CrossRef]
- David, A.S.; Seabloom, E.W.; May, G. Plant host species and geographic distance affect the structure of aboveground fungal symbiont communities, and environmental filtering affects belowground communities in a coastal dune ecosystem. Microb. Ecol. 2016, 71, 912–926. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J. Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef]
- Pereira, E.; Vázquez de Aldana, B.R.; San Emeterio, L.; Zabalgogeazcoa, I. A survey of culturable fungal endophytes from Festuca rubra subsp. pruinosa, a grass from marine cliffs, reveals a core microbiome. Front. Microbiol. 2019, 9, 3321. [Google Scholar] [CrossRef] [PubMed]
- Yagame, T.; Funabiki, E.; Yukawa, T.; Nagasawa, E. Identification of mycobionts in an achlorophyllous orchid, Cremastra aphylla (Orchidaceae), based on molecular analysis and basidioma morphology. Mycoscience 2018, 59, 18–23. [Google Scholar] [CrossRef]
- Gao, Y.; Peng, S.; Hang, Y.; Xie, G.; Ji, N.; Zhang, M. Mycorrhizal fungus Coprinellus disseminatus influences seed germination of the terrestrial orchid Cremastra appendiculata (D. Don) Makino. Sci. Hortic. 2022, 293, 110724. [Google Scholar] [CrossRef]
- Potvin, L.R.; Richter, D.L.; Jurgensen, M.F.; Dumroese, R.K. Association of Pinus banksiana Lamb. and Populus tremuloides Michx. seedling fine roots with Sistotrema brinkmannii (Bres.) J. Erikss. (Basidiomycotina). Mycorrhiza 2012, 22, 631–638. [Google Scholar] [CrossRef]
- Hallenberg, N. Speciation and distribution in Corticiaceae (Basidiomycetes). Plant Syst. Evol. 1991, 177, 93–110. [Google Scholar] [CrossRef]
- Nikoh, N.; Fukatsu, T. Interkingdom host jumping underground: Phylogenetic analysis of entomoparasitic fungi of the genus Cordyceps. Mol. Biol. Evo. 2000, 17, 629–638. [Google Scholar] [CrossRef]
- Lei, Y.; Hussain, A.; Guan, Z.; Wang, D.; Jaleel, W.; Lyu, L.; He, Y. Unraveling the Mode of Action of Cordyceps fumosorosea: Potential Biocontrol Agent against Plutella xylostella (Lepidoptera: Plutellidae). Insects 2021, 12, 179. [Google Scholar] [CrossRef]
- Vega, F.E. The use of fungal entomopathogens as endophytes in biological control: A review. Mycologia 2018, 110, 4–30. [Google Scholar] [CrossRef]
- Raman, A.; Suryanarayanan, T.S. Fungus–plant interaction influences plant-feeding insects. Fungal Ecol. 2017, 29, 123–132. [Google Scholar] [CrossRef]
- Gundel, P.E.; Rudgers, J.A.; Ghersa, C.M. Incorporating the process of vertical transmission into understanding of host-symbiont dynamics. Oikos 2011, 120, 1121–1128. [Google Scholar] [CrossRef]
- Shade, A.; Jacques, M.S.; Barret, M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 2017, 37, 15–22. [Google Scholar] [CrossRef]
- Di Francesco, A.; Di Foggia, M.; Corbetta, M.; Baldo, D.; Ratti, C.; Baraldi, E. Biocontrol Activity and Plant Growth Promotion Exerted by Aureobasidium pullulans Strains. J. Plant Growth Regul. 2021, 40, 1233–1244. [Google Scholar] [CrossRef]
- Islam, M.T. Current Status and Future Prospects of Cladosporium sp., a Biocontrol Agent for Sustainable Plant Protection. Biocontrol Sci. 2022, 27, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Fávaro, L.C.dL.; Sebastianes, F.L.dS.; Araújo, W.L. Epicoccum nigrum P16, a Sugarcane Endophyte, Produces Antifungal Compounds and Induces Root Growth. PLoS ONE 2012, 7, e36826. [Google Scholar] [CrossRef] [PubMed]
- Tichý, L.; Axmanová, I.; Dengler, J.; Guarino, R.; Jansen, F.; Midolo, G.; Nobis, M.P.; Van Meerbeek, K.; Aćić, S.; Attorre, F.; et al. Ellenberg-type indicator values for European vascular plant species. J. Veg. Sci. 2023, 34, e13168. [Google Scholar] [CrossRef]
- Wemheuer, F.; Kaiser, K.; Karlovsky, P.; Daniel, R.; Vidal, S.; Wemheuer, B. Bacterial endophyte communities of three important agricultural grass species differ in their response towards management regimes. Sci. Rep. 2017, 7, 40914. [Google Scholar] [CrossRef]
- Mandic-Mulec, I.; Stefanic, P.; van Elsas, J.D. Ecology of Bacillaceae. Microbiol. Spectr. 2015, 3, 1–24. [Google Scholar] [CrossRef]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for Plant Growth Promotion and Stress Resilience: What Have We Learned? Plants 2022, 11, 2482. [Google Scholar] [CrossRef]
- Coy, R.M.; Held, D.W.; Kloepper, J.W. Rhizobacterial Colonization of Bermudagrass by Bacillus spp. in a Marvyn Loamy Sand Soil. Appl. Soil Ecol. 2019, 141, 10–17. [Google Scholar] [CrossRef]
- Li, Q.; Hou, Z.; Zhou, D.; Jia, M.; Lu, S.; Yu, J. A plant growth-promoting bacteria Priestia megaterium JR48 induces plant resistance to the crucifer black rot via a salicylic acid-dependent signaling pathway. Front. Plant. Sci. 2022, 13, 1046181. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.; Singh, V.K.; Mishra, A.; Jha, B. Plant growth promoting rhizobacterium Stenotrophomonas maltophilia BJ01 augments endurance against N2 starvation by modulating physiology and biochemical activities of Arachis hypogea. PLoS ONE 2019, 14, e0222405. [Google Scholar] [CrossRef]
- Sharma, P.; Pandey, R.; Chauhan, N.S. Biofertilizer and biocontrol properties of Stenotrophomonas maltophilia BCM emphasize its potential application for sustainable agriculture. Front. Plant. Sci. 2014, 15, 1364807. [Google Scholar] [CrossRef]
- Kodama, K.; Kimura, N.; Komagata, K. Two new species of Pseudomonas: P. oryzihabitans isolated from rice paddy and clinical specimens and P. luteola isolated from clinical specimens. Int. J. Syst. Bacteriol. 1985, 35, 467–474. [Google Scholar] [CrossRef]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhu, J.N.; Liu, Q.F.; Liu, Y.; Liu, M.; Liu, L.; Zhang, Q. Comparison of the diversity of root-associated bacteria in Phragmites australis and Typha angustifolia L. in artificial wetlands. World J. Microbiol. Biotechnol. 2013, 29, 1499–1508. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Guo, J.; Bowatte, S.; Hou, F. Diversity of endophytic bacteria and fungi in seeds of Elymus nutans growing in four locations of Qinghai Tibet Plateau, China. Plant Soil 2021, 459, 49–63. [Google Scholar] [CrossRef]

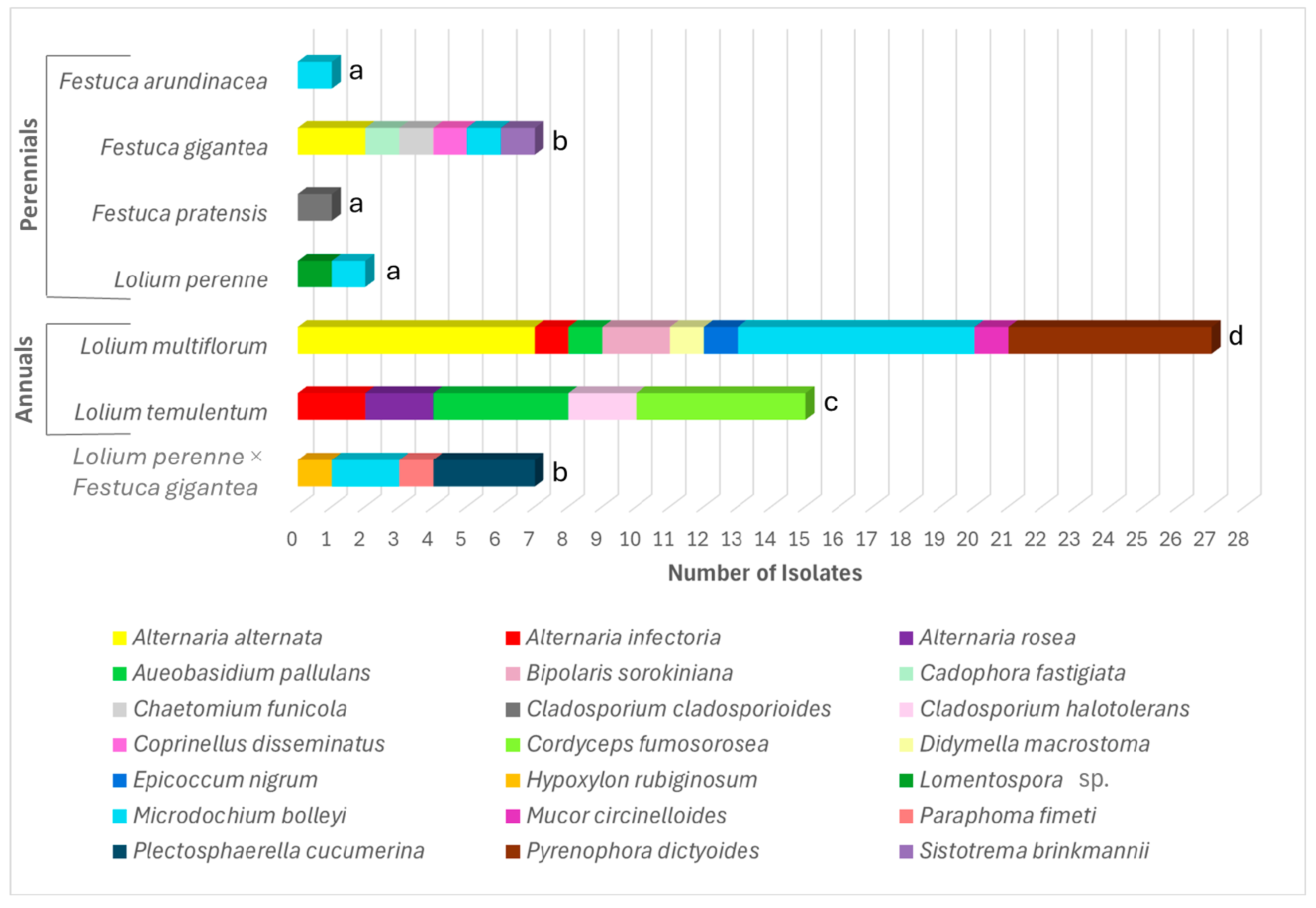
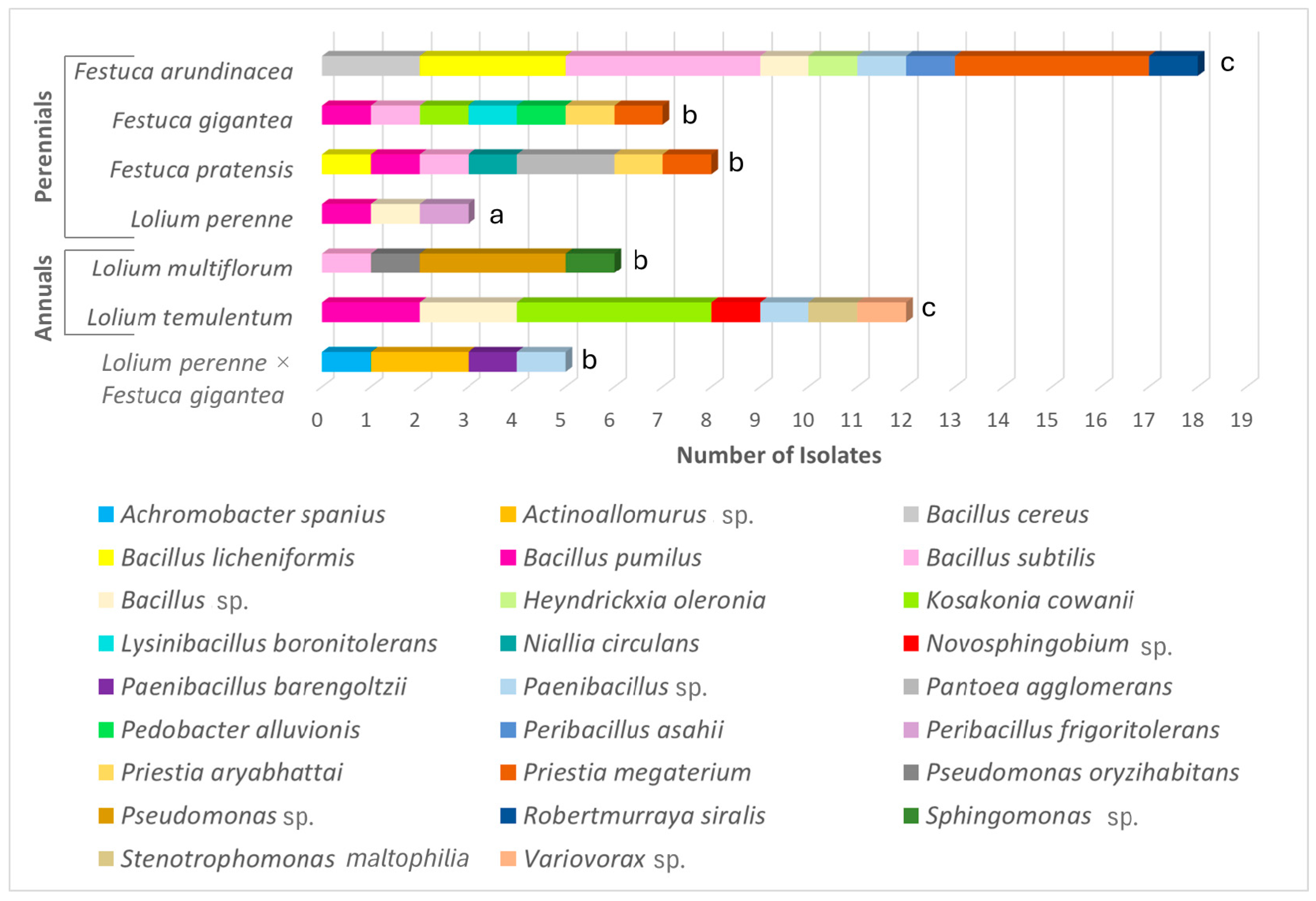
| Species | Ploidy Level | Growth Type | Habitat Type | Accession Origin |
|---|---|---|---|---|
| Festuca arundinacea ‘Monas’ | 2n = 6x = 42 | Perennial | Open-grassland | LCAFS, IA |
| Festuca gigantea | 2n = 6x = 42 | Perennial | Forest sites | VU BG, Kairėnai, Vilnius, LT; Vingis Park, Vilnius, LT |
| Festuca pratensis ‘Alanta’ | 2n = 2x = 14 | Perennial | Open-grassland | LCAFS, IA |
| Lolium perenne ‘Veja’ | 2n = 2x = 14 | Perennial | Open-grassland | LCAFS, IA |
| Lolium multiflorum ‘Grazer’ | 2n = 2x = 14 | Annual | Open-grassland | LCAFS, IA |
| Lolium temulentum | 2n = 2x = 14 | Annual | Open-grassland | IPK Leibniz Institute, Gatersleben, DE |
| Lolium perenne × Festuca gigantea | 2n = 4x = 28 | Perennial | Open-grassland | Laboratory-produced, VU BG laboratory collection |
| Locus | Primers | Primer Sequences (5′–3′) | Tm °C | Reference |
|---|---|---|---|---|
| Primers for fungal DNA | ||||
| ITS | ITS1 ITS4 | TCCGTAGGTGAACCTGCGG TCCTCCGCTTATTGATATGC | 54 | [34] |
| TEFa | EF1-278F EF-2 | CATCGAGAAGTTCGAGAAGG GGARGTACCAGTSATCATGTT | 54 | [29] |
| SSU | NS1 NS4 | GTAGTCATATGCTTGTCTC CTTCCGTCAATTCCTTTAAG | 49 | [34] |
| RPB2 | RPB2-5F2 fRPB2-7cR | GGGGWGAYCAGAAGAAGGC CCCATRGCTTGYTTRCCCAT | 58 | [35] |
| Primers for bacterial DNA | ||||
| 16S rDNA | 27f CM 1492R | AGAGTTTGATCMTGGCTCAG TACGGYTACCTTGTTACGACTT | 52 | [28] |
| 16S rDNA | 704F 765R | GTAGCGGTGAAATGCGTAGA CTGTTTGCTCCCCACGCTTTC | 56 | [28] |
| 16S rDNA | S-D-Bact-0341-b-S-17 S-D-Bact-0785-a-A-21 | CCTACGGGNGGCWGCAG GACTACHVGGGTATCTAATCC | 56 | [36] |
| Plant Species | Endophytic Fungi | Endophytic Bacteria | Total Microbial Endophyte Abundance | |||||
|---|---|---|---|---|---|---|---|---|
| * No. Isolates | No. Species | ** D | No. Isolates | No. Species | D | No. Isolates | No. Species | |
| Perennial species | ||||||||
| Festuca arundinacea | 1 | 1 | 1.00 | 18 | 9 | 2.77 | 19 | 10 |
| Festuca gigantea | 7 | 6 | 2.57 | 7 | 7 | 3.08 | 14 | 13 |
| Festuca pratensis | 1 | 1 | 1.00 | 8 | 7 | 2.89 | 9 | 8 |
| Lolium perenne | 2 | 2 | 1.44 | 3 | 3 | 1.82 | 5 | 5 |
| Lolium perenne × Festuca gigantea | 7 | 4 | 1.54 | 5 | 4 | 1.86 | 12 | 8 |
| Annual species | ||||||||
| Lolium multiflorum | 27 | 9 | 2.43 | 6 | 4 | 1.67 | 33 | 13 |
| Lolium temulentum | 15 | 5 | 1.48 | 12 | 7 | 2.41 | 27 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stakelienė, V.; Pašakinskienė, I.; Matijošiūtė, S.; Martūnas, J.; Štukėnienė, G. Identifying Root-Associated Endophytic Fungi and Bacteria in Festuca and Lolium Grasses from a Site in Lithuania. Microorganisms 2025, 13, 799. https://doi.org/10.3390/microorganisms13040799
Stakelienė V, Pašakinskienė I, Matijošiūtė S, Martūnas J, Štukėnienė G. Identifying Root-Associated Endophytic Fungi and Bacteria in Festuca and Lolium Grasses from a Site in Lithuania. Microorganisms. 2025; 13(4):799. https://doi.org/10.3390/microorganisms13040799
Chicago/Turabian StyleStakelienė, Violeta, Izolda Pašakinskienė, Saulė Matijošiūtė, Justas Martūnas, and Gitana Štukėnienė. 2025. "Identifying Root-Associated Endophytic Fungi and Bacteria in Festuca and Lolium Grasses from a Site in Lithuania" Microorganisms 13, no. 4: 799. https://doi.org/10.3390/microorganisms13040799
APA StyleStakelienė, V., Pašakinskienė, I., Matijošiūtė, S., Martūnas, J., & Štukėnienė, G. (2025). Identifying Root-Associated Endophytic Fungi and Bacteria in Festuca and Lolium Grasses from a Site in Lithuania. Microorganisms, 13(4), 799. https://doi.org/10.3390/microorganisms13040799






