Genome Analysis and In Vitro Assay of Probiotic Properties of Bacillus paranthracis YC03 with Urate-Lowering Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Samples
2.2. Isolation of the Bacterial Strain with Uric Acid Biodegrading Ability
2.3. Determination of Uric Acid and Nucleoside Biodegradation Ability of YC03
2.4. Genomic DNA Extraction, Sequencing, Assembly and CDS Prediction
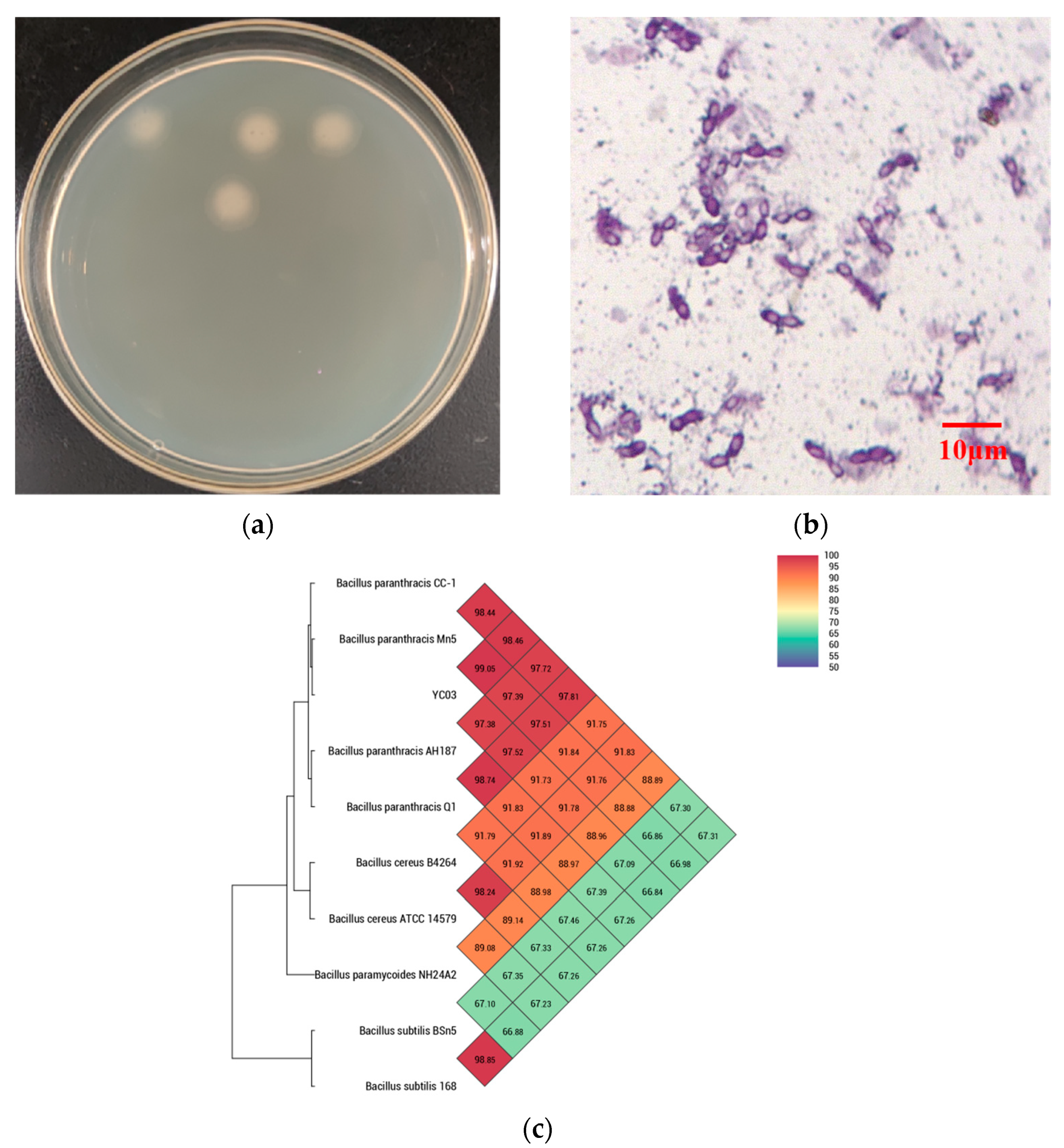
2.5. Identification of YC03
2.6. Determination of Nucleosides Biodegradation Ability of YC03’s Cell-Free Extracts
2.7. Analysis of Uric Acid, Inosine and Guanosine and Biodegradation Products by HPLC
2.8. In Vitro Probiotic Properties
2.8.1. Evaluation of the Acid and Bile Salt Tolerance
2.8.2. Auto-/Co-Aggregation Assay
2.8.3. Cell Surface Hydrophobicity
2.9. Safety Assessment
2.9.1. Antibiotic Resistance Test
2.9.2. Hemolysis Assay
2.9.3. Biogenic Amine Production Assay
2.10. Statistics Analyses
3. Results and Discussion
3.1. Isolation and Identification of Uric Acid Biodegrading Strain
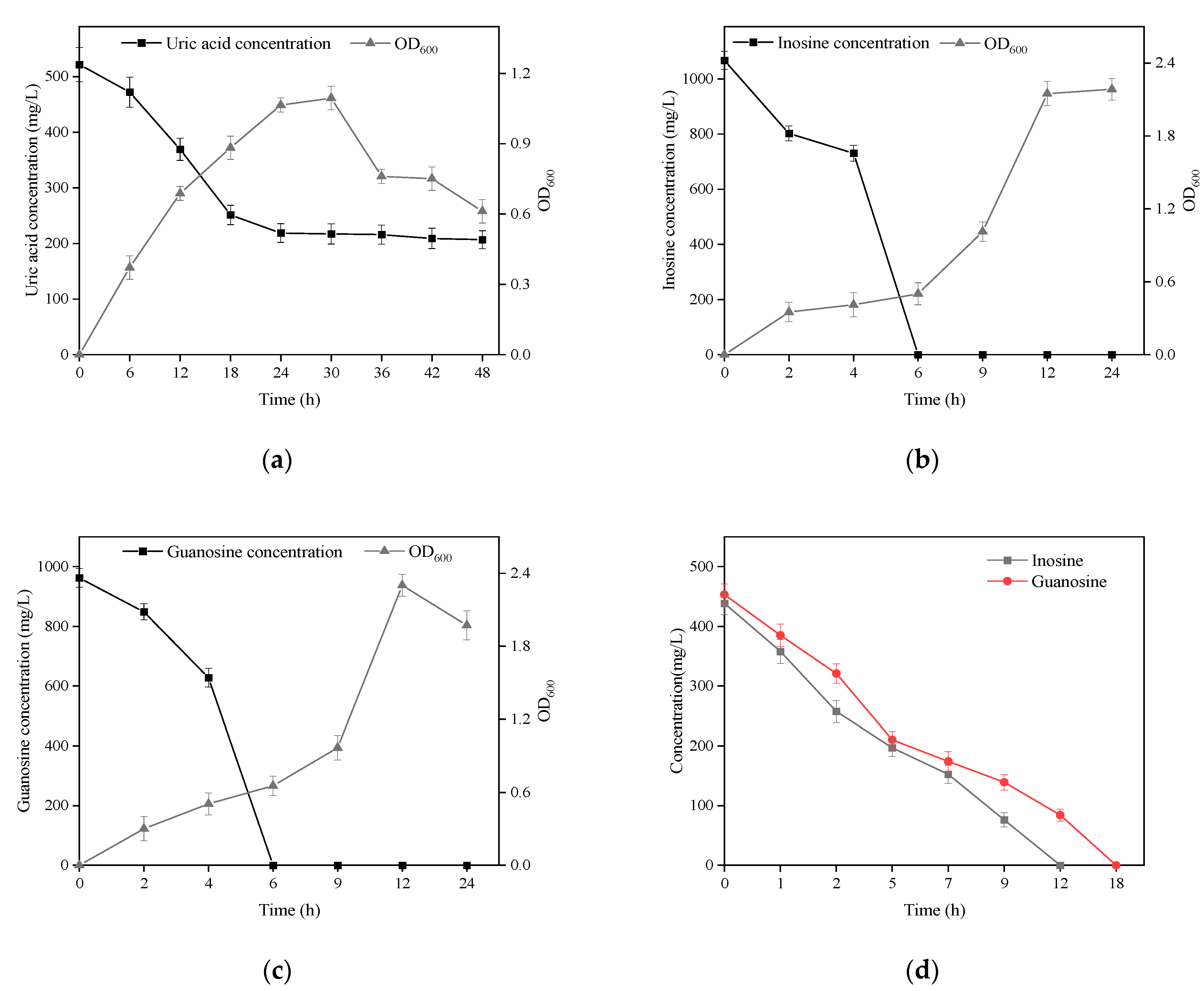
3.2. Biodegradation of Uric Acid and Nucleoside by B. paranthracis YC03
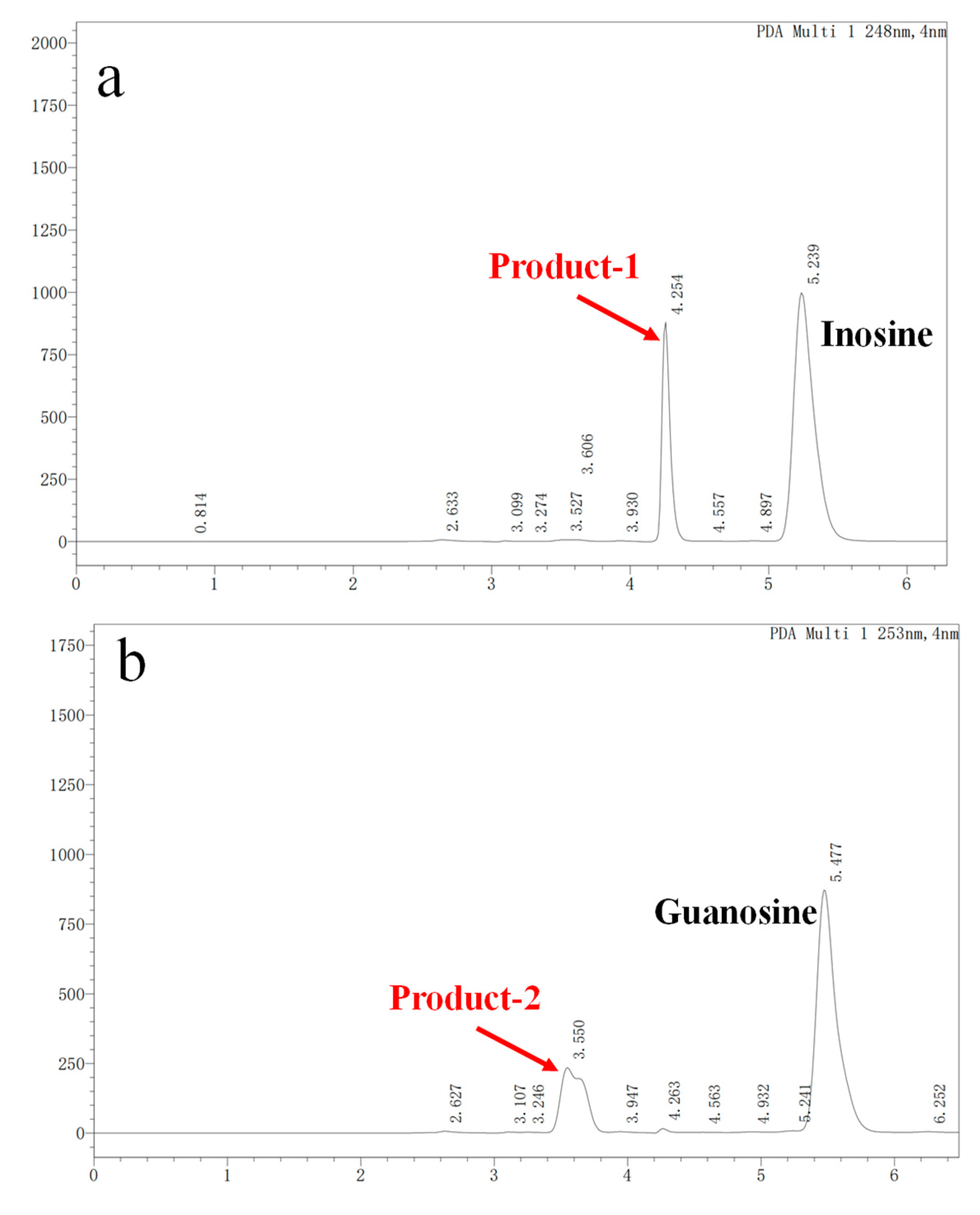
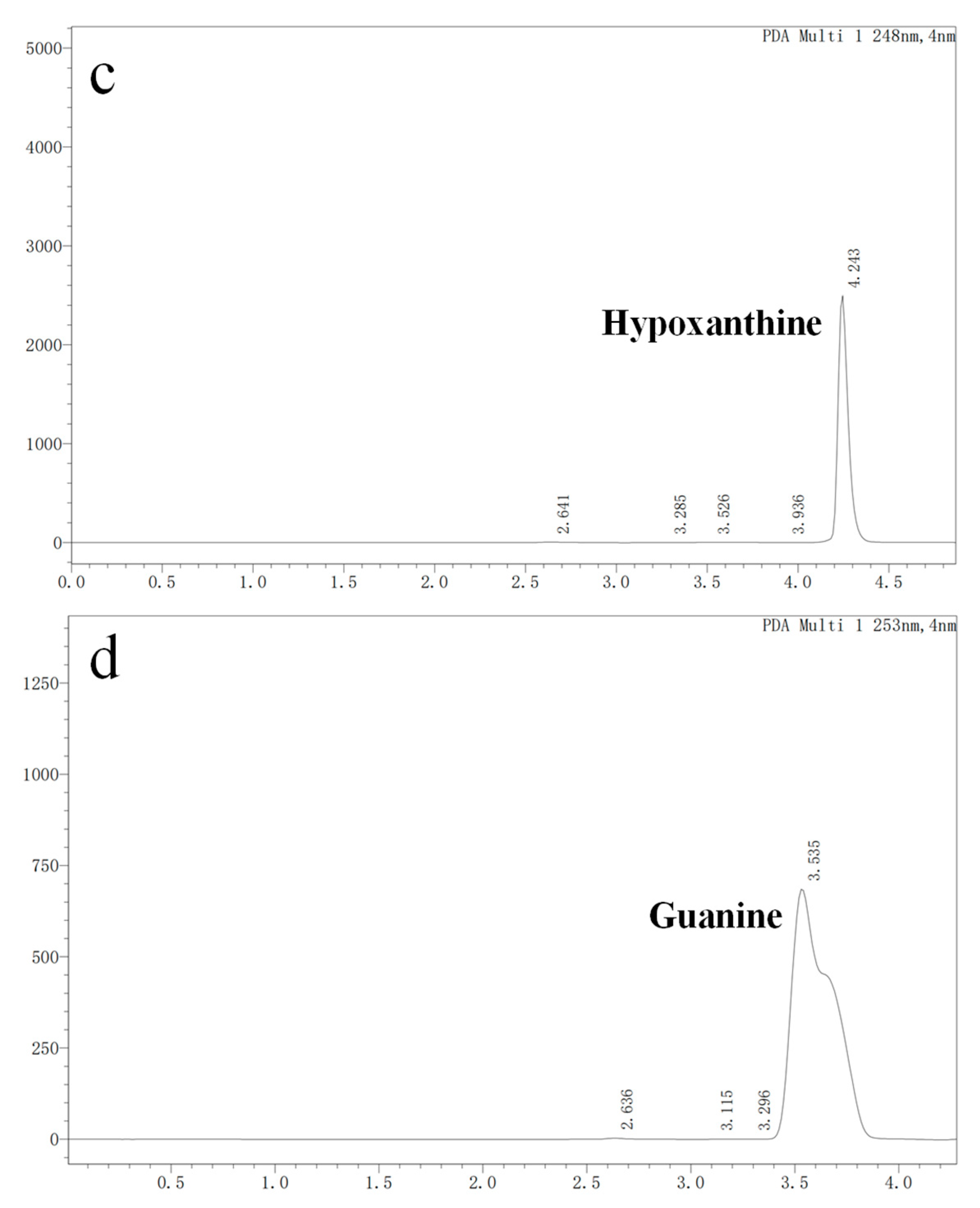
3.3. Nucleoside Biodegradation Products Identification
3.4. Overview of Genome Analysis
3.5. Genetic Features of Probiotic Properties in YC03
3.6. In Vitro Assay of Probiotic Properties of YC03
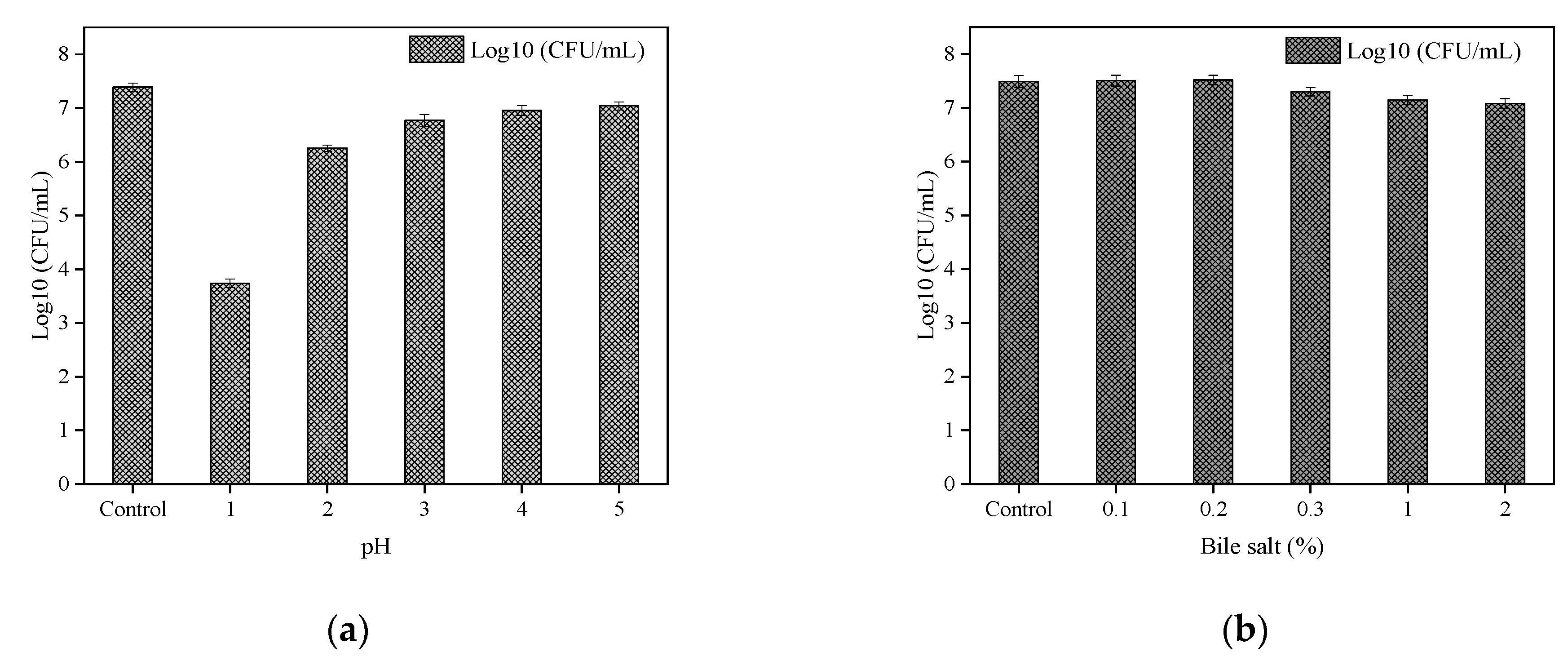
3.6.1. Acid and Bile Salt Tolerance
3.6.2. Auto-/Co-Aggregation and Cell Surface Hydrophobicity
3.7. Antibiotic Resistance
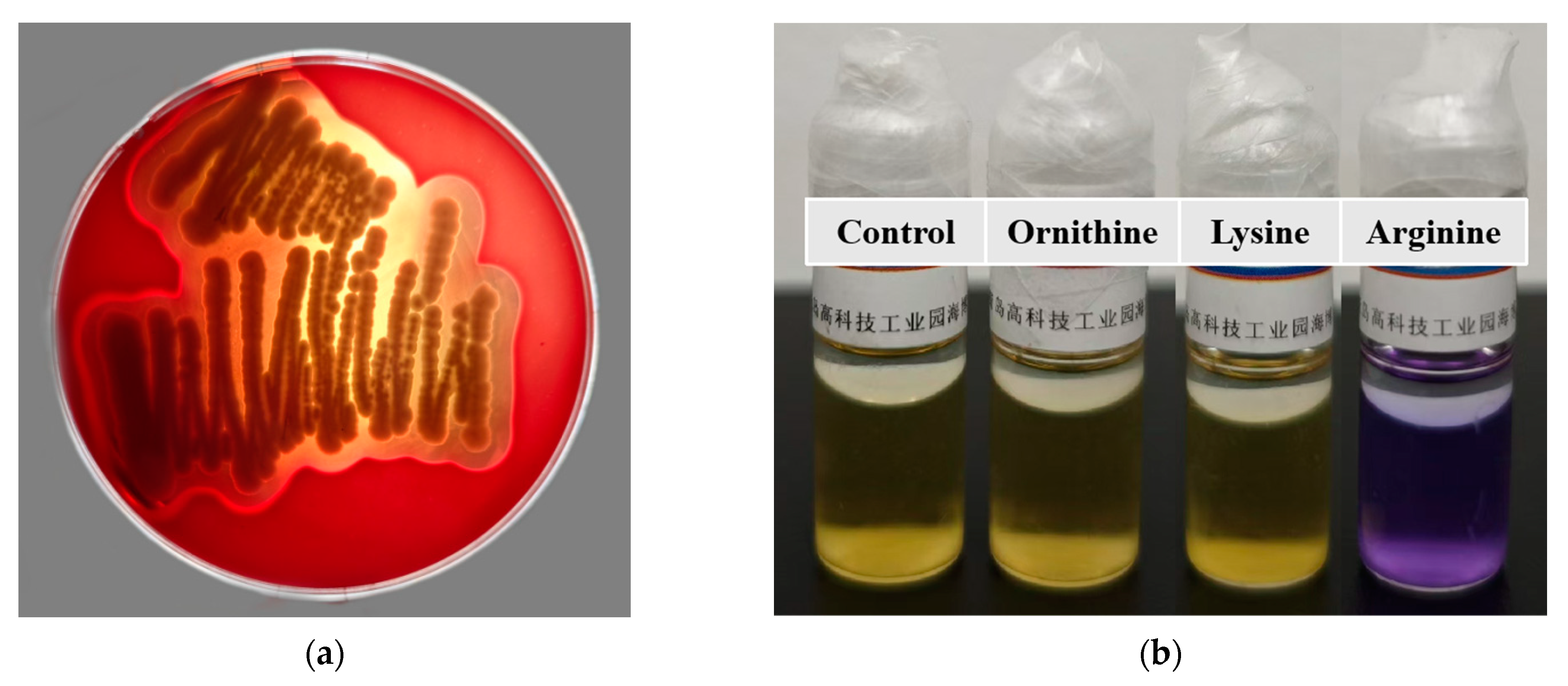
3.8. Hemolysis Assay
3.9. Biogenic Amine Production
3.10. Metabolism Pathway for Uric Acid, Inosine and Guanosine Biodegradation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bardin, T.; Richette, P. Definition of hyperuricemia and gouty conditions. Curr. Opin. Rheumatol. 2014, 26, 186–191. [Google Scholar] [CrossRef]
- Sun, L.; Ni, C.; Zhao, J.; Wang, G.; Chen, W. Probiotics, bioactive compounds and dietary patterns for the effective management of hyperuricemia: A review. Crit. Rev. Food Sci. Nutr. 2022, 64, 2016–2031. [Google Scholar] [CrossRef]
- Ragab, G.; Elshahaly, M.; Bardin, T. Gout: An old disease in new perspective—A review. J. Adv. Res. 2017, 8, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Dubey, A.; Nolkha, N.; Singh, J.A. Hyperuricemia, urate-lowering therapy, and kidney outcomes: A systematic review and meta-analysis. Ther. Adv. Musculoskelet. Dis. 2021, 13, 1759720X211016661. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, M.; Wang, C.; Zhang, S.; Yun, C.; Chen, S.; Cui, L.; Wu, S.; Xue, H. Early onset of hyperuricemia is associated with increased cardiovascular disease and mortality risk. Clin. Res. Cardiol. 2021, 110, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Mortada, I. Hyperuricemia, Type 2 Diabetes Mellitus, and Hypertension: An Emerging Association. Curr. Hypertens. Rep. 2017, 19, 69. [Google Scholar] [CrossRef]
- Huang, J.; Ma, Z.F.; Zhang, Y.; Wan, Z.; Li, Y.; Zhou, H.; Chu, A.; Lee, Y.Y. Geographical distribution of hyperuricemia in mainland China: A comprehensive systematic review and meta-analysis. Glob. Health Res. Policy 2020, 5, 52. [Google Scholar] [CrossRef]
- Chen-Xu, M.; Yokose, C.; Rai, S.K.; Pillinger, M.H.; Choi, H.K. Contemporary Prevalence of Gout and Hyperuricemia in the United States and Decadal Trends: The National Health and Nutrition Examination Survey, 2007–2016. Arthritis Rheumatol. 2019, 71, 991–999. [Google Scholar] [CrossRef]
- Nielsen, S.M.; Zobbe, K.; Kristensen, L.E.; Christensen, R. Nutritional recommendations for gout: An update from clinical epidemiology. Autoimmun. Rev. 2018, 17, 1090–1096. [Google Scholar] [CrossRef]
- Ge, H.; Jiang, Z.; Li, B.; Xu, P.; Wu, H.; He, X.; Xu, W.; Huang, Z.; Xiong, T.; Wang, P.; et al. Dendrobium officinalis Six Nostrum Promotes Intestinal Urate Underexcretion via Regulations of Urate Transporter Proteins in Hyperuricemic Rats. Comb. Chem. High Throughput Screen. 2023, 26, 848–861. [Google Scholar] [CrossRef]
- Maiuolo, J.; Oppedisano, F.; Gratteri, S.; Muscoli, C.; Mollace, V. Regulation of uric acid metabolism and excretion. Int. J. Cardiol. 2016, 213, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Kakutani-Hatayama, M.; Kadoya, M.; Okazaki, H.; Kurajoh, M.; Shoji, T.; Koyama, H.; Tsutsumi, Z.; Moriwaki, Y.; Namba, M.; Yamamoto, T. Nonpharmacological Management of Gout and Hyperuricemia: Hints for Better Lifestyle. Am. J. Lifestyle Med. 2017, 11, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Li, W.; Zhang, J.; Ke, Q.; Zhu, X.; Long, L.; Li, C. Safety and tolerability of available drugs for hyperuricemia: A critical review and an update on recent developments. J. Chin. Pharm. Sci. 2022, 31, 397–411. [Google Scholar] [CrossRef]
- Zhao, H.; Lu, Z.; Lu, Y. The potential of probiotics in the amelioration of hyperuricemia. Food Funct. 2022, 13, 2394–2414. [Google Scholar] [CrossRef]
- Dewulf, J.P.; Marie, S.; Nassogne, M.C. Disorders of purine biosynthesis metabolism. Mol. Genet. Metab. 2022, 136, 190–198. [Google Scholar] [CrossRef]
- Zhang, H.; Xiang, S.; Zhai, R.; Li, X.; Hu, M.; Wang, T.; Zhang, H.; Pan, L. Analysis of microbial and metabolic diversity in Jiangshui from Northwest China. Food Sci. Technol. 2023, 43, e107222. [Google Scholar] [CrossRef]
- Wu, Y.; Ye, Z.; Feng, P.; Li, R.; Chen, X.; Tian, X.; Han, R.; Kakade, A.; Liu, P.; Li, X. Limosilactobacillus fermentum JL-3 isolated from “Jiangshui” ameliorates hyperuricemia by degrading uric acid. Gut Microbes 2021, 13, 1–18. [Google Scholar] [CrossRef]
- Zhao, S.; Feng, P.; Hu, X.; Cao, W.; Liu, P.; Han, H.; Jin, W.; Li, X. Probiotic Limosilactobacillus fermentum GR-3 ameliorates human hyperuricemia via degrading and promoting excretion of uric acid. iScience 2022, 25, 105198. [Google Scholar] [CrossRef]
- Smith, D.L.; Lemieux, E.N.; Barua, S. Correction in Bicinchoninic Acid (BCA) Absorbance Assay to Analyze Protein Concentration. Nano LIFE 2018, 8, 1850005. [Google Scholar] [CrossRef]
- Du, X.; Jiang, Y.; Sun, Y.; Cao, X.; Zhang, Y.; Xu, Q.; Yan, H. Biodegradation of Inosine and Guanosine by Bacillus paranthracis YD01. Int. J. Mol. Sci. 2023, 24, 14462. [Google Scholar] [CrossRef]
- Diale, M.O.; Kayitesi, E.; Serepa-Dlamini, M.H. Genome In Silico and In Vitro Analysis of the Probiotic Properties of a Bacterial Endophyte, Bacillus Paranthracis Strain MHSD3. Front. Genet. 2021, 12, 672149. [Google Scholar] [CrossRef]
- Ait Seddik, H.; Bendali, F.; Cudennec, B.; Drider, D. Anti-pathogenic and probiotic attributes of Lactobacillus salivarius and Lactobacillus plantarum strains isolated from feces of Algerian infants and adults. Res. Microbiol. 2017, 168, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Çetin, B.; Aktaş, H. Monitoring probiotic properties and safety evaluation of antilisterial Enterococcus faecium strains with cholesterol-lowering potential from raw Cow’s milk. Food Biosci. 2024, 61, 104532. [Google Scholar] [CrossRef]
- Li, T.; Lyu, L.; Zhang, Y.; Dong, K.; Li, Q.; Guo, X.; Zhu, Y. A newly isolated E. thailandicus strain d5B with exclusively antimicrobial activity against C. difficile might be a novel therapy for controlling CDI. Genomics 2021, 113, 475–483. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Borgmeier, C.; Garvey, S.M.; Spears, J.L. Preclinical Safety Assessment of Bacillus subtilis BS50 for Probiotic and Food Applications. Microorganisms 2022, 10, 1038. [Google Scholar] [CrossRef]
- Fu, X.; Lyu, L.; Wang, Y.; Zhang, Y.; Guo, X.; Chen, Q.; Liu, C. Safety assessment and probiotic characteristics of Enterococcus lactis JDM1. Microb. Pathog. 2022, 163, 105380. [Google Scholar] [CrossRef]
- Liu, Y.; Du, J.; Lai, Q.; Zeng, R.; Ye, D.; Xu, J.; Shao, Z. Proposal of nine novel species of the Bacillus cereus group. nt. J. Syst. Evol. Microbiol. 2017, 67, 2499–2508. [Google Scholar] [CrossRef]
- Fukuda, D.; Nolasco-Hipolito, C.; Gill, S.R. Draft Genome Sequence of Bacillus paranthracis Strain DB-4, Isolated from Nukadoko, Fermented Rice Bran for Japanese Pickles. Microbiology Resource Announcements 2021, 10, e0070521. [Google Scholar] [CrossRef]
- Bukharin, O.V.; Perunova, N.B.; Andryuschenko, S.V.; Ivanova, E.V.; Bondarenko, T.A.; Chainikova, I.N.; Rasko, D. Genome Sequence Announcement of Bacillus paranthracis Strain ICIS-279, Isolated from Human Intestine. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef]
- Lu, L.; Liu, T.; Liu, X.; Wang, C. Screening and identification of purine degrading Lactobacillus fermentum 9-4 from Chinese fermented rice-flour noodles. Food Sci. Hum. Wellness 2022, 11, 1402–1408. [Google Scholar] [CrossRef]
- Lee, M.J.; Khang, A.R.; Kang, Y.H.; Yun, M.S.; Yi, D. Synergistic Interaction between Hyperuricemia and Abdominal Obesity as a Risk Factor for Metabolic Syndrome Components in Korean Population. Diabetes Metab. J. 2022, 46, 756–766. [Google Scholar] [CrossRef]
- Yamada, N.; Saito-Iwamoto, C.; Nakamura, M.; Soeda, M.; Chiba, Y.; Kano, H.; Asami, Y. Lactobacillus gasseri PA-3 Uses the Purines IMP, Inosine and Hypoxanthine and Reduces their Absorption in Rats. Microorganisms 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.P.; Hu, Y.S.; Wei, M.; Wang, K.M.; Wang, Y.Y.; Wang, S.L.; Hu, Q.; Wei, H.; Zhang, Z.H. Amelioration of hyperuricemia by Lactobacillus acidophilus F02 with uric acid-lowering ability via modulation of NLRP3 inflammasome and gut microbiota homeostasis. J. Funct. Foods 2023, 111, 105903. [Google Scholar] [CrossRef]
- Kuo, Y.W.; Hsieh, S.H.; Chen, J.F.; Liu, C.R.; Chen, C.W.; Huang, Y.F.; Ho, H.H. Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ 2021, 9, e11209. [Google Scholar] [CrossRef]
- Li, M.; Wu, X.; Guo, Z.; Gao, R.; Ni, Z.; Cui, H.; Zong, M.; Van Bockstaele, F.; Lou, W. Lactiplantibacillus plantarum enables blood urate control in mice through degradation of nucleosides in gastrointestinal tract. Microbiome 2023, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.K.; Mount, D.B. The molecular physiology of uric acid homeostasis. Annu. Rev. Physiol. 2015, 77, 323–345. [Google Scholar] [CrossRef]
- Chung, W.-H.; Kang, J.; Lim, M.Y.; Lim, T.-j.; Lim, S.; Roh, S.W.; Nam, Y.-D. Complete Genome Sequence and Genomic Characterization of Lactobacillus acidophilus LA1 (11869BP). Front. Pharmacol. 2018, 9, 83. [Google Scholar] [CrossRef]
- Li, P.; Tian, W.N.; Jiang, Z.; Liang, Z.H.; Wu, X.Y.; Du, B. Genomic Characterization and Probiotic Potency of Bacillus sp DU-106, a Highly Effective Producer of L-Lactic Acid Isolated From Fermented Yogurt. Front. Microbiol. 2018, 9, 2216. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S.; Wang, L.; Lu, H.; Zhang, T.; Zeng, W. Characterization of Genomic, Physiological, and Probiotic Features of Lactiplantibacillus plantarum JS21 Strain Isolated from Traditional Fermented Jiangshui. Foods 2024, 13, 1082. [Google Scholar] [CrossRef]
- Khalil, E.S.; Manap, M.Y.; Mustafa, S.; Amid, M.; Alhelli, A.M.; Aljoubori, A. Probiotic characteristics of exopolysaccharides-producingLactobacillusisolated from some traditional Malaysian fermented foods. CyTA J. Food 2018, 16, 287–298. [Google Scholar] [CrossRef]
- Pan, M.; Kumaree, K.K.; Shah, N.P. Physiological Changes of Surface Membrane in Lactobacillus with Prebiotics. J. Food Sci. 2017, 82, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Gołaś-Prądzyńska, M.; Łuszczyńska, M.; Rola, J.G. Dairy Products: A Potential Source of Multidrug-Resistant Enterococcus faecalis and Enterococcus faecium Strains. Foods 2022, 11, 4116. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ying, W.; Dunlap, K.A.; Lin, G.; Satterfield, M.C.; Burghardt, R.C.; Wu, G.; Bazer, F.W. Arginine Decarboxylase and Agmatinase: An Alternative Pathway for De Novo Biosynthesis of Polyamines for Development of Mammalian Conceptuses1. Biol. Reprod. 2014, 90, 84. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Zhao, Z.; Li, L.; Min, Y.; Zhang, D.; Ji, A.; Jiang, C.; Wei, X.; Wu, X. A comprehensive review of spermidine: Safety, health effects, absorption and metabolism, food materials evaluation, physical and chemical processing, and bioprocessing. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2820–2842. [Google Scholar] [CrossRef]
- Cao, X.; Cai, J.; Zhang, Y.; Liu, C.; Song, M.; Xu, Q.; Liu, Y.; Yan, H. Biodegradation of Uric Acid by Bacillus paramycoides-YC02. Microorganisms 2023, 11, 1989. [Google Scholar] [CrossRef]


| Function | Gene | Product |
|---|---|---|
| Adhesion | lspA | Lipoprotein signal peptidase |
| dltD | Protein DltD | |
| dltC | D-alanyl carrier protein | |
| dltA | D-alanine-D-alanyl carrier protein ligase | |
| Acid stress | atp | ATP synthase |
| aspS | Aspartate-tRNA (Asp/Asn) ligase | |
| aspA | Aspartate ammonia-lyase | |
| recA | Protein RecA | |
| soda | Superoxide dismutase [Mn] | |
| relA | GTP pyrophosphokinase | |
| groL | 60 kDa chaperonin | |
| groS | 10 kDa chaperonin | |
| Acid stress/bile resistance | dnaA | Chromosomal replication initiator protein DnaA |
| dnaC | Replicative DNA helicase | |
| dnaE | DNA polymerase III subunit alpha | |
| dnaI | Primosomal protein DnaI | |
| dnaK | Chaperone protein DnaK | |
| dnaJ | Chaperone protein DnaJ | |
| dnaG | DNA primase | |
| dnaD | DNA replication protein DnaD | |
| Bile resistance | nagA | N-acetylglucosamine-6-phosphate deacetylase |
| nagB | Glucosamine-6-phosphate deaminase | |
| pyrG | CTP synthase | |
| argS | Arginine-tRNA ligase | |
| rps | 30S ribosomal protein | |
| rpl | 50S ribosomal protein | |
| Antimicrobial properties | ||
| mdh | alsT | Amino-acid carrier protein AlsT |
| alsS | Acetolactate synthase | |
| alsD | Alpha-acetolactate decarboxylase | |
| Transcriptional regulator | sig | RNA polymerase |
| ctsR | Transcriptional regulator CtsR |
| Antibiotic Substance | EFSA MIC (mg/L) Resistance Threshold | MIC (mg/L) |
|---|---|---|
| Chloramphenicol | 8 | 4, S |
| Clindamycin | 4 | 2, S |
| Tetracycline | 8 | 32, R |
| Gentamicin | 4 | 0.25, S |
| Kanamycin | 8 | 0.5, S |
| Vancomycin | 4 | 2, S |
| Erythromycin | 4 | 1, S |
| Ampicillin | 16 | 64, R |
| Gene ID | Gene | Product |
|---|---|---|
| ctg00001-00729 | auaG | FAD-dependent urate hydroxylase |
| ctg00007-03248 | ribF | riboflavin kinase/FMN adenylyltransferase |
| ctg00003-01568 | bglH | hydroxyisourate hydrolase |
| ctg00005-02479, ctg00026-05485 | hpt | hypoxanthine-guanine phosphoribosyltransferase [EC:2.4.2.8] |
| ctg00001-00080, ctg00032-05730, ctg00002-00970, ctg00001-00531 | rihA, rihB | purine nucleosidase [EC:3.2.2.1] |
| ctg00013-04301, ctg00012-04009 | punA, deoD | purine-nucleoside phosphorylase [EC:2.4.2.1] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Zhang, Y.; Xu, Q.; Yan, H. Genome Analysis and In Vitro Assay of Probiotic Properties of Bacillus paranthracis YC03 with Urate-Lowering Potential. Microorganisms 2025, 13, 798. https://doi.org/10.3390/microorganisms13040798
Cao X, Zhang Y, Xu Q, Yan H. Genome Analysis and In Vitro Assay of Probiotic Properties of Bacillus paranthracis YC03 with Urate-Lowering Potential. Microorganisms. 2025; 13(4):798. https://doi.org/10.3390/microorganisms13040798
Chicago/Turabian StyleCao, Xiaoyu, Yu Zhang, Qianqian Xu, and Hai Yan. 2025. "Genome Analysis and In Vitro Assay of Probiotic Properties of Bacillus paranthracis YC03 with Urate-Lowering Potential" Microorganisms 13, no. 4: 798. https://doi.org/10.3390/microorganisms13040798
APA StyleCao, X., Zhang, Y., Xu, Q., & Yan, H. (2025). Genome Analysis and In Vitro Assay of Probiotic Properties of Bacillus paranthracis YC03 with Urate-Lowering Potential. Microorganisms, 13(4), 798. https://doi.org/10.3390/microorganisms13040798





